Abstract
Picobirnaviruses (PBVs) are small, non-enveloped, 35–41 nm virion with bisegmented double-stranded RNA genome. PBVs are widespread and were detected in feces of humans and a wide variety of animals. Domestic pig, one of the ubiquitous farm animal reported incessant association with a variety of viral zoonoses. The objective of our study is to find out the incidence of PBV infection in healthy domestic pigs. The study was conducted by collecting feces of healthy/asymptomatic pigs from a piggery located in an urban slum at Kolkata, India to detect PBV infections. All the 11 fecal samples were tested by polyacrylamide gel electrophoresis and reverse transcription–polymerase chain reaction assay. In this study, we report the first incidence of detection and molecular characterization of porcine PBV (BG-Por-2/2010 and BG-Por-7/2010) in feces of domestic pigs from India using the human PBV genogroup I specific primer pair: PicoB25(+) and PicoB43(−). Sequence comparison and phylogenetic analysis of partial RNA-dependent RNA polymerase gene of genome segment 2 revealed genetic relatedness to hitherto reported porcine, murine and human genogroup I PBVs from different geographical regions. This warrants a stringent global surveillance to study the potential zoonotic and emerging PBV infections.
Electronic supplementary material
The online version of this article (doi:10.1007/s13337-012-0106-z) contains supplementary material, which is available to authorized users.
Keywords: Picobirnaviruses (PBVs), Genogroup I PBVs, Domestic pigs, Genomic diversity, Zoonoses
Picobirnavirus (PBV), a non-enveloped, bisegmented double-stranded RNA (dsRNA) virus, with spherical virion of 35–41 nm in diameter. PBV is the only genus in the Picobirnaviridae family [6]. PBV infect vertebrates and is thought to be transmitted via the fecal-oral route. When PBV is shed in large quantity in the feces, its genome segments are visible by polyacrylamide gel electrophoresis (PAGE) and silver staining. The size of the large genome segment ranges from 2.3 to 2.6 kbp, while the small genome segment is 1.5–1.9 kbp [35]. Genome patterns can be classified into large and small profiles [15].
To date, only one complete nucleotide sequence of a PBV strain; the human Hy005102 strain has been published. This strain was detected in a stool specimen of an infant with acute non-bacterial gastroenteritis from Thailand [37]. The genomic segment 1 has two open reading frames (ORF1 and ORF2). ORF1 codes for a hydrophilic 224 amino acids protein of unknown function. ORF2 encodes for the 552 amino acids capsid protein. The ORF1 gene, in the segment 2, encodes for the viral RNA-dependent RNA polymerase (RdRp) (534 amino acids). The virion structure consists of an outer capsid and a simple core inside with distinctive icosahedral arrangement [10].
The serendipitous detection of PBV dates back to late 1980s while rotavirus surveillance during gastroenteritis outbreaks as well as analyzing feces of free-living rats by PAGE assay [32, 33]. Thereafter, PBVs were detected from different countries in feces of humans, a wide range of mammals, birds, reptiles and even from environmental samples [1–5, 7, 8, 12, 14, 16–18, 20–23, 25, 27–30, 34, 36]. PBVs were detected from immunocompromised patients [24] in which implied to be “opportunistic pathogens” and have also been detected from asymptomatic hosts [11]. The question whether PBV is an ‘innocuous agent’ of the intestine remains to be investigated [2, 7, 19, 31, 35].
The detection of bisegmented dsRNA genome of PBV by PAGE and silver staining [26], is one of the standard and reliable laboratory diagnosis. The reverse transcription–polymerase chain reaction (RT–PCR) protocol developed by Rosen et al. [35] enabled the detection and molecular characterization of PBV studies, worldwide. With the two sets of primer pairs that specifically amplify small fragments within the RdRp gene of segment 2, PBV strains have been classified into two genogroups (genogroup I with 201 bp amplicon and genogroup II with 369 bp amplicon) represented by prototype strains 1-CHN-97 and 4-GA-91 isolated in China and USA, respectively.
