Abstract
An isolate of Cucumber mosaic virus (CMV) retrieved from tomato in India was characterized based on its transmission by sap inoculations, Western blot immuno-assay and sequence analysis of RNA3 and 2b genome. The RNA3 genome was of 2,220 nucleotides (nt) which contained two ORFs: movement protein of 852 nt translating 283 amino acids and coat protein of 657 nt translating 218 amino acids. The complete sequence of RNA3 geneome (Acc. EF153734) shared highest 98–99% identities with P1-1, Tfn, and Nt9 strains of CMV infecting tomato reported from abroad. The 333 nucleotides long RNA2b gene (Acc. EF710773) also showed highest 98% identities with P1-1 and 97% with Tfn and NT9 strains of CMV but only 83–84% identities with Indian strains of CMV. Therefore, the isolate under study was identified as a new isolate of CMV of subgroup IB based on highest sequence similarities and closer affinity to European or East Asian isolates of CMV.
Keywords: Tomato, Cucumber mosaic virus, Molecular characterization, Distinct isolate
Cucumber mosaic virus (CMV) is a type member of genus Cucumovirus, family Bromoviridae. Due to its wide host range (more than 1,000 plant species) and its economic impact, CMV has been considered as one of the most important viruses [13]. CMV genome is organized into three single-stranded messenger-sense genomic RNAs (RNAs 1, 2, and 3). RNAs 1 and 2 codes for components of the replicase complex, and RNA2 also codes for the 2b protein, involves in the suppression of gene silencing [3]. RNA3 encodes 3a protein, essential for the virus movement, and the coat protein (CP), which is expressed from subgenomic RNA4. The CP has an important role not only in the formation of the viral particles but also in virus movement, transmission by aphid vectors, and symptom expression [13]. Some CMV strains also harbor satellite RNA molecules, which are small, linear, noncoding and single-stranded RNA molecules that depends entirely on the helper virus (CMV) for replication, encapsidation, and transmission but has almost no sequence similarity with the helper CMV genome. Based on their biological, serological, and molecular properties, CMV strains can be divided in two subgroups, I and II, the former being further divided into subgroups IA and IB. The subgroup IB is suggested to contain the ‘Asian strains’ whereas other members of subgroup I have been kept under subgroup IA. The nucleotide sequence identity between CMV subgroup II and I strains ranges from 69 to 77%, while above 90% within subgroup [13].
Tomato (Solanum lycopersicum L., family Solanaceae) is one of the most important vegetable crops grown commercially worldwide. Tomato crop is susceptible to infection of many plant viruses including CMV which causes immense economic loss to its productivity. Various symptoms like yellowing, mottling, twisting, shoestring of leaves, leaf distortion and fern-like appearance of the leaves on tomato infected with CMV have been described earlier [4, 5]. In India, CMV infecting tomato was characterized on the basis of biological and serological level by Kiranmai et al. [6, 7]. Restriction fragment length polymorphism analysis of the PCR product of the CP region of a subgroup II CMV strain causing severe mosaic on tomato in south India was also reported [25]. The association of CMV with shoestring symptoms of tomato for the first time in India was identified by us [16]. In this communication, we further report biological and molecular characterization of the CMV isolate causing shoestring in tomato.
The surveys were conducted in 2007 in tomato fields cultivated nearby the bank of Gomati River, Lucknow to observe virus like symptoms on tomato plants. The natural occurrence of viral disease on tomato (Solanum lycopersicum L.) was observed in tomato growing areas in and around Lucknow. The infected tomato plants exhibited mild to severe mosaic accompanied with shoestring symptoms, which resulted in reduction in fruit quality and in tomato production. The disease incidence was about 20–25% in all four fields surveyed.
