Abstract
Newcastle disease (ND) is a fatal and contagious disease that poses a constant threat to the poultry industry around the globe. Due to the complex clinico-pathological picture and high genetic variability, the efficient diagnosis of NDV strains is a challenge. In an emerging wave of ND in the north of Pakistan, samples from six outbreaks in commercial poultry and two from healthy backyard poultry flocks were screened for NDV. A real-time PCR based on the fusion and polymerase genes of NDV detected all six isolates whereas a validated real-time PCR based on the matrix gene failed to detect any of these isolates, most likely due to substantial mismatches in the probe-binding site. All isolates have shown ICPI and MDT values similar to the velogenic form of NDV strains. The cleavage site in the F protein was found to be 112RRQKR↓F117, typical of virulent NDV. Phylogenetic reconstruction, based on fusion and matrix genes, provided enough evidences to consider these isolates as a new subgenotype within genotype VII. This study raised concerns about the genetic variability of NDV circulating in Pakistan, and sensitivity of the assays for the detection of the NDV isolates in clinical samples.
Keywords: Backyard poultry, Commercial poultry, Newcastle disease, Pakistan, Real time PCR, Sequencing
Newcastle disease (ND) is a highly contagious and fatal disease, which can infect a broad rage of domestic and wild host species [1, 8]. However, the severity of the disease varies from undetectable to rapidly fatal, and in clinical cases is characterized by sneezing, respiratory distress, green diarrhoea and oedema of several of the body regions [1]. In some cases, the nervous system signs such as circling, backward progression and convulsive somersaulting, head tremor, wing and leg paralysis may develop. With such a variable expression of the disease, definitive diagnosis of ND as a single clinico-pathological entity is impossible. Therefore, for simplicity, ND is divided into lentogenic, mesogenic and velogenic, which represent avirulent, mild virulent and highly virulent forms of the disease, respectively. Several laboratories assess the pathogenicity of ND based on MDT (mean death time), IVPI (intravenous pathogenicity index) and ICPI (intracerebral pathogenicity index) as numerical evaluation criteria. In addition to these, OIE has established a general standard to distinguish virulent from avirulent NDV strains. The presence of one basic amino acid at residues 116 and 115 of the F protein cleavage site as well as a phenylalanine at residue 117 along with a basic amino acid (R) at amino acid position 113 are considered markers of a virulent pathotype and such isolates are notifiable to OIE [17].
There is extensive genetic variability among ND strains, despite the fact that all belong to same serotype (APMV-1). Based on F gene phylogenetic reconstruction, NDV has been divided into two classes: Class I and II. Class II is comprised of pathogenic strains and is divided into 11 genotypes (I–XI), with those arising from the period between 1930 and 1960 (I, II, III, IV, IX) being considered as “early” or “old” and those appearing after 1960 (V, VI, VII, VIII, X, XI) as “late” or “recent” [10, 12]. The genotypes VI and VII are subdivided into eight (a–h) and five (a–e) subgenotypes, respectively. Class I consists of viruses that have the largest genome (1,5198 nt), are avirulent for chickens except for one strain, and are usually isolated from waterfowl and shore birds [6, 9, 12].
Since the clinical picture of the disease is complicated, the NDV diagnosis historically relies on virus isolation, which is a mandatory assay for international trade and is regarded as the “gold standard” to validate new assays [16]. Molecular techniques are predominantly described not only for characterization but also for pathotyping of NDV [18, 21]. It is generally believed that the replication machinery of NDV introduces errors when it copies the genome during replication, in common with other RNA viruses. As a consequence, the viruses evolve very rapidly and quickly adapt to different environmental pressures. Such a phenomenon may have implications in disease severity and diagnosis, which has recently been reviewed for NDV [5]. In an effort to characterize NDV isolates from emerging outbreaks of NDV in commercial poultry farms in the north of Pakistan, the efficacy of two validated real-time PCRs was determined in order to facilitate prompt detection of ND in Pakistan.