Pigs are increasingly recognized to harbor a wide range of viruses that apparently establish long-term persistence and of emerging zoonotic potential [9]. In this study, we carried out the detection and molecular characterization assays for porcine PBV in feces of asymptomatic domestic pigs in Kolkata, India. Out of 11 samples, two showed very faint PBV positive (large genome profile) by PAGE and both the samples showed positive for genogroup I PBV by RT–PCR using genogroup specific primer pair developed by Rosen et al. [35]. Further characterization by sequencing and phylogenetic analyses revealed that the genogroup I porcine PBV strain detected during this study clustered with hitherto reported porcine, murine and human PBV strains reported from various countries.
Eleven fecal specimens were collected from domestic pigs (Sus scrofa domesticus) without diarrhea from a piggery located at an urban slum at Kolkata, India during November 2010 as part of an ongoing epidemiological study on PBV infections. The age of hosts ranged from 1 to 6 years. Ten percent clarified fecal suspensions were prepared using 10 mM phosphate buffered saline (PBS) solution (pH 7.4) and stored at 4 °C refrigerator till use as previously described [17]. Briefly, an aliquot of the fecal sample was diluted with 10 mM (1×) PBS solution, vortexed thoroughly and centrifuged at 3,000 rpm (704×g) for 15 min at 4 °C for preliminary clarification. The supernatant was transferred to a fresh microfuge tube and centrifuged again at 7,000 rpm (3,834×g) for 15 min. The supernatant was finally saved in a fresh microfuge tube as clarified fecal suspension and stored at 4 °C.
The dsRNA was extracted from fecal suspension using phenol–chloroform-isoamyl alcohol mixture for PAGE assays as previously described [3] and subsequent visualization of dsRNA migration patterns after PAGE and silver staining was performed according to Herring et al. [26].
Molecular biology grade viral RNA extraction was carried out using the commercially available QIAGEN QIAamp® Viral RNA mini kit (QIAGEN GmbH, Hilden, Germany) as per manufacturer’s instructions.
The primer pairs described by Rosen et al. [35] were used for molecular characterization experiments. RT–PCR was carried out following the protocol of Bhattacharya et al. [3]. The purification of PCR products, sequencing and sequence analysis were performed as previously described [16].
The sequence data of 201 bp amplicon of one PBV positive sample (nucleotide sequence fragment covering partial RdRp gene of genomic segment 2 of genogroup I porcine PBV strain: genogroup I PBV/Pig/India/BG-Por-2/2010) analyzed during this study was submitted to the DNA Data Bank of Japan (DDBJ; http://www.nig.ac.jp/) DDBJ/EMBL/GenBank nucleotide sequence databases with the following accession number: AB669584.
Picobirnavirus was detected from fecal specimens of asymptomatic pigs using a combination of molecular methods. By PAGE assay, all the 11 porcine fecal samples were negative for rotavirus, except two, which showed very faint PBV positive. Subsequently, all the samples were subjected for RT–PCR targeting RdRp gene with genogroup I specific primers [PicoB25(+) and PicoB43(−)] and genogroup II specific primers [PicoB23(+) and PicoB24(−)]. Genotyping RT–PCR results confirmed the presence of only two genogroup I PBV strains with the amplicon of 201 bp in BG-Por-2 and BG-Por-7. The genogroup nature of strain was further confirmed by direct sequencing in both directions separately. Sequence analysis of one of the PBV strain (BG-Por-2) showed close relatedness to hitherto reported porcine, murine and human PBV strains. Based on the recently proposed nomenclature by the Brazilian research group [13], the genogroup I PBV strain detected and sequenced during this study is designated as: genogroup I PBV/Pig/India/BG-Por-2/2010. The other PBV RT–PCR positive sample (BG-Por-7/2010) did not give a clear sequence even after repetition of assay, so not included for analyses.