For virus transmission mechanical inoculations were done using the leaf sap of infected young leave macerated in 0.1 M potassium phosphate buffer, pH 6.8 (supplemented with 0.1% Na2SO3) in a ratio of 1:10 (w/v) on different host species as described in Table 1. The sap inoculations resulted in chlorotic or necrotic local lesion on Chenopodium album, C.amaranticolor, and Spinacia oleracia at 5–7 days post inoculation (dpi) and various systemic symptoms on a number of plant species viz. Nicotiana glutinosa, N. benthamiana, Hyoscyamus muticus, Solanum lycopersicum,Datura innoxia and Cucumis sativus at 15–20 dpi (Table 1). However, Capsicum annuum, Cucurbita maxima, Petunia hybrida, N. tabaccum cv. Samsun NN, N. tabaccum cv. White Burley and N. rustica neither develop any local nor systemic symptoms even at 30 dpi. The response of virus strain under study (CMV-Ts) on different host species was also compared with other CMV strains viz. CMV-Ch [9], CMV-A [24], CMV-D [17], and CMV-H [21] reported earlier from India, which showed a clear cut distinct identity of the virus isolate (Table 1).
Table 1.
Comparison of symptoms (local and systemic)* induced by Cucumber mosaic virus tomato isolate under study (CMV-Ts) with CMV strains of Amaranths (CMV-A) [24], Datura (CMV-D) [17] Henbane (CMV-H) [21] and Chrysanthemum (CMV-Ch) [9] reported earlier from India
| Host plant | CMV-Ts | CMV-Ch | CMV-A | CMV-D | CMV-H | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| L | S | L | S | L | S | L | S | L | S | |
| Amaranthaceae | ||||||||||
| Amaranthus tricolor | – | – | – | – | MChl | SM, Crk & StG | – | MChl | – | MM |
| Asteraceae | ||||||||||
| Chrysanthemum morifolium | – | – | CLL | M & RS | – | – | – | – | – | – |
| Chenopodeaceae | ||||||||||
| Chenopodium album | NLL | – | NLL | – | NLL | – | NLL | – | NLL | – |
| C. amaranticolor | CLL | – | CLL | – | NLL | – | NLL | – | NLL | – |
| Spinacia oleracia | CLL | CLL | – | – | Chl | NLL | – | – | – | |
| Cucurbitaceae | ||||||||||
| Cucurbita maxima | – | – | MChl | MM | MChl | MM | – | – | – | – |
| Cucumis sativus | – | SM | – | SM | – | SM | – | M | – | M |
| Solanaceae | ||||||||||
| Capsicum annuum | – | – | – | MM | – | – | – | LD | – | – |
| Datura innoxia | – | SM, & SL | CLL | SM, TLL & SL | CLL | – | – | SM & TLL | NLL | – |
| Hyoscyamus muticus | – | MM | – | SM & B | – | – | – | – | MChl | SM |
| Nicotiana benthamiana | – | LD & Crk | MChl | SM, LSR, D | MChl | SM | – | – | – | LSR |
| N. glutinosa | – | SM, SL | CLL | SM, SL & StG | CLL | SM, LD & StG | – | MM | BNL | MM |
| N. tabaccum cv. Samsun NN | – | – | – | SM & LD | Chl | SM | – | MM | – | MM |
| N. tabaccum cv. White Burley | – | – | CR | SM & CR | Chl | SM | – | MM | – | MM |
| N. rustica | – | – | CLL | MM, LD & Crk | N | SM, B & LD | Chl | LN & LD | – | LD & Crk |
| Pitunia hybrida | – | – | – | SM, B & Crk | – | MM | – | – | CLL | MM |
| Solanum lycopersicum | MM | SM & SL | MChl | SM & SL | MChl | MM | – | MM | MChl | SM & TLL |
* Local symptoms as observed 7 dpi and systemic symptoms at 20 dpi
B blistering, BNL brown necrotic lesions, Chl chlorosis, CLL chlorotic local lesions, CR chlorotic rings, Crk crinkling, D death of inoculated plants, LD leaf deformation, LN leaf necrosis, LSR reduction in leaf size, MChl mild chlorosis, MM mild mosaic, N necrosis leading to death of tissue, NLL necrotic local lesions, NLL necrotic local lesions of expanding nature, SL shoestring of leaf, SM severe mosaic, StG stunted growth, TLL thinning of leaf lamina
– no symptoms observed
Molecular weight of the CP subunits of virus isolate was determined by SDS-PAGE and comparing its mobility with a protein marker on a 12% Poly acrylamide gel under denaturing conditions. The protein bands were blotted on nitrocellulose membrane and Western blot immuno assay was done as described by Renart and Sandoval [19] using antiserum of CMV (PVAS-242a) as primary antibody and anti rabbit IgG-alkaline phosphatase conjugate (Sigma Aldrich, USA) as secondary antibody. Finally, the protein bands were elucidated by color development reaction on adding BCIP/NBT (Sigma Aldrich, USA) in the dark. The reaction was terminated by adding sterile water and the blot was dried. Positive bands were obtained corresponding to the size of 24 kDa, which closely matched the molecular weight of CP of CMV-Glad [18] taken as a positive control, confirming the association of CMV with shoestring disease of tomato plants and the strain was designated as CMV-Ts.