The clinical material, including tracheal and cloacal swabs, was collected from dead birds while blood samples were collected from alive but sick birds. Samples were collected from ND outbreaks in six commercial poultry farms. Additionally, samples from two domestic healthy backyard poultry flocks were also collected to screen for the presence of subclinical ND infection (Table 1). It is worth mentioning that commercial farm attendants have frequent contact with these backyard poultry flocks. The samples were passaged in 10 day old specific pathogen free (SPF) embryonated eggs. The allantoic fluids were used for the assessment of pathogenicity (ICPI, MDT), detection efficacy for validated real-time PCRs and for sequence analysis. For the latter two purposes, QIAcard FTA Indicator Four Spots (Qiagen, Hilden, Germany) were impregnated with allantoic fluids and brought to the Swedish University of Agricultural Sciences, Uppsala, Sweden, for processing. The eluted RNA was used for both real-time PCR and for the amplification of the M and F genes of each isolate, as described previously [15]. Six isolates from commercial as well as two isolates from backyard poultry were phylogenetically characterized and analyzed. The trees were constructed using the Neighbor-joining method (Kimura 2 parameter) with 2,000 bootstrap replicates in Molecular Evolutionary Genetics Analysis (MEGA) version 5 (http://www.megasoftware.net/).
Table 1.
Information of the samples included in this study
| Sample name | Age of birds | Number of birds at farm | Location | Country | Cleavage site | Accession number (M gene) | Accession number (F gene) |
|---|---|---|---|---|---|---|---|
| Chicken/CP/Attock/2010 | 18 days | 3,000 | Attock | Pakistan | 112RRQKR↓F117 | JN682192 | JN682188 |
| Chicken/CP/Islamabad1/2010 | 16 days | 17,000 | Islamabad | Pakistan | 112RRQKR↓F117 | JN682196 | JN682186 |
| Chicken/CP/Rawalpindi1/2010 | 22 days | 28,000 | Rawalpindi | Pakistan | 112RRQKR↓F117 | JN682199 | JN682185 |
| Chicken/CP/Islamabad2/2010 | 1 year | 5,000 | Islamabad | Pakistan | 112RRQKR↓F117 | JN682195 | JN682190 |
| Chicken/CP/Islamabad3/2010 | 100 weeks | 4,000 | Islamabad | Pakistan | 112RRQKR↓F117 | JN682193 | JN682191 |
| Chicken/BYP/Rawalpindi/2010 | 3 months | 07 | Rawalpindi | Pakistan | 112RRQKR↓F117 | JN682197 | JN682187 |
| Chicken/BYP/Lahore/2010 | 6 months | 18 | Lahore | Pakistan | 112RRQKR↓F117 | JN682198 | JN682184 |
| Chicken/CP/Rawalpindi2/2010 | 9 weeks | 3,350 | Rawalpindi | Pakistan | 112RRQKR↓F117 | JN682194 | JN682189 |
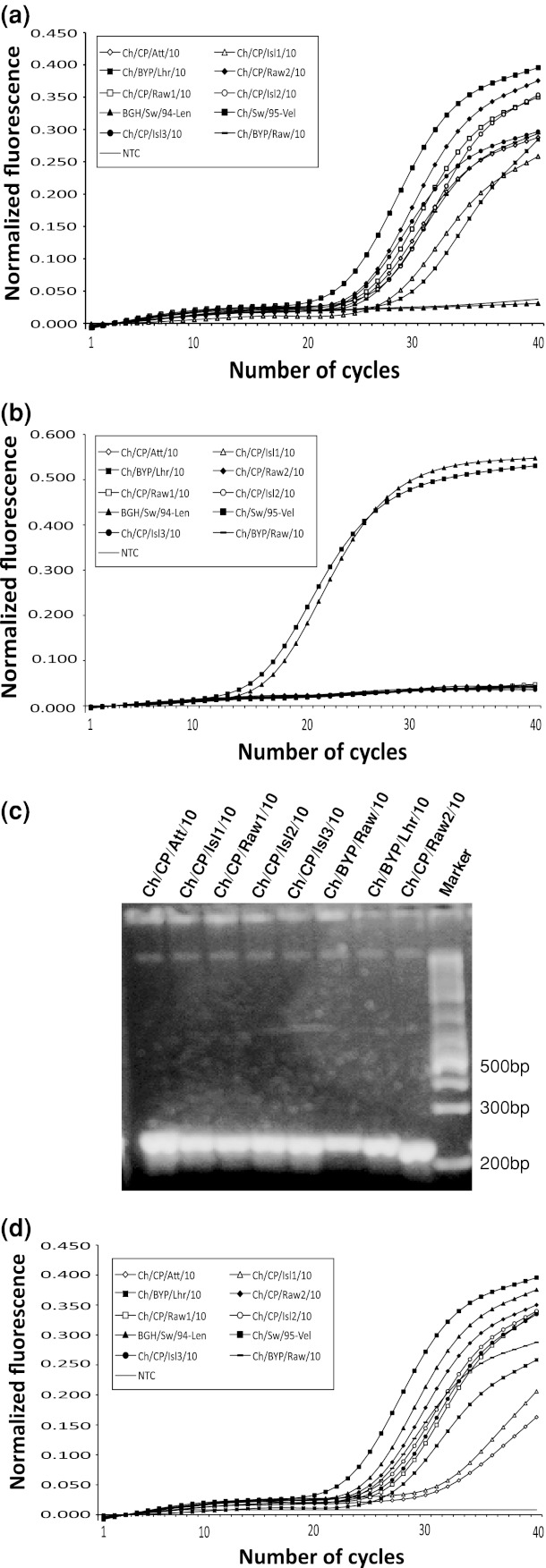