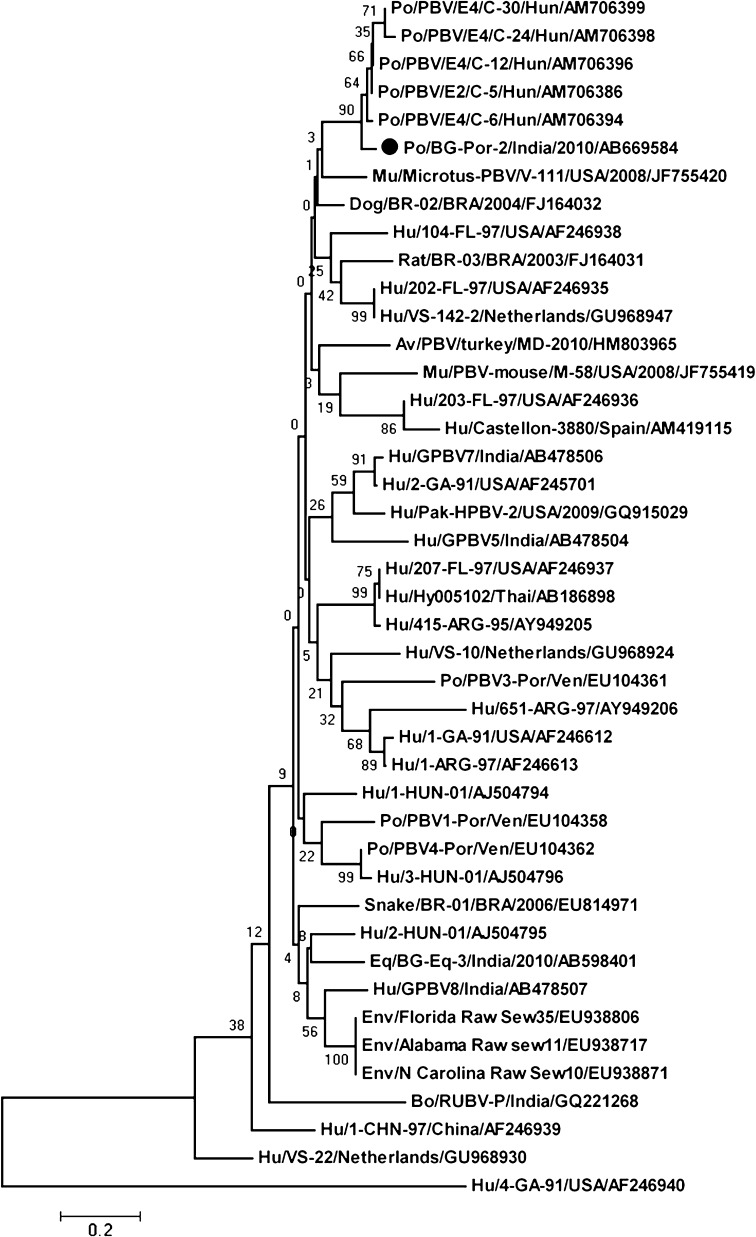
The comparison of percentage nucleotide identity (170 bp) and the partial length deduced amino acid identity (given in parentheses) of gene segment 2 (stretch of 56 amino acids) among the porcine genogroup I PBV strain (PBV/Pig/India/BG-Por-2/2011) detected in Kolkata and hitherto reported PBV strains from various hosts and PBV prototype strains were compared (Supplementary Table 1). The comparison of deduced stretch of 56 amino acids of porcine PBV strain BG-Por-2 and hitherto reported PBVs from various hosts showed that 14 amino acids were conserved, whereas distinct amino acid changes were observed in other positions (Supplementary Table 2). The comparison of nucleotide sequence of Indian isolate of Porcine PBV with that of a few randomly selected Hungarian PBV strains from human and porcine hosts was shown in the Supplementary Table 3. The phylogenetic tree (Fig. 1) also indicated that the detected genogroup I porcine PBV strain was distinct as it clustered on separate branches showing close homology to other porcine PBV strains reported from Hungary.