Total RNA was isolated from 100 mg young leaf tissue of naturally infected tomato as described earlier [14]. The reverse transcription-polymerase chain reaction (RT-PCR) was performed using RNA3 genome specific primers designed from the sequence data of CMV-Fny (Acc. D10538). First strand cDNA synthesis was done using 200U of Revert Aid H-minus MMuLV reverse transcriptase (MBI Fermentas, USA) and the reverse primer. PCR was carried out in thermal cycler PTC-200 (MJ research) using 10 ng cDNA as template, forward and reverse primers and 1.5U of Pfu DNA polymerase [16]. RT-PCR was also performed using the primers specific for 2b gene of CMV. The sequence of upstream primer: 5′-ATGGAATTGAACGCCGGAGGCGCAATG-3′ and downstream primer: 5′-TCAAAACACCCT(CT)CCGCCCACTCGTT-3′ resulting in ~300 bp amplicons. PCR condition for reaction was as follow: initial denaturation at 94°C for 5 min followed by 30 cycles consisting of denaturation at 94°C for 1 min, annealing at 62°C for 30 s, extension of 45 s at 72°C and a final extension for 10 min at 72°C. The PCR products were assessed on 1% agarose gel with DNA marker (100 bp DNA ladder, Genei Pvt. Ltd., India).
During RT-PCR the expected size amplicons of ~2,200 and ~300 bp were obtained with RNA3 and RNA2b specific primers, respectively. Both the amplicons were eluted using QIAquick gel extraction kit (Qiagen, USA) and ligated separately into pGEM-T easy vector system-1 (Promega Corporation, USA). Competent cells of Escherichia coli (strain DH5α) were transformed by following standard molecular biology procedures [22]. Three positive clones were got sequenced from both the ends (MWG, Germany) and nucleotide sequences data was analyzed by the Entrez program using BLAST (http://www.ncbi.nlm.nih.gov/BLAST) at NCBI for knowing consensus sequence and submitted in GenBank database under accession numbers: RNA3 (Acc.EF153734) and RNA2b (Acc. EF710773).
To find out the two ORFs in RNA3 genome, the gene was translated into amino acid residues utilizing the expasy (http://www.expasy.org/tools/dna.html). The nucleotide sequence analysis of RNA3 of CMV-Ts isolate showed the complete length of RNA3 (Acc.EF153734) was of 2,220 nucleotides and consisted of two ORFs. The first frame start codon (AUG) in the ORF was at position 122–124 nucleotide and termination codon (UAG) at 971–973 nucleotide position and identified as MP gene of 852 nucleotides translating 283 amino acids. The second frame ORF started (AUG) at position 1,261–1,263 nucleotide and terminated (UAG) at 1,915–1,917 nucleotide position and identified as CP gene of 657 nucleotides translating 218 amino acids. The ORFs were separated by 288 nucleotides long intergenic region (IR) and flanked by 5′ un-translated region (5′ UTR) and 3′ UTR of 121 and 303 nucleotides, respectively. The sequence data of RNA2b gene (Acc. EF710773) was also analyzed and found to be of 333 nucleotides, starting from ATG at first nucleotide and terminated at 333th nucleotide. The 2b gene translated 110 amino acid residues.
