Abstract
Rabies in domestic and wild animals continues to be a major public health threat in India. Rapid and accurate diagnosis of rabies in animals is therefore of utmost importance as the individuals who were in contact with the rabid animals are at a greater risk. A significant amount of diagnostic tissue samples submitted to our laboratory are often autolysed and the WHO recommended direct fluorescent antibody test (FAT) for rabies diagnosis cannot be used in such samples. In this pilot study we have evaluated three different diagnostic primer sets for rapid sensitive and specific detection of rabies genome from the brain samples of different species of animals. We have validated a sensitive RT-PCR assay using brain tissue samples from different species of animals such as cat, cattle, dog, mouse and human, for routine diagnosis of rabies. Our results show the potential of this assay as a confirmatory test when the FAT results are unreliable and also as an alternative diagnostic test in circumstances when the diagnostic samples are unsuitable for use in FAT. Furthermore the nucleotide sequence of nucleoprotein gene amplified using this assay can also be used for the molecular epidemiological study of the rabies viruses in India.
Keywords: Rabies, India, Diagnosis, FAT, RT-PCR
Animal and human rabies are endemic throughout the Indian subcontinent with the exception of Andaman and Nicobar Islands. The causative agent, rabies virus (family: Rhabdoviridae; genus: Lyssavirus) is largely maintained in two ecologically inter-related disease cycles; urban and sylvatic (wildlife). Although, dog is primarily responsible for rabies transmission to humans in India [3], wildlife plays a much lesser but not insignificant role in transmission [15]. It has been reported that more than 96 % of rabies incidence in India are the result of contact with infected dogs. In addition, rabies was reported as a result of contact with infected jackals in 1.7 %, cats in 0.8 %, monkeys in 0.4 %, mongooses in 0.4 %, and foxes in 3 % of the cases [3]. Therefore ante and post mortem diagnosis of rabies in animals is of utmost importance as the animals and human beings who come in contact with these rabid animals are at a greater risk of contracting the disease. The most widely used diagnostic test for rabies is the fluorescent antibody test (FAT), which is recommended by both WHO and OIE [17]. This ‘gold standard’ test may be used directly on a brain tissue impression smear and can also be used to confirm the presence of rabies antigen in cell culture or in brain tissue of mice that have been inoculated for diagnostic purposes. Although, the FAT gives reliable results on fresh specimens within a few hours in more than 95–99 % of the cases [17], autolysed tissue samples can reduce the sensitivity of this test and often are unsuitable for confirming the presence of rabies antigen. Classical reverse transcription-polymerase chain reaction (RT-PCR) assays on the other hand has been reported to be a sensitive and specific tool for routine diagnostic purposes [18, 19]. These assays were also used to detect rabies virus in decomposed samples [1, 6] and archival specimens [4, 14, 20]. Few of the brain tissue samples submitted for diagnosing rabies to our laboratory are often autolysed due to faulty transport conditions. In addition, the laboratory also frequently receives cerebrospinal fluid samples for ante-mortem rabies diagnosis. In this study, the sensitivity and specificity of the RT-PCR assay was evaluated for the routine and rapid laboratory diagnosis of rabies.
The brain tissue samples used in this study were collected from animals suspected to have rabies and submitted to Rabies Laboratory, Department of Animal Biotechnology, Madras Veterinary College, Chennai between the years 2005 and 2007. In addition, brain samples from mouse which was intra-cerebrally inoculated with Challenge virus standard-11 (CVS-11) strain of rabies virus was used as positive control. Brain samples from rabies free cattle, sheep, goat, mouse and dog were used as negative controls. The details of the test samples are summarised in Table 1.
Table 1.