Fig. 1.
Phylogenetic tree showing the porcine PBV strain (genogroup I PBV/Pig/India/BG-Por-2/2010) with cognate stretch of hitherto reported human, porcine, bovine, canine, murine, avian, serpentine and environmental genogroup I PBV strains based on partial amino acid sequence [56 amino acids (aa)] partial RdRp gene of genomic segment 2. The phylogenetic tree was constructed by the neighbor-joining method using the MEGA software (Version 4.1). Phylogenetic distances were measured by the Kimura two-parameter model, and the tree was statistically supported by bootstrapping with 1,000 replicates. The genogroup I porcine PBV strain BG-Por-2 is denoted with a filled circle symbol. The tree was rooted with cognate stretch of gene segment 2 of genogroup II prototype strain Hu/4-GA-91 (USA) defined as the outgroup strain. Bar 0.2 substitutions per nucleotide. Abbreviations: Hu human, Bo bovine, Po porcine, Eq equine, Av avian, Mu murine, Env environmental, ARG Argentina, BRA Brazil, Hun Hungary, Thai Thailand, USA United States of America, Ven Venezuela
Detection of PBV in feces of domestic pigs has been reported for the first time in Kolkata, India. The presence of PBV in asymptomatic swine of 1 and 6 years of age might be consistent with a previous onset of infection and subsequent establishment of viral persistence.
A research group in Argentina published a study on systematic sampling of fecal specimens from 150 various animals and birds in captivity from a zoo [30]. In that study, PBV was detected by PAGE and silver staining in 3.70 % fecal samples (19 out of 513) among mammals and birds. The authors suggested a lack of etiological relation of PBV with disease; since, none of the host species showed any signs of diarrhea. These authors proposed that factors like stress due to captivity and/or isolations might favor the PBV replicative cycle in these hosts.
Likewise, another research group from the same region conducted a study in farm animals in Argentina [29] reported dissimilar excretion of PBV in fecal specimens of pigs at different age groups. Also varied physiological characteristics were observed among pigs during the period of lactation and final stage of pregnancy. The research group reported their observations stating that the conditions for higher PBV excretion was attributed to particular physiological status of the hosts, especially farrowing and lactation. They also concluded that the higher detection rates of PBV in the study population might be due to the stress conditions generated by pig farming practices during these stages [29]. They have also suggested that PBV is acquired early in life and establishes a persistent infection among the hosts which exhibit a unique pattern of virus excretion with periods of high viral activity intermingled with periods of silence. Thus concluding that the host infected with PBV could remain life-long asymptomatic carriers and serve as reservoirs of infection [29].
Picobirnaviruses present wide genetic diversity and are evolving rapidly. However, the PAGE assay detects the PBVs readily as it is not dependent on their genomic sequences. PBVs are known to cause chronic diarrhoea with prolonged shedding of the virus in humans [24] and various animals [30] besides frequent infections among piglets [5, 29]. Moreover, the presence of genogroup I PBVs in humans and different animals, rodents and reptiles, suggests that any specific genogroup is not restricted to specific host [2, 3, 5, 12, 16, 19, 31].
Evidence for genetic relatedness was reported between human and animal PBVs or vice versa [2, 5, 16–18, 22] from around the globe. Genogroup I PBVs detected and sequenced from pigs in Hungary [2] and Venezuela and Argentina [5, 22], were observed to be closely related to human genogroup I PBVs. These results strongly suggest that PBV strains may circulate in the shared environment where humans and pigs live. The question if PBV is transmitted directly through contact between different host species or by using the same sources (e.g. Water) is an objective of future studies [19].
These findings on detection of PBV in Indian (Kolkata) pigs extend our knowledge about their widespread geographical occurrence and reinforce the need to survey parallel PBV infections in humans and animals.