BLASTn analysis of nucleotide sequences of the RNA3 (EF153734) showed highest 99–98% identities with CMV-Tfn (Y16926), Nt9 (D28780) and P1-1 (AM183116) isolates of Italy, Taiwan and Spain, respectively. It also showed 97–96% with CMV strains of India (JF279606, GU111229) and China (EF216867, EF216865, FJ268746, DQ412732, HQ283393). However, the identities were 93% with other CMV strains of Indian origin (EF178298, EF593023, EF593024, EF593026, EF153733 and EF593025) of subgroup IB. BLAST analysis of RNA2b (EF710773) also showed highest 97% nucleotide sequence identity with Tfn, P1-1, and Nt9 strains and 87% identity with Indian strains of CMV.
The matrix for pair wise alignments was obtained using the DiAlign 2 program [11]. Genomatix Dialign analysis of complete RNA3 of sequence data of CMV-Ts isolate (EF153734) with some selected strains of CMV belonging to subgroups I and II, showed highest 99–97% similarities with Tfn, Nt9, P1-1, and Tfr-In strains of CMV of subgroup IB reported from Italy, Taiwan, Spain, and India, respectively. The isolate also showed 96–94% similarities with T-In, Cb7, YN, Cah 1, Phy, T1-2, PE strains reported from India and China. However, CMV-Ts showed only 92% nucleotide similarities with other strains of CMV belonging to subgroup IB reported from India (Table 2). RNA2b ORF showed highest 98% similarities with P1-1 and 97% with Tfn and NT9 strains of CMV, however, only 83–84% sequence similarities with Indian strain of CMV (EU417842, EU006067, EU118810) of subgroup IB.
Table 2.
RNA3 sequence based percent similarities* of CMV tomato isolate (CMV-Ts, Acc. EF153734) at nucleotide (nt) and amino acid (aa) level with respect to various isolates of CMV reported from India and abroad obtained Genomatix DiAlign program
| CMV strains | Country | Host | Accession | Subgroup | RNA3 | 5′ UTR | MP | CP | 3′ UTR | IR | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nt aa | nt aa | |||||||||||
| Tfn | Italy | Solenum lycopersicum | Y16926 | IB | 99 | 99 | 99 | 98 | 99 | 96 | 97 | 98 |
| Nt9 | Taiwan | – | D28780 | IB | 99 | 99 | 98 | 98 | 99 | 96 | 97 | 98 |
| P1-1 | Spain | S. lycopersicum | AM183116 | IB | 97 | 97 | 98 | 98 | 98 | 95 | 97 | 98 |
| Tfr-In | India | S. lycopersicum | JF279606 | IB | 97 | 98 | 98 | 98 | 98 | 96 | 100 | 94 |
| T-In | India | S. lycopersicum | GU111229 | IB | 96 | 95 | 93 | 89 | 99 | 97 | 97 | 96 |
| Cb7 | China | S. lycopersicum | EF216867 | IB | 96 | 97 | 97 | 98 | 96 | 96 | 97 | 92 |
| YN | China | Brassica campestris | EF216865 | IB | 96 | 97 | 96 | 98 | 96 | 96 | 95 | 94 |
| Phy | China | – | DQ412732 | IB | 96 | 97 | 96 | 98 | 96 | 95 | 96 | 94 |
| Cah1 | China | Canna sp. | FJ268746 | IB | 95 | 97 | 96 | 98 | 94 | 96 | 97 | 94 |
| T1-2 | China | S. lycopersicum | HQ283393 | IB | 95 | 96 | 96 | 98 | 96 | 96 | 95 | 93 |
| PE | China | Passion flower | AF268597 | IB | 94 | 98 | 93 | 94 | 95 | 93 | 96 | 86 |
| Ia | Indonesia | – | AB042294 | IB | 93 | 96 | 93 | 95 | 92 | 96 | 94 | 91 |