Comparison between FAT and RT-PCR assays for the detection of rabies virus in brain samples of different species of animals suspected for rabies
| Sl. No. | Host | Place of origin | FAT | RT-PCR | ||
|---|---|---|---|---|---|---|
| Primers BB2 and JW6 | Primers N-For and N-Rev | Primers JW12 and JW6dpl | ||||
| 1 | Dog | Tamilnadu | + | + | + | + |
| 2 | Dog | Tamilnadu | – | – | – | – |
| 3 | Dog | Tamilnadu | – | – | – | – |
| 4 | Dog | Tamilnadu | + | + | + | + |
| 5 | Dog | Tamilnadu | + | + | + | + |
| 6 | Dog | Tamilnadu | – | – | – | – |
| 7 | Dog | Tamilnadu | + | + | + | + |
| 8 | Dog | Tamilnadu | – | – | – | – |
| 9 | Dog | Tamilnadu | + | + | + | + |
| 10 | Dog | Tamilnadu | + | + | + | + |
| 11 | Dog | Tamilnadu | + | + | + | + |
| 12 | Dog | Tamilnadu | + | + | + | + |
| 13 | Dog | Tamilnadu | + | + | + | + |
| 14 | Cow | Tamilnadu | + | + | + | + |
| 15 | Cow | Tamilnadu | + | + | + | + |
| 16 | Cow | Tamilnadu | + | + | + | + |
| 17 | Cow | Tamilnadu | – | – | – | – |
| 18 | Goat | Tamilnadu | + | + | + | + |
| 19 | Goat | Tamilnadu | + | + | + | + |
| 20 | Goat | Tamilnadu | + | + | + | + |
| 21 | Goat | Tamilnadu | + | + | + | + |
| 22 | Cat | Tamilnadu | – | + | + | + |
| 23 | Human | Tamilnadu | + | + | + | + |
| 24 | Mouse | Tamilnadu | + | + | + | + |
| 25 | Mouse | Tamilnadu | + | + | + | + |
| 26 | Mouse | CVS11 strain | + | + | + | + |
| 27 | Dog | Karnataka | – | + | + | + |
| 28 | Dog | Karnataka | + | + | + | + |
| 29 | Dog | Karnataka | + | + | + | + |
| 30 | Dog | Karnataka | + | + | + | + |
| 31 | Dog | Karnataka | + | + | + | + |
| 32 | Dog | Karnataka | # | + | + | + |
| 33 | Dog | Karnataka | + | + | + | + |
| 34 | Dog | Karnataka | # | + | + | + |
| 35 | Dog | Karnataka | # | + | + | + |
| 36 | Dog | Karnataka | + | + | + | + |
| 37 | Dog | Karnataka | # | + | + | + |
| 38 | Mouse | Karnataka | + | + | + | + |
| 39 | Mouse | Karnataka | + | + | + | + |
| 40 | Mouse | Karnataka | + | + | + | + |
| 41 | Dog | Kerala | + | + | + | + |
| 42 | Dog | Kerala | + | + | + | + |
| 43 | Dog | Kerala | + | + | + | + |
| 44 | Dog | Kerala | + | + | + | + |
| 45 | Dog | Kerala | – | + | + | + |
| 46 | Dog | Kerala | – | + | + | + |
| 47 | Dog | Kerala | + | + | + | + |
| 48 | Dog | Kerala | + | + | + | + |
| 49 | Dog | Kerala | + | + | + | + |
| 50 | Dog | Kerala | + | + | + | + |
| 51 | Dog | Kerala | + | + | + | + |
| 52 | Dog | Kerala | + | + | + | + |
+ indicates the presence of rabies virus antigen/genome
− indicates no rabies virus antigen/genome detected
# indicates samples unsuitable for use in FAT
Monolayers of baby hamster kidney (BHK)-21 epithelial cells were maintained in Eagle’s minimum essential medium (Gibco BRL, Life Technologies, Paisley, United Kingdom) supplemented with 10 % heat inactivated foetal calf serum (Gibco BRL, Life Technologies, Paisley, United Kingdom), 2 mM l-glutamine (Sigma, Poole, United Kingdom), and antibiotics at 35 °C. The CVS-11 strain of rabies virus was propagated and titrated on BHK-21 cells as described previously [17].
Impression smears were made from the brain samples on a clean glass slide and fixed in acetone for 30 min at −20 °C and tested for the presence of viral antigen by direct FAT [7]. The staining was done using anti rabies nucleocapsid rabbit immunoglobulin (IgG) conjugated with fluorescein isothiocyanate (BioRad, India).