Initially, a USDA-validated real-time PCR, based on the F gene (F-based assay) of NDV, was employed as recommended [21]. This assay is commonly used for the detection and differentiation of lentogenic, mesogenic and velogenic strains of NDV based on changes in the F protein cleavage site, which is a well-recognized marker for pathogenicity. All the Pakistani isolates used in this study were detected positive by the F-based assay. Additionally, both lentogenic and mesogenic Swedish NDV strains were also detected as positive (Fig. 1a). This assay has previously been described to be less sensitive since it appeared to give false-negative results due to a significant variability in the probe binding site [9, 11, 16, 18], raising a concern to develop assays that do not require probes such as ligase chain reaction. There are reports that modified primers and probes in this already existing USDA-validated protocol can improve the screening of the genetically divergent NDV strains [18].
Fig. 1.
Results for the real-time PCR assay applied to detect Newcastle disease virus. RNA from all eight isolates was used along with two Swedish lentogenic and mesogenic/velogenic isolates named here as BGH/Sweden/94 and Chicken/Sweden/95, respectively. A no template control (NTC) was also included. a Results of real-time PCR based on the fusion (F) gene and b matrix (M) gene of NDV. c The real-time PCR products of the assay targeting the M gene. Only PCR products from eight Pakistani isolates were run on a gel. d Normalized fluorescent signals for the real-time PCR based on the large (L) gene of NDV
Due to the great wealth of concerns regarding the suitability of F-based assays, efforts have been made to target a relatively conserved region near the 5′ end of the M gene [19]. In parallel, each of the isolates were also screened for this USDA-validated assay based on the M gene of NDV (M-based assay) [21]. This assay has been validated for efficient detection of all the genotypes within class II in clinical samples. Interestingly, none of the Pakistani NDV isolates were detected positive in this assay but all of the Swedish isolates were found positive, in contrast to the F-based assay (Fig. 1b). Notably, the NDV strains isolated from healthy backyard poultry flocks were also negative. The consequences of this escaped diagnosis can’t be realized in case this assay would have been applied to screen the healthy birds populations (domestic or wild) for ND. The gel picture from the real-time PCR demonstrated adequate performance of the primers and tolerance of the mismatches (Fig. 1c). Since this assay is meant for clinical diagnosis of ND, failure in the assays has an unacceptable implication in the disease diagnosis. This assay has been reported to show false-negative results for class I strains of NDV (note that the isolates in the current study belong to class II) [9]. There are few reports of this failure for class II strains, which warrants revisiting this assay’s design and validation. Moreover, it is not realistic to rely on a single test to detect such a diverse group of viruses. Recently, a real-time PCR targeting the polymerase gene (L) of NDV has been demonstrated [7]. With an aim to detect all genotypes within class I and II, a single primer set and 2 different hydrolysis probes were included in this assay. Testing demonstrated that all the Pakistani isolates and Swedish isolates in the current study were positively detected (Fig. 1d).