| Rs | India | Rauvolfia serpentina | EF593025 | IB | 92 | 97 | 92 | 95 | 91 | 94 | 97 | 90 |
| CTL | China | Brassica chinensis | EF213025 | IB | 92 | 99 | 94 | 96 | 93 | 96 | 91 | 89 |
| Ban | India | Musa paradisiaca | EF178298 | IB | 92 | 96 | 92 | 95 | 91 | 94 | 91 | 90 |
| J | India | Jatropha curcas | EF593026 | IB | 92 | 97 | 92 | 95 | 91 | 91 | 97 | 90 |
| B2 | Indonesia | Musa sapientum | AB046951 | IB | 92 | 96 | 92 | 95 | 93 | 96 | 90 | 91 |
| Ch | India | Chrysanthemum morifolium | EF153733 | IB | 92 | 97 | 92 | 95 | 90 | 91 | 97 | 90 |
| Ix | Phillippines | S. lycopersicum | U20219 | IB | 92 | 95 | 93 | 96 | 92 | 91 | 89 | 83 |
| D | India | Datura innoxia | EF593024 | IB | 92 | 97 | 92 | 94 | 91 | 91 | 97 | 90 |
| A | India | Amaranthus tricolor | EF593023 | IB | 92 | 97 | 92 | 94 | 90 | 92 | 97 | 90 |
| Mf | South Korea | – | AJ276481 | IA | 91 | 88 | 93 | 96 | 94 | 96 | 87 | 83 |
| Fny | USA | Cucurbita pepo | D10538 | IA | 90 | 88 | 92 | 96 | 94 | 96 | 88 | 83 |
| Sny | USA | C. pepo | U66094 | IA | 90 | 82 | 93 | 95 | 94 | 96 | 88 | 82 |
| Leo | Hunagry | S. lycopersicum | AM114273 | IA | 90 | 87 | 92 | 96 | 94 | 96 | 88 | 83 |
| Y | Japan | – | D12499 | IA | 90 | 82 | 93 | 97 | 94 | 94 | 88 | 84 |
| Pepo | Japan | C. pepo | AF103991 | IA | 89 | 89 | 93 | 96 | 92 | 98 | 90 | 81 |
| LI | India | Lilum longifolium | AJ831578 | IA | 82 | 91 | 76 | 77 | 92 | 96 | 77 | 81 |
| Q | Australia | – | M21464 | II | 66 | 54 | 76 | 83 | 73 | 65 | 59 | 57 |
| Tss-In | India | S. lycopersicum | JF279605 | II | 66 | 80 | 70 | 61 | 75 | 65 | 59 | 59 |
| Trk7 | Hungary | – | L15336 | II | 65 | 54 | 76 | 83 | 70 | 63 | 61 | 57 |
| Car | India | Daucus carota | EU642567 | II | 62 | 53 | 74 | 81 | 68 | 52 | 59 | 57 |
| TAV | India | C. morifolium | EF153735 | Outgroup | 42 | 45 | 52 | 63 | 36 | 11 | 48 | 46 |
| PSV | USA | Vigna unguiculata | NC_002040 | Outgroup | 39 | 33 | 56 | 42 | 40 | 18 | 56 | 39 |
* Percent similarities denote the number of identical nucleotide and amino acids (in percent of shorter sequence)
To know the sequence variability, complete RNA3 sequences of CMV-Ts isolate (EF153734) were aligned with CMV strains of subgroup IB, IA and II. Sequence alignments revealed that CMV-Ts shared all the common features of Cucumoviruses. CMV-Ts had 12 unique nucleotide substitutions in its entire genome and two substitutions in MP and IR, five substitutions in CP and three substitutions in 3′ UTR region but no substitution in 5′ UTR. The multiple alignment of amino acid sequences of CP revealed five unique substitutions in C-terminal region at positions 149 (S/P), 150(A/V), 151(M/L), 156 (T/A), and 158 (T/S). However, MP ORF showed only one unique substitution at positions 272 (S/R), suggesting high conservation of amino acid sequence required for its functionality in co-ordination with MP. The RNA2b ORF showed three unique substitutions at positions 5 (A/E), 69 (R/H), and 88 (V/A).