Total RNA was extracted directly from the infected brain tissues or rabies virus infected BHK-21 cell monolayer using the TRIzol reagent according to the manufacturer’s instructions (Gibco BRL, Life Technologies, Paisley, United Kingdom). Reverse transcription was performed with 2 μg of the total brain RNA using the RevertAid™ cDNA synthesis kit (Fermentas, India) as per manufacturer’s instructions. Briefly, RNA was denatured at 70 °C for 5 min, cooled on ice which was then added to a final volume of 20 μl containing Moloney Murine Leukemia Virus (M-MLV) reverse transcription buffer, 10 mM dNTPs, 20 U of RNase inhibitor, 10 pmol of primer JW12 (Table 2) and 200 U of MMLV reverse transcriptase. This mixture was incubated at 42 °C for 60 min in a water bath and heated at 70 °C for 5 min to terminate the reaction. Amplification of 5 μl of the cDNA template was carried out using the Hotmaster-Taq mix (Eppendorf, India) containing 10 pmol each of forward and reverse primers (Table 2) in a 50 μl reaction volume. The amplifications were performed on a PTC100 Thermal Cycler (MJ Research, USA). After denaturation at 95 °C for 10 min, the reactions were cycled five times at 95 °C for 60 s, 50 °C for 60 s and 72 °C for 60 s and then 30 times at 95 °C for 30 s, 45 °C for 30 s, 72 °C for 60 s. This was followed by a final elongation step at 72 °C for 10 min. On completion, the samples were analysed on 1 % agarose gel electrophoresis.
Table 2.
Oligonucleotide primers for the RT-PCR amplification of partial N gene of Rabies viruses used in this study
The analytical sensitivity of the three different sets of primers in the RT-PCR assay was evaluated by extracting the total cellular RNA from one ml of the serial dilutions of BHK cells passaged CVS-11 virus suspension containing 102 50 % fluorescent focus forming doses (FFD50) per ml. The diagnostic sensitivity and specificity of the RT-PCR assay was calculated using FAT as the gold standard. Agreement between the FAT and RT-PCR assay was measured by calculating the Cohen’s kappa coefficient (κ).
Of the fifty-two samples tested, thirty-nine were found to be positive for the presence of the rabies antigen using the direct FAT. Four out of these fifty-two samples were autolysed and hence were found unsuitable for preparation of impression smears and were subsequently not tested in FAT. An impression smear from CVS-11 infected mouse brain and from the corresponding rabies free animal (dog/cattle/buffalo/sheep/goat) were included as positive and negative controls, each time the FAT was performed.
All the 52 samples were also tested using the RT-PCR assay using three different primer pairs which were previously shown to amplify nucleoprotein gene of rabies virus (Table 2). The results of both the FAT and RT-PCR assay are summarised in Table 1. The four autolysed tissue samples that were unsuitable for FAT, tested positive for the presence of rabies genome in the RT-PCR assay using all the three primer sets (Fig 1). There was no difference in the diagnostic sensitivity of the three primer pairs in the RT-PCR assay. Host genomic amplicons of the same size as that of the target amplicons were commonly observed on the agarose gels when a nested RT-PCR was used for the detection of lyssa viruses [13]. However the amplification in our assay was N gene specific and no non specific-false positive amplification products were observed in any of the brain samples from dog, cattle, buffalo, sheep, goat and mouse.
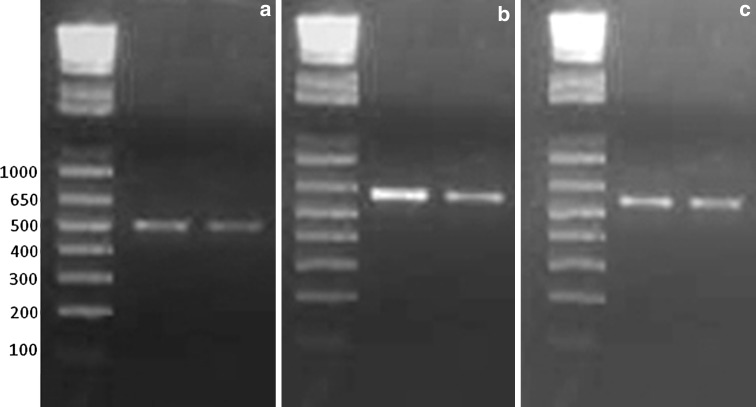
Fig. 1.
RT-PCR assay using different primer pairs to detect the rabies virus genome. a BB2–JW6 primers showing 506 bp amplification ,b N For–N Rev primers showing 533 bp amplification, c JW12–JW6dpl primers showing 600 bp amplification. Molecular weight marker: 1 Kb plus DNA ladder (Invitrogen)
The sensitivity and specificity of the RT-PCR assay were found to be 100 % in comparison with FAT. The kappa coefficient for the agreement between the two assays is 0.97 indicating almost complete agreement. The analytical sensitivity of the RT-PCR assay was found to be 1 FFD50 using the primers BB2 and JW6, 0.1 FFD50 using primers N-For and N–Rev and 0.01 FFD50 using JW12–JW6dpl. Although there is a significant difference in the analytical sensitivity of these three primer pairs, all of them detected the presence of rabies genome in 100 % of the brain samples that were positive in FAT and also from the four autolysed samples that were unsuitable for use in FAT. This is probably due to the presence of higher quantities of the viral genome in the post mortem brain samples. The primer pair which showed a higher analytical sensitivity can therefore be used for routine diagnosis of rabies to detect samples with a lesser viral load.