Electronic supplementary material
Acknowledgments
We are grateful to Tapan Dutta, Debshis Khatua, Md. Azim and Dipak Das who helped in organizing, collection and transport of porcine fecal specimens to the Virology laboratory for processing. We sincerely acknowledge the technical assistance of Bimal Bera, Khokon Sen, Mussaraf Hossain, Neh Nupur, Chandrima Ghosh, Durga Poddor, Srinwanthi Bhattacharya, Arpan Kahali and the staff members of Division of Virology for their whole-hearted support. The study was financially supported by Indian Council of Medical Research (ICMR, Govt. of India), Japan International Co-operation Agency (JICA, Govt. of Japan) and Program of Founding Research Centre for Emerging and Reemerging Infectious Disease (Okayama University—NICED, India) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
References
- 1.Bányai K, Jakab F, Reuter G, Bene J, Uj M, Melegh B, Szücs G. Sequence heterogeneity among human picobirnaviruses detected in a gastroenteritis outbreak. Arch Virol. 2003;148:2281–2291. doi: 10.1007/s00705-003-0200-z. [DOI] [PubMed] [Google Scholar]
- 2.Bányai K, Martella V, Bogdán A, Forgách P, Jakab F, Meleg E, Bíró H, Melegh B, Szucs G. Genogroup I picobirnaviruses in pigs: evidence for genetic diversity and relatedness to human strains. J Gen Virol. 2008;89:534–539. doi: 10.1099/vir.0.83134-0. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharya R, Sahoo GC, Nayak MK, Rajendran K, Dutta P, Mitra U, Bhattacharya MK, Naik TN, Bhattacharya SK, Krishnan T. Detection of genogroup I and II human picobirnaviruses showing small genomic RNA profile causing acute watery diarrhoea among children in Kolkata, India. Infect Genet Evol. 2007;7:229–238. doi: 10.1016/j.meegid.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Browning GF, Chalmers RM, Snodgrass DR, Batt RM, Hart CA, Ormarod SE, Leadon D, Stoneham SJ, Rossdale PD. The prevalence of enteric pathogens in diarrhoeic thoroughbred foals in Britain and Ireland. Equine Vet J. 1991;23:405–409. doi: 10.1111/j.2042-3306.1991.tb03751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carruyo GM, Mateu G, Martínez LC, Pujol FH, Nates SV, Liprandi F, Ludert JE. Molecular characterization of porcine picobirnaviruses and development of a specific reverse transcription–PCR assay. J Clin Microbiol. 2008;46:2402–2405. doi: 10.1128/JCM.00655-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carstens EB, Ball LA. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses. Arch Virol. 2009;154:1181–1188. doi: 10.1007/s00705-009-0400-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandra R. Picobirnavirus, a novel group of undescribed viruses of mammals and birds: a minireview. Acta Virol. 1997;41:59–62. [PubMed] [Google Scholar]
- 8.Chasey D. Porcine picobirnavirus in UK? Vet Rec. 1990;126:465. [PubMed] [Google Scholar]
- 9.Drew TW. The emergence and evolution of swine viral diseases: to what extent have husbandry systems and global trade contributed to their distribution and diversity? Rev Sci Tech. 2011;30:95–106. doi: 10.20506/rst.30.1.2020. [DOI] [PubMed] [Google Scholar]
- 10.Duquerroy S, Da Costa B, Henry C, Vigouroux A, Libersou S, Lepault J, Navaza J, Delmas B, Rey FA. The picobirnavirus crystal structure provides functional insights into virion assembly and cell entry. EMBO J. 2009;28:1655–1665. doi: 10.1038/emboj.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 2008;4:e1000011. doi: 10.1371/journal.ppat.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fregolente MC, de Castro-Dias E, Martins SS, Spilki FR, Allegretti SM, Gatti MS. Molecular characterization of picobirnaviruses from new hosts. Virus Res. 2009;143:134–136. doi: 10.1016/j.virusres.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Fregolente MC, Gatti MS. Nomenclature proposal for picobirnavirus. Arch Virol. 2009;154:1953–1954. doi: 10.1007/s00705-009-0537-z. [DOI] [PubMed] [Google Scholar]
- 14.Gallimore C, Lewis D, Brown D. Detection and characterization of a novel bisegmented double-stranded RNA virus (picobirnavirus) from rabbit faeces. Arch Virol. 1993;133:63–73. doi: 10.1007/BF01309744. [DOI] [PubMed] [Google Scholar]
- 15.Gallimore CI, Appleton H, Lewis D, Green J, Brown DW. Detection and characterisation of bisegmented double-stranded RNA viruses (picobirnaviruses) in human faecal specimens. J Med Virol. 1995;45:135–140. doi: 10.1002/jmv.1890450204. [DOI] [PubMed] [Google Scholar]
- 16.Ganesh B, Nataraju SM, Rajendran K, Ramamurthy T, Kanungo S, Manna B, Nagashima S, Sur D, Kobayashi N, Krishnan T. Detection of closely related picobirnaviruses among diarrhoeic children in Kolkata: evidence of zoonoses? Infect Genet Evol. 2010;10:511–516. doi: 10.1016/j.meegid.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Ganesh B, Nagashima S, Ghosh S, Nataraju SM, Rajendran K, Manna B, Ramamurthy T, Niyogi SK, Kanungo S, Sur D, Kobayashi N, Krishnan T. Detection and molecular characterization of multiple strains of picobirnavirus causing mixed infection in a diarrhoeic child: emergence of prototype genogroup II-like strain in Kolkata, India. Int J Mol Epidemiol Genet. 2011;2:61–72. [PMC free article] [PubMed] [Google Scholar]
- 18.Ganesh B, Banyai K, Masachessi G, Mladenova Z, Nagashima S, Ghosh S, Nataraju SM, Pativada M, Kumar R, Kobayashi N. Genogroup I picobirnavirus in diarrhoeic foals: can the horse serve as a natural reservoir for human infection? Vet Res. 2011;42:52. doi: 10.1186/1297-9716-42-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganesh B, Banyai K, Martella V, Jakab F, Masachessi G, Kobayashi N. Picobirnavirus infections: viral persistence and zoonotic potential. Rev Med Virol. 2012;22:245–256. doi: 10.1002/rmv.1707. [DOI] [PubMed] [Google Scholar]
- 20.Gatti MS, de Castro AF, Ferraz MM, Fialho AM, Pereira HG. Viruses with bisegmented double-stranded RNA in pig faeces. Res Vet Sci. 1989;47:397–398. [PubMed] [Google Scholar]
- 21.Ghosh S, Kobayashi N, Nagashima S, Naik TN. Molecular characterization of full-length genomic segment 2 of a bovine picobirnavirus strain: evidence for high genetic diversity with genogroup I picobirnaviruses. J Gen Virol. 2009;90:2519–2524. doi: 10.1099/vir.0.013987-0. [DOI] [PubMed] [Google Scholar]
- 22.Giordano MO, Martinez LC, Masachessi G, Barril PA, Ferreyra LJ, Isa MB, Valle MC, Massari PU, Nates SV. Evidence of closely related picobirnavirus strains circulating in humans and pigs in Argentina. J Infect. 2011;62:45–51. doi: 10.1016/j.jinf.2010.09.031. [DOI] [PubMed] [Google Scholar]
- 23.Green J, Gallimore CI, Clewley JP, Brown DW. Genomic characterisation of the large segment of a rabbit picobirnavirus and comparison with the atypical picobirnavirus of Cryptosporidium parvum. Arch Virol. 1999;144:2457–2465. doi: 10.1007/s007050050658. [DOI] [PubMed] [Google Scholar]
- 24.Grohmann GS, Glass RI, Pereira HG, Monroe SS, Hightower AW, Weber R, Bryan RT. Enteric viruses and diarrhea in HIV-infected patients. Enteric Opportunistic Infections Working Group. N Engl J Med. 1993;329:14–20. doi: 10.1056/NEJM199307013290103. [DOI] [PubMed] [Google Scholar]
- 25.Hamza IA, Jurzik L, Uberla K, Wilhelm M. Evaluation of pepper mild mottle virus, human picobirnavirus and Torque teno virus as indicators of fecal contamination in river water. Water Res. 2011;45:1358–1368. doi: 10.1016/j.watres.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 26.Herring AJ, Inglis NF, Ojeh CK, Snodgrass DR, Menzies JD. Rapid diagnosis of rotavirus infection by direct detection of viral nucleic acid in silver-stained polyacrylamide gels. J Clin Microbiol. 1982;16:473–477. doi: 10.1128/jcm.16.3.473-477.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludert JE, Hidalgo M, Gil F, Liprandi F. Identification in porcine faeces of a novel virus with a bisegmented double stranded RNA genome. Arch Virol. 1991;117:97–107. doi: 10.1007/BF01310495. [DOI] [PubMed] [Google Scholar]
- 28.Malik YS, Chandrashekar KM, Sharma K, Haq AA, Vaid N, Chakravarti S, Batra M, Singh R, Pandey AB. Picobirnavirus detection in bovine and buffalo calves from foothills of Himalaya and Central India. Trop Anim Health Prod. 2011;43:1475–1478. doi: 10.1007/s11250-011-9834-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martínez LC, Masachessi G, Carruyo G, Ferreyra LJ, Barril PA, Isa MB, Giordano MO, Ludert JE, Nates SV. Picobirnavirus causes persistent infection in pigs. Infect Genet Evol. 2010;10:984–988. doi: 10.1016/j.meegid.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Masachessi G, Martínez LC, Giordano MO, Barril PA, Isa BM, Ferreyra L, Villareal D, Carello M, Asis C, Nates SV. Picobirnavirus (PBV) natural hosts in captivity and virus excretion pattern in infected animals. Arch Virol. 2007;152:989–998. doi: 10.1007/s00705-006-0900-2. [DOI] [PubMed] [Google Scholar]
- 31.Nates SV, Gatti MSV, Ludert JE. The picobirnavirus: an integrated view on its biology, epidemiology and pathogenic potential. Future Virol. 2011;6:223–235. doi: 10.2217/fvl.10.76. [DOI] [Google Scholar]
- 32.Pereira HG, Fialho AM, Flewett TH, Teixeira JM, Andrade ZP. Novel viruses in human faeces. Lancet. 1988;2:103–104. doi: 10.1016/S0140-6736(88)90032-3. [DOI] [PubMed] [Google Scholar]
- 33.Pereira HG, Flewett TH, Candeias JA, Barth OM. A virus with a bisegmented double-stranded RNA genome in rat (Oryzomys nigripes) intestines. J Gen Virol. 1988;69:2749–2754. doi: 10.1099/0022-1317-69-11-2749. [DOI] [PubMed] [Google Scholar]
- 34.Pongsuwanna Y, Taniguchi K, Chiwakul M, Urasawa T, Wakasugi F, Jayavasu C, Urasawa S. Serological and genomic characterization of porcine rotaviruses in Thailand: detection of a G10 porcine rotavirus. J Clin Microbiol. 1996;34:1050–1057. doi: 10.1128/jcm.34.5.1050-1057.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosen BI, Fang ZY, Glass RI, Monroe SS. Cloning of human picobirnavirus genomic segments and development of an RT–PCR detection assay. Virology. 2000;277:316–329. doi: 10.1006/viro.2000.0594. [DOI] [PubMed] [Google Scholar]
- 36.Symonds EM, Griffin DW, Breitbart M. Eukaryotic viruses in wastewater samples from the United States. Appl Environ Microbiol. 2009;75:1402–1409. doi: 10.1128/AEM.01899-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wakuda M, Pongsuwanna Y, Taniguchi K. Complete nucleotide sequences of two RNA segments of human picobirnavirus. J Virol Methods. 2005;126:165–169. doi: 10.1016/j.jviromet.2005.02.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.