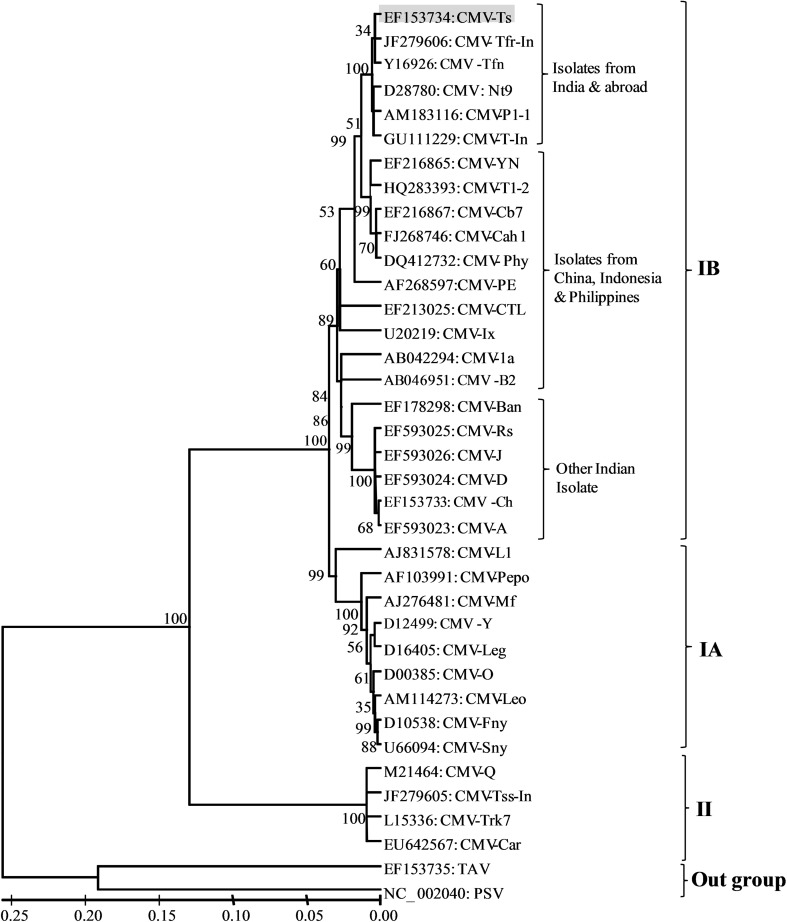
The phylogeny estimation and molecular evolutionary relationships was established using Molecular Evolutionary Genetics Analysis software (MEGA 4.1 version). Tree was rooted on ER-PSV and TAV strains, the other members of genus Cucumovirus. Complete RNA3 sequences of CMV strains of subgroup IA, IB and II which showed highest homology with CMV-Ts strain (EF153734) were considered for phylogenetic analysis along with the other member of genus Cucumovirus: PSV (Acc.NC_002040) and TAV (Acc.EU163411) as out group. RNA3 based phylogenetic analysis clearly divided CMV strains into three clusters of subgroup IA, IB, and II (Fig. 1). CMV-Ts clad within subgroup IB and showed closest relationships with Tfn, Nt9, P1-1, Tfr-In, and T-In CMV isolates reported from abroad and India. While it showed distinct relationships with other CMV strains of Indian origin: CMV-D (EF593024), CMV-A (EF503023), CMV-Ban (EF178298), CMV-J (EF593026) and CMV-Rs (EF593025). The Ts isolate clearly indicated a closer affinity to European or East Asian CMV isolates (Tfn and Nt9) than to other isolates of IB group previously characterized in India. Phylogenetic analysis of RNA2b gene (EF710773) was also carried out with the selected and reported strains of CMV belonging to subgroup IA, IB, and II. During analysis at amino acid level, the CMV-Ts isolate revealed closest relationships with the subgroup IB strains and clustered with Tfn, Nt9, and P1-1 strains of CMV of subgroup IB together with Indian strains of CMV.
Fig. 1.
Phylogenetic analysis of CMV-Ts isolate virus under study (highlighted bygrey shadowing) at nucleotide level of complete RNA3 by MEGA v4.1. The evolutionary history was inferred using the Neighbor-Joining method with 100 replicate bootstraps
The virus associated with shoestring disease of tomato was transmitted successfully by sap inoculations to a number of host plants including Cucumis sativus, a diagnostic host of CMV described by Francki et al. [4], the causal virus was suspected as CMV. Remarkable differences in symptoms were observed with CMV tomato isolate such as: it did not produce any symptoms on N. tabaccum cv. White Burley, cv. Samsun NN, whereas previously characterized Indian CMV strains: CMV-A [24], CMV-H [21], CMV-D (EF593024), CMV-Ban (EF178298) and CMV-Ch [9] induced systemic mosaic on these cultivars of N. tabaccum. In contrast, it did not produce any symptom on chilli, petunia, chrysanthemum and amaranths. The symptom comparisons well distinguished the CMV tomato isolate from other strains of CMV reported earlier from India. This variation in symptoms produced might be due to the invasion of an exotic strain of CMV in India.
Positive reaction of the virus with antiserum of CMV (PVAS 242a) and 24 kDa protein band hybridized with the CMV antiserum during WIBA, which closely matched with the molecular weight of CP of CMV-Glad [18] taken as a positive control, confirming virus isolate under study was CMV. The isolate was further confirmed by RT-PCR using RNA3 specific primers which resulted in positive amplification of the expected size band of ~2.2 kb. The sequencing of PCR amplicons resulted in complete RNA3 genome comprised of 2,220 nucleotides (Acc. EF153734). The sequence analysis of the data revealed highest 98–99% identities with P1-1, Tfn, and Nt9 strains of CMV reported from abroad and 96–97% identities with two Indian strains (Tfr-In and T-In) reported recently, while identities were only 92% with other CMV strains reported earlier from India.
Sequence alignments of complete RNA3 sequences of CMV-Ts isolate (EF153734) revealed that CMV-Ts shared all the common features of Cucumoviruses viz. presence of conserved TG tract in 5′ UTR (as reported earlier by Boccard and Baulcombe [1], the motif ‘CAA CAG TCC TC’ and GGT TCA ATT CC in the 3a and IR gene, respectively and a highly conserved region ‘TCC AGC TTA CGG CTA AAA TGG TCA GTC G’ in 3′ UTR, like in other strains of CMV strains was also obtained [10]. However, no substitutions were observed in 5′ UTR and IR regions. No change was found in the core promoter region within IR region which explains the extreme conservation of this region essentially required for the virus replication as suggested earlier [3]. Five unique amino acid substitutions in C-terminal region at positions 149 (S/P), 150(A/V), 151(M/L), 156 (T/A) and 158 (T/S) of CP of the virus isolate under study were found. The mutation of threonine (T) at position 156 by alanine (A) has also been reported to affect significantly in transmission of CMV by aphid and virion stability [12, 15, 23]. Alterations at these positions might suggest some functional importance to provide selective biological advantages for survival of CMV strains of Indian subcontinent.
Phylogenetic analysis of RNA3 and CP level of CMV-Ts showed the closest affinity to European or East Asian CMV isolates than to other CMV isolates previously characterized in India. The analysis placed CMV-Ts along with other Indian strains in subgroup IB and suggested to be derived from subgroup I as proposed earlier by [20] but separate cluster formed by Indian strains indicated that the Indian strains may have an ancient lineage to IB. Alternatively they could be diverged members of subgroup IB that are evolving independently.
There is a record on a CMV strain causing shoestring disease in tomato from India based on sequence analysis of RNA3 [16]. RNA3 genome based characterization of three distinct tomato strains of CMV have been reported recently from India [8, 26]. We further report, the biological and molecular characterization of CMV isolate retrieved from tomato which has closer affinity to European or East Asian CMV isolates than to other isolates previously characterized in India.
Acknowