Abstract
Yellow mosaic disease of cultivated legumes in South-East Asia, is caused by Mungbean yellow mosaic India virus (MYMIV) and Mungbean yellow mosaic virus (MYMV) belonging to the genus Begomovirus of the family Geminiviridae. Efforts to engineer resistance against the genus Begomovirus are focused mainly on silencing of complementary-sense virus genes involved in virus replication. Here we have targeted a complementary-sense gene (ACI) encoding Replication initiation Protein (Rep) to develop resistance against soybean isolate of Mungbean yellow mosaic India virus-[India:New Delhi:Soybean 2:1999], a bipartite begomovirus prevalent throughout the Indian subcontinent. We show that the legume host plants co-agroinoculated with infectious constructs of soybean isolate of Mungbean yellow mosaic India virus [India:New Delhi:Soybean 2:1999] along with this antisense Rep gene construct show resistance to the virus.
Keywords: MYMIV isolate, Agroinoculation, Legumes
MYMIV infects six important pulse crops, blackgram, mungbean, French bean, pigeonpea and soybean. Yield loss in blackgram, mungbean and soybean together is estimated to be around US$ 300 million in a year [16]. If the plants are infected at the seedling stage, it can lead to 85–100% yield loss [11]. Geminiviruses comprise an important group of plant ssDNA viruses capable of replicating in the host nuclei using dsDNA intermediates via the rolling circle mechanism. Members of the family Geminivirideae have circular, ssDNA genomes enecapsidated in twinned icosahedral particles [15]. Geminiviruses are divided into four genera, genus Mastrevirus, Curtovirus, Topocuvirus and Begomovirus, based on the viral genome structure, host range and type of insect vector [6]. Members of the genus Begomovirus are either monopartite (one component DNA A, ~2.8 kb) or bipartite (two components, ~2.7 kb, DNA A and DNA B). They are transmitted by whiteflies and infect dicotyledonous plants.
Pathogen-derived resistance (PDR) is a very effective genetic engineering approach to control plant viruses [3]. Geminiviruses encode specific gene essential for replication, encapsidation of their genome, cell to cell movement and systemic movement. Many of these genes are excellent candidates for developing host resistance based on PDR. Replication-initiation protein (Rep) is an important protein analyzed for PDR in geminiviruses. Rep is a multifunctional protein involved in viral replication, regulation of its own transcription and activation and recruitment of host-encoded proteins related to host DNA synthesis [7]. The central portion of Rep has a role in oligomerization [12].
The AntiRep spanning from 2613 to 1973 nt co-ordinate of MYMV when inoculated onto tobacco [14] inhibited replication of MYMV DNA A. When blackgram plants infected with MYMV were bombarded with hairpin construct containing intergenic region sequences they recovered from infection [13]. They hypothesized that the viral DNA is targeted and methylated by RNA dependent DNA polymerase resulting in reversion of symptoms. To achieve RNAi mediated resistance against yellow mosaic viruses, in the present study, a construct targetting conserved region of replication initiation protein, was made. This construct spans, 2206 nt co-ordinate to 1640 nt co-ordinate (total length 566 bp) of AC1 gene of MYMIV [India:New Delhi:Soybean 2:1999].
From the genomic component DNA A (Accession No. AY049772) of MYMIV [India:New Delhi:Soybean 2:1999] which has been already cloned and sequenced in Advanced Centre for Plant Virology, Division of Plant Pathology, IARI, the truncated replication initiation protein (Rep) gene was sub cloned in an antisense orientation under 35S promoter in the plant transformation vector pBinAR (Fig. 1). 100 ng of plasmid DNA of MYMIV-Sb was subjected to PCR amplification with primers RepSoyV (5′-ATTGGATCCAATCAATTCGAGAGCGTC-3′) and RepSoyC (5′-GGCTCTAGAGCCTACACAGGCATATGAG-3′). The BamHI and XbaI restriction sites are shown in the bold, italics. The PCR fragment of 566 bp length was restricted with BamHI and XbaI and ligated with BamHI and XbaI linearised vector pBinAR. The transformants were selected on kanamycin plate and confirmed by colony PCR (Fig. 2) and sequencing (ABI Prism sequencer, Delhi University, South Campus). One of the selected colonies (AntiRep construct) was mobilized into Agrobacterium tumefaciens EHA 105 [8] through triparental mating system [5].
Fig. 1.
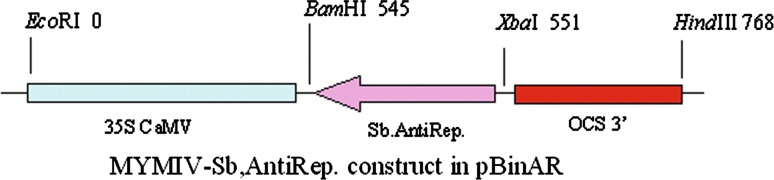
Diagrammatic representation of the Rep gene construct derived from MYMIV-Sb in antisense orientation in plant transformation vector, pBinAR. 35S CaMV promoter from Cauliflower mosaic virus, OCS opaline synthase terminator
Fig. 2.
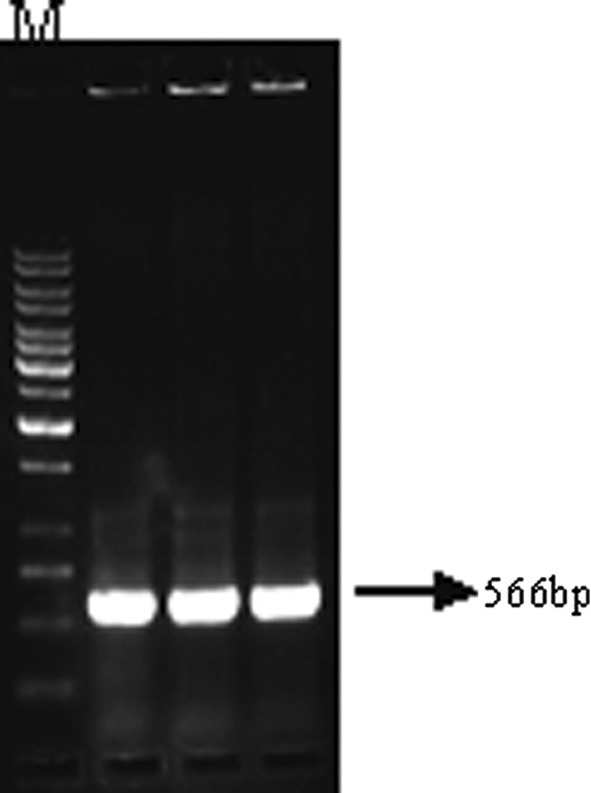
Agarose gel electrophoresis of PCR amplicons using RepSoyV/RepSoyC primers. Plasmid DNA of the clone used as a template. M = 1 Kb DNA ladder
Agrobacterium containing AntiRep and MYMIV-Sb, DNA A and DNA B constructs were selected against rifampicin (50 μg/ml) and kanamycin (50 μg/ml). The distinct colonies that appeared were confirmed to be trans conjugants through colony PCR using SoyRepV/SoyRepC primers for the AntiRep and full length abutting primers for MYMIV-Sb constructs used for agroinoculation studies on different legume hosts.
The AntiRep construct (ARC) was co-inoculated with the infectious clones DNA A and DNA B of MYMIV-Sb (against which the construct is made) on three legume hosts i.e., cowpea (var.Pusa komal), mungbean (var.PS 16) and blackgram (var.T 9), by seed-sprout method, described earlier by Mandal et al. [10]. The plants were maintained in growth chambers at National Phytotron Facility at 27 + 2°C, RH 85 and day light 18000 Lux and were monitored for symptom development till 30 days post-inoculation. The symptoms were recorded every day after the day of inoculation. Following were the parameters considered: (a) Number of infected plants/Number of plants inoculated to calculate percent infectivity. (b) Type of symptoms. (c) Reduction in leaf lamina size.
In conclusion, the plants inoculated with infectious clones, showed 77, 64 and 48% in cowpea, mungbean and blackgram respectively. However the plants co-inoculated with AntiRep construct, the symptom severity as well as the percentage of infection was almost negligible. It was only 17.8, 20 and 18% in cowpea, mungbean and blackgram plants respectively. Typical yellow mosaic symptom were seen in all the three blackgram, cowpea and mungbean plants inoculated with infectious clone, MYMIV-Sb (Fig. 3). Symptoms were seen in 7–10 days after inoculation (DAI). In co-inoculated plants unlike wild type inoculated plants which showed golden mosaic and dark yellow mosaic type of symptoms, the symptoms were less severe, less yellow mosaic and in some leaves only restricted spots of yellow mosaic were seen. The cowpea, mungbean and blackgram co-inoculated with antisense construct showed 59.2, 44 and 30% reduction in infectivity respectively. The symptom were attenuated, plants almost appeared healthy. The experiment was repeated three times and the efficiency of the construct was proved. The pooled data of three experiments are given in Table 1.
Fig. 3.

Blackgram plants showing disease inhibition by soybean AntiRep construct through co-agroinoculation. a Blackgram healthy. b Blackgram inoculated with MYMIV-Sb. c Blackgram inoculated with MYMIV-Sb + AntiRep construct showing inhibition of the disease development
Table 1.
Disease inhibition by MYMIV-Sb + AntiRep construct through co-agroinoculation
| Constructs inoculated | Hosts | No. of plants infected/No. of plants inoculated | Percent infectivity (%) | Percent reduction in disease (%) | Symptom severity | Symptoms | Days taken for symptom expression (days after inoculation |
|---|---|---|---|---|---|---|---|
| MYMIV-Sb | Cowpea | 35/45 | 77 | – | 4 | YM,GM | 7 |
| Mungbean | 32/50 | 64 | – | 4 | YM | 8 | |
| Black gram | 24/50 | 48 | – | 3 | YM | 8 | |
| MYMIV-Sb + AntiRep. | Cowpea | 8/45 | 17.8 | 59.2 | 1 | YM | 10 |
| Mungbean | 10/50 | 20 | 44 | 2 | YM | 12 | |
| Black gram | 9/50 | 18 | 30 | 2 | YM | 12 |
YM yellow mosaic, GM golden mosaic (1: <25%; 2: 25–50%; 3: 50–75%; 4: 75–100% mosaic area of leaf lamina)
Such antisense RNA mediated resistance originate from breakdown of dsRNA transcripts in the case of DNA viruses. For begomoviruses sense and antisense versions mainly targeting AC1 gene have been used with various success rates [1, 2, 4, 9]. A detailed study on the siRNA generated and the nature of the ds RNA formed using the AntiRep construct prepared in the present study would reveal the intricacies of RNA mediated resistance to MYMIV and pave way for effective management strategies.
Acknowledgements
The financial support given by Indian Council of Agricultural Research (ICAR) Government of India is gratefully acknowledged. We are thankful to the Head, Division of Plant Pathology, Indian Agricultural Research Institute, New Delhi-110012, India, for providing necessary facilities. We are also grateful to the scientists and staff at National Phytotron Facility, Indian Agricultural Research Institute, New Delhi-110012, India, for their guidance and support to grow plants under controlled conditions.
References
- 1.Aragao FJL, Ribeiro SG, Barros LMG, Brasileiro ACM, Maxwell DP, Rech EL, Faria JC. Transgenic beans (Phaseolus vulgaris L.) engineered to express viral antisense RNAs show delayed and attenuated symptoms in bean golden mosaic geminivirus. Mol Breed. 1998;4:491–499. doi: 10.1023/A:1009613607559. [DOI] [Google Scholar]
- 2.Bendahmane M, Gronenborn B. Engineering resistance against tomato yellow leaf curl virus (TYLCV) using antisense RNA. Plant Mol Biol. 1997;33:351–357. doi: 10.1023/A:1005715805000. [DOI] [PubMed] [Google Scholar]
- 3.Chellappan P, Masona MV, Vanitharani R, Taylor NJ, Fauquet CM. Broad spectrum resistance to ssDNA viruses associated with transgene-induced gene silencing in cassava. Plant Mol Biol. 2004;56:601–611. doi: 10.1007/s11103-004-0147-9. [DOI] [PubMed] [Google Scholar]
- 4.Day AG, Bejarano ER, Buck KW, Burrell M, Lichtenstein CP. Expression of an antisense viral gene in transgenic tobacco confers resistance to the DNA virus tomato golden mosaic virus. Proc Natl Acad Sci USA. 1991;88:6721–6725. doi: 10.1073/pnas.88.15.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ditta G, Stanfield S, Corbin D, Helinksi OR. Broad host range DNA cloning system for gram negative bacteria Construction of gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fauquet CM, Briddon RW, Brown JK, Moriones E, Stanley J, Zerbini M, Zhou X. Geminivirus strain demarcation and nomenclature. Arch Virol. 2008;153:783–821. doi: 10.1007/s00705-008-0037-6. [DOI] [PubMed] [Google Scholar]
- 7.Hanley-Bowdoin L, Settlage SB, Orozco BM, Nagar S, Robertson D. Geminiviruses: models for plant DNA replication, transcription and cell cycle regulation. Crit Rev Plant Sci. 1999;18:71–106. doi: 10.1016/S0735-2689(99)00383-4. [DOI] [PubMed] [Google Scholar]
- 8.Hood EE, Gelvin SB, Melchers S, Hoekema A. New Agrobacterium helper plasmid for gene transfer to plants (EHA 105) Transgenic Res. 1993;2:208–218. doi: 10.1007/BF01977351. [DOI] [Google Scholar]
- 9.Lucioli A, Noris E, Brunetti A, Tavazza R, Ruzza V, Castillo AG, Bejarano ER, Accotto GP, Tavazza M. Tomato yellow leaf curl Sardinia virus Rep-derived resistance to homologous and heterologous geminiviruses occurs by differential mechanisms and is overcome if virus-mediated transgene silencing is activated. J Virol. 2003;77:6785–6798. doi: 10.1128/JVI.77.12.6785-6798.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mandal B, Varma A, Malathi VG. Systemic infection of Vigna mungo using the cloned DNAs of the blackgram isolate mungbean yellow mosaic geminivirus through agroinoculation and transmission of the progeny virus by whiteflies. J Phytopathol. 1997;145:503–510. doi: 10.1111/j.1439-0434.1997.tb00358.x. [DOI] [Google Scholar]
- 11.Nene YL. Viral disease of some warm weather pulse crops in India. Plant Dis Rep. 1973;57:463–467. [Google Scholar]
- 12.Orozco BM, Miller AB, Settlage SB, Hanley-Bowdoin L. Functional domains of a geminivirus replication protein. J Biol Chem. 1997;272:9840–9846. doi: 10.1074/jbc.272.15.9840. [DOI] [PubMed] [Google Scholar]
- 13.Pooggin M, Shivaprasad PV, Veluthambi K, Hohn T. RNAi targeting of DNA virus in plants. Natl Biotechnol. 2003;21:131–132. doi: 10.1038/nbt0203-131b. [DOI] [PubMed] [Google Scholar]
- 14.Shivaprasad PV, Thillaichidambaram P, Balaji V, Veluthambi K. Expression of full-length and truncated Rep genes from Mungbean yellow mosaic virus-Vigna inhibits viral replication in transgenic tobacco. Virus Genes. 2006;33:365–374. doi: 10.1007/s11262-006-0077-5. [DOI] [PubMed] [Google Scholar]
- 15.Stanley J, Bisaro DM, Briddon RW, Brown JK, Fauquet CM, Harrison BD, Rybicki EP, Stenger DC. Geminiviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus taxonomy, VIIIth report of the ICTV. London: Elsevier/Academic Press; 2005. pp. 301–326. [Google Scholar]
- 16.Varma A, Malathi VG. Emerging geminivirus problems: a serious threat to crop production. Ann Appl Biol. 2003;142:145–164. doi: 10.1111/j.1744-7348.2003.tb00240.x. [DOI] [Google Scholar]