The primer pairs BB2–JW6 and JW12–JW6dpl amplify the 238–507 bp region of nucleoprotein gene (position based on Pasteur virus genome NC_001542) which is commonly used for the molecular epidemiological study of rabies viruses in India [2, 16]. Therefore the significant advantage with this RT-PCR is that the amplified products can be sequenced to confirm the identity of the virus. This genome sequence data will be useful in molecular epidemiological studies to trace the origin and movement of the viruses.
Molecular tools based on the detection of the viral genome are becoming widely accepted and accessible for the diagnosis of rabies [8]. Although the FAT is a simple and rapid technique, the evaluation of results can be subjective. An important factor influencing the interpretation of the FAT results is the strong background fluorescence of the visible field caused by the immunofluorescent conjugate [12]. Moreover for numerous diagnostic laboratories in rabies-endemic regions of the developing world, cost and simplicity are critical factors in disease diagnosis which cannot be neglected, even when the principal consideration is for rapid diagnosis [8]. RT-PCR is a suitable alternative for FAT in rabies diagnosis as the cost incurred per diagnostic sample is lesser than that of FAT. Although recent techniques such as the real time RT-PCR has proven much more sensitive in detecting rabies virus than a classical RT-PCR assay, the expensive instrumentation and the higher cost incurred per diagnostic sample are the major hurdles for its use as routine diagnostic test.
At present, no recommended standard protocol for rabies diagnosis using RT-PCR has been published by the OIE or WHO. Obstacles to the general use of PCR for rabies diagnosis are the lack of standardised procedures, quality issues like contamination or false-negative results and the varying reliability of PCR results in many laboratories, especially in developing countries [11]. This pilot study clearly demonstrates the potentiality of this assay in routine diagnosis of rabies when used in conjunction with FAT. Although the WHO does not currently recommend RT-PCR for routine post mortem diagnosis of rabies, this method is approved for epidemiological surveys in laboratories with strict quality control procedures in place and that have experience and expertise with these techniques [21]. Moreover, this assay can be the method of choice when ante-mortem diagnostic samples such as the cerebrospinal fluid, saliva and skin biopsies that contain significantly lower quantities of the viral genome than the brain tissue are to be tested. It can further be used in instances when the conventional virological assays cannot be used due to the poor quality of the diagnostic sample.
Contributor Information
R. P. Aravindh Babu, Email: aravindh.babu@iah.ac.uk
S. Manoharan, Email: ulagaimano@yahoo.com
References
- 1.Araujo DB, Langoni H, Almeida MF, Megid J. Heminested reverse-transcriptase polymerase chain reaction (hnRT-PCR) as a tool for rabies virus detection in stored and decomposed samples. BMC Res Notes. 2008;1:17. doi: 10.1186/1756-0500-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravindh Babu RP, Manoharan S, Ramadass P, Chandran NDJ. Rabies in south Indian cows: an evidence of Sri Lankan rabies virus variant Infection based on the analysis of partial nucleoprotein gene. Indian J Virol. 2011;22:138–141. doi: 10.1007/s13337-011-0050-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhatia R, Ichhpujani RL, Madhusudana SN, Hemachudha T. Rabies in South and Southeast Asia. Proceedings of the WHO expert consultation on rabies. Geneva; 2004.
- 4.Biswal M, Ratho R, Mishra B. Usefulness of reverse transcriptase-polymerase chain reaction for detection of rabies RNA in archival samples. Jpn J Infect Dis. 2007;60:298–299. [PubMed] [Google Scholar]
- 5.Black EM, McElhinney LM, Lowings JP, Smith J, Johnstone P, et al. Molecular methods to distinguish between classical rabies and the rabies-related European bat lyssaviruses. J Virol Methods. 2000;87:123–131. doi: 10.1016/S0166-0934(00)00159-2. [DOI] [PubMed] [Google Scholar]
- 6.David D, Yakobson B, Rotenberg D, Dveres N, Davidson I, et al. Rabies virus detection by RT-PCR in decomposed naturally infected brains. Vet Microbiol. 2002;87:111–118. doi: 10.1016/S0378-1135(02)00041-X. [DOI] [PubMed] [Google Scholar]
- 7.Dean DJ, Abelseth MK, Athanasiu P. The fluorescent antibody test. In: Meslin FX, Kaplan MM, Kowprowski H, editors. Laboratory techniques in rabies. Geneva: World Health Organization; 1996. pp. 88–93. [Google Scholar]
- 8.Fooks AR, Johnson N, Freuling CM, Wakeley PR, Banyard AC, et al. Emerging technologies for the detection of rabies virus: challenges and hopes in the 21st century. PLoS Negl Trop Dis. 2009;3:e530. doi: 10.1371/journal.pntd.0000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta PK, Singh RK, Sharma RN, Rao YU, Butchaiah G. Preliminary report on a single-tube, non-interrupted reverse transcription-polymerase chain reaction for the detection of rabies virus in brain tissue. Vet Res Commun. 2001;25:239–247. doi: 10.1023/A:1006437810594. [DOI] [PubMed] [Google Scholar]
- 10.Heaton PR, Johnstone P, McElhinney LM, Cowley R, O’Sullivan E, et al. Heminested PCR assay for detection of six genotypes of rabies and rabies-related viruses. J Clin Microbiol. 1997;35:2762–2766. doi: 10.1128/jcm.35.11.2762-2766.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann B, Freuling CM, Wakeley PR, Rasmussen TB, Leech S, et al. Improved safety for molecular diagnosis of classical rabies viruses by use of a TaqMan real-time reverse transcription-PCR “double check” strategy. J Clin Microbiol. 2010;48:3970–3978. doi: 10.1128/JCM.00612-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hostnik P, Bidovec A, Maganja DB, Grom J. Rabies: doubtful and discordant results in fluorescent antibody test. Med Pregl. 2001;54(Suppl 1):29–31. [PubMed] [Google Scholar]
- 13.Hughes GJ, Kuzmin IV, Schmitz A, Blanton J, Manangan J, et al. Experimental infection of big brown bats (Eptesicus fuscus) with Eurasian bat lyssaviruses Aravan, Khujand, and Irkut virus. Arch Virol. 2006;151:2021–2035. doi: 10.1007/s00705-005-0785-0. [DOI] [PubMed] [Google Scholar]
- 14.Kulonen K, Fekadu M, Whitfield S, Warner CK. An evaluation of immunofluorescence and PCR methods for detection of rabies in archival Carnoy-fixed, paraffin-embedded brain tissue. Zentralbl Veterinarmed B. 1999;46:151–155. doi: 10.1046/j.1439-0450.1999.00200.x. [DOI] [PubMed] [Google Scholar]
- 15.Matha IS, Salunke SR. Immunogenicity of purified vero cell rabies vaccine used in the treatment of fox-bite victims in India. Clin Infect Dis. 2005;40:611–613. doi: 10.1086/427700. [DOI] [PubMed] [Google Scholar]
- 16.Nagarajan T, Nagendrakumar SB, Mohanasubramanian B, Rajalakshmi S, Hanumantha NR, et al. Phylogenetic analysis of nucleoprotein gene of dog rabies virus isolates from Southern India. Infect Genet Evol. 2009;9:976–982. doi: 10.1016/j.meegid.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 17.OIE. Rabies. In: manual of diagnostic tests and vaccines for terrestrial animals. Paris; 2011. pp. 2.1.13.
- 18.Tordo N, Bourhy H, Sacramento D. PCR technology for lyssavirus diagnosis. In: Clewley JP, editor. Polymerase chain reaction (PCR) for human viral diagnosis. Boca Raton: CRC; 1995. pp. 125–145. [Google Scholar]
- 19.Tordo N, Sacramento D, Bourhy H. The polymerase chain reaction (PCR) technique for diagnosis, typing and epidemiological studies. In: Meslin FX, Kaplan MM, Koprowski H, editors. Laboratory techniques in rabies. Geneva: World Health Organisation; 1996. pp. 157–170. [Google Scholar]
- 20.Whitby JE, Johnstone P, Sillero-Zubiri C. Rabies virus in the decomposed brain of an Ethiopian wolf detected by nested reverse transcription-polymerase chain reaction. J Wildl Dis. 1997;33:912–915. doi: 10.7589/0090-3558-33.4.912. [DOI] [PubMed] [Google Scholar]
- 21.WHO. WHO expert consultation on rabies. World Health Organ Tech Rep Ser 931: 2005. 1–88, back cover. [PubMed]