To further investigate the reasons for this failure, the complete M genes of all the isolates were sequenced using the primers and protocol we previously described [13, 14], using One-Step RT-PCR kit (Qiagen, Hilden, Germany). The primers and probe in the USDA-validated real-time PCR for the M gene were aligned and compared with sequences of the lentogenic, mesogenic and velogenic strains of NDV. Comparison indicated that there were two and three nucleotide mismatches in the forward and reverse primers, respectively (data not shown). The probe-binding site shows four nucleotide mismatches, including three at the 5′-end, which is considered crucial for oligonucleotide hybridization. This indicates that the lack of fluorescent signals in real-time PCR was due to failure in probe binding. This can further be justified by the fact that this assay failed to detect class I NDV isolates carrying less than four mismatches [9]. Apart from this, several other isolates within class II have three mismatches and thus are more likely to escape detection. Such scenarios will be even complication if 1–2 more mismatches occur at this binding site, which is not less likely in RNA viruses. This protocol has been used for surveillance in the European wild bird population [2] and for screening samples from Europe, Africa and Asia [3–5]. Pakistan is a major exporter of fancy birds to Europe, especially to Italy, Spain, Portugal and Greece [20], and recently the NDV is identified in birds exported to Italy [20]. Such facts questioned the application of this assay (M-based) for the detection of NDV from all subgroups and emerging new genetic groups.
Pathotyping indicated that all the NDV isolates were velogenic based on their ICPI, MDT and the sequence of the cleavage site in the F protein. The MDT and ICPI were recorded to be <60 h and ≤1.5, respectively. The cleavage site was found to be 112RRQKR↓F117, identical to that of virulent strains of NDV. Notably, the NDV strains isolated from healthy backyard poultry flocks also showed the characteristics of pathogenicity, raising concern about of contribution of domestic poultry in the epizootiology of NDV especially in countries where control measures and biosecurity levels are sub-optimal.
Six isolates from commercial as well as two isolates from backyard poultry were phylogenetically characterized. The phylogenetic reconstruction was performed based on the complete open reading frame of the F gene and was compared to the available sequences in GenBank. All the Pakistani isolates clustered with viruses of genotype VII. However, all of these isolates were significantly different from the rest of the isolates and can tentatively be regarded as a distinct genetic group within genotype VII (Fig. 2a). Phylogenetic analysis of the complete open reading frame of the matrix gene has revealed a similar branching pattern of NDV isolates. Similarly, the matrix gene-based analysis showed that Pakistani isolates cluster in a separated group (Fig. 2b). For exact comparative purposes, the matrix and fusion gene sequences were phylogenetically analyzed from the same isolates of which the complete genomes (n = 54) are available in the GenBank.
Fig. 2.
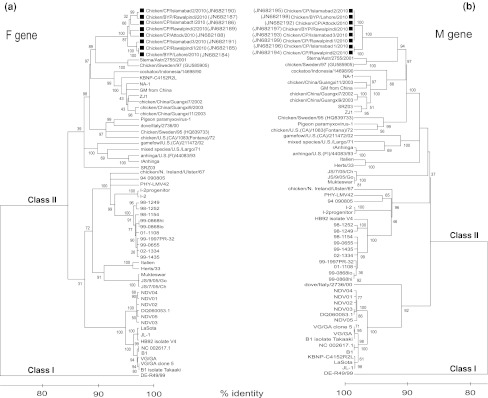
Phylogenetic analysis of the fusion and matrix genes of Pakistani isolates compared to previously characterized NDV strains. The trees were constructed using the Neighbour-joining method (Kimura 2 parameter) with 2,000 bootstrap replicates. a Phylogenetic tree based on the complete open reading frame of the fusion (F) gene, b and of the matrix (M) gene from corresponding isolates. A black square indicates the isolates presented in this study
Currently, there is great wealth of reports discussing the error prone genome transcription during replication of RNA viruses such as NDV. This process helps viruses escape host immune defences, alter pathogenicity and host range, and evade diagnostic tests. To properly understand these mechanisms, it is of paramount importance to fully characterize the viruses in order to study within-host dynamics and genetic variation, relate dynamics and variation to transmission, and to reconstruct transmission trees at high resolution. The results in this report highlight similar mechanisms in the NDV strains circulating in Pakistan. A re-visited assay for the detection of novel strains of NDV would not only help to detect these isolates within the region but will also improve the diagnostic possibilities if these viruses escape to other parts of the world. This is of special concern with NDV, which has been described historically as having great mobility and the strains of NDV from European, American and Asian origin have gained worldwide distribution [12]. This will further help to understand the nature of viruses and to build a basis for novel diagnostic tests and vaccine development. The extensive genetic variability within avian paramyxoviruses 1 has placed deep interest in targeting more stable genes such as the polymerase (L) or nucleoprotein (NP) genes for diagnostic purposes [5].
Acknowledgments
Authors like to thank Martí Cortey for his help in phylogenetic analysis and Claudia Baule for critical reading of the manuscript. We also like to thank Jenna Anderson for revising the manuscript for linguistic errors.
References
- 1.Alexander DJ. Newcastle disease. In: Saif YM, Glisson JR, Fadly AM, McDougal LR, Swayne DE, editors. Diseases of poultry. Ames: Iowa State University Press; 2003. [Google Scholar]
- 2.Camenisch G, Bandli R, Hoop R. Monitoring of wild birds for Newcastle disease virus in Switzerland using real time RT-PCR. J Wildl Dis. 2008;44:772–776. doi: 10.7589/0090-3558-44.3.772. [DOI] [PubMed] [Google Scholar]
- 3.Cattoli G, De Battisti C, Marciano S, Ormelli S, Monne I, Terregino C, Capua I. False-negative results of a validated real-time PCR protocol for diagnosis of newcastle disease due to genetic variability of the matrix gene. J Clin Microbiol. 2009;47:3791–3792. doi: 10.1128/JCM.00895-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cattoli G, Fusaro A, Monne I, Molia S, Le Menach A, Maregeya B, Nchare A, Bangana I, Maina AG, Koffi JN, Thiam H, Bezeid OE, Salviato A, Nisi R, Terregino C, Capua I. Emergence of a new genetic lineage of Newcastle disease virus in West and Central Africa: implications for diagnosis and control. Vet Microbiol. 2010;142:168–176. doi: 10.1016/j.vetmic.2009.09.063. [DOI] [PubMed] [Google Scholar]
- 5.Cattoli G, Susta L, Terregino C, Brown C. Newcastle disease: a review of field recognition and current methods of laboratory detection. J Vet Diagn Invest. 2011;23:637–656. doi: 10.1177/1040638711407887. [DOI] [PubMed] [Google Scholar]
- 6.Czegledi A, Ujvari D, Somogyi E, Wehmann E, Werner O, Lomniczi B. Third genome size category of avian paramyxovirus serotype 1 (Newcastle disease virus) and evolutionary implications. Virus Res. 2006;120:36–48. doi: 10.1016/j.virusres.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 7.Fuller CM, Brodd L, Irvine RM, Alexander DJ, Aldous EW. Development of an L gene real-time reverse-transcription PCR assay for the detection of avian paramyxovirus type 1 RNA in clinical samples. Arch Virol. 2010;155:817–823. doi: 10.1007/s00705-010-0632-1. [DOI] [PubMed] [Google Scholar]
- 8.Higgins DA, Shortridge KF. Newcastle disease in tropical and developing countries. In: Alexander DJ, editor. Newcastle disease. Boston: Kulwer Academic Publishers; 1988. [Google Scholar]
- 9.Kim LM, King DJ, Suarez DL, Wong CW, Afonso CL. Characterization of class I newcastle disease virus isolates from Hong Kong live bird markets and detection using real-time reverse transcription-PCR. J Clin Microbiol. 2007;45:1310–1314. doi: 10.1128/JCM.02594-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lomniczi B, Wehmann E, Herczeg J, Ballagi-Pordany A, Kaleta EF, Werner O, Meulemans G, Jorgensen PH, Mante AP, Gielkens AL, Capua I, Damoser J. Newcastle disease outbreaks in recent years in western Europe were caused by an old (VI) and a novel genotype (VII) Arch Virol. 1998;143:49–64. doi: 10.1007/s007050050267. [DOI] [PubMed] [Google Scholar]
- 11.Mia Kim L, Suarez DL, Afonso CL. Detection of a broad range of class I and II Newcastle disease viruses using a multiplex real-time reverse transcription polymerase chain reaction assay. J Vet Diagn Invest. 2008;20:414–425. doi: 10.1177/104063870802000402. [DOI] [PubMed] [Google Scholar]
- 12.Miller PJ, Decanini EL, Afonso CL. Newcastle disease: evolution of genotypes and the related diagnostic challenges. Infect Genet Evol. 2010;10:26–35. doi: 10.1016/j.meegid.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Munir M, Linde AM, Zohari S, Stahl K, Baule C, Holm K, Engstrom B, Berg M. Complete genome analysis of an avian paramyxovirus type 1 strain isolated in 1994 from an asymptomatic black-headed gull (Larus ridibundus) in southern Sweden. Avian Dis. 2010;54:923–930. doi: 10.1637/9086-092409-RESNOTE.1. [DOI] [PubMed] [Google Scholar]
- 14.Munir M, Linde AM, Zohari S, Stahl K, Baule C, Engstrom B, LH MR, Berg M. Whole genome sequencing and characterization of a virulent Newcastle disease virus isolated from an outbreak in Sweden. Virus Genes. 2011;43:261–271. doi: 10.1007/s11262-011-0636-2. [DOI] [PubMed] [Google Scholar]
- 15.Munir M, Zohari S, Saeed A, Khan QM, Abubakar M, Leblanc N, Berg M. Detection and phylogenetic analysis of peste des petits ruminants virus isolated from outbreaks in Punjab, Pakistan. Transbound Emerg Dis. 2012;59:85–93. doi: 10.1111/j.1865-1682.2011.01245.x. [DOI] [PubMed] [Google Scholar]
- 16.OIE. Chapter 2.3.14 In: Manual of diagnostic tests and vaccines for terres-trial animals, 6th edn. OIE, Paris, France. 2008; pp. 76–589.
- 17.OIE. Newcastle disease. In: Manual of Standards for Diagnostic Tests and Vaccines 4th edn. Paris; 2000. pp 221–232.
- 18.Rue CA, Susta L, Brown CC, Pasick JM, Swafford SR, Wolf PC, Killian ML, Pedersen JC, Miller PJ, Afonso CL. Evolutionary changes affecting rapid identification of 2008 Newcastle disease viruses isolated from double-crested cormorants. J Clin Microbiol. 2010;48:2440–2448. doi: 10.1128/JCM.02213-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seal BS, King DJ, Meinersmann RJ. Molecular evolution of the Newcastle disease virus matrix protein gene and phylogenetic relationships among the paramyxoviridae. Virus Res. 2000;66:1–11. doi: 10.1016/S0168-1702(99)00119-7. [DOI] [PubMed] [Google Scholar]
- 20.Trust WP. Deadly Newcastle Disease Discovered in Parrots and other Birds Imported from Pakistan to Italy; 2004.
- 21.Wise MG, Suarez DL, Seal BS, Pedersen JC, Senne DA, King DJ, Kapczynski DR, Spackman E. Development of a real-time reverse-transcription PCR for detection of newcastle disease virus RNA in clinical samples. J Clin Microbiol. 2004;42:329–338. doi: 10.1128/JCM.42.1.329-338.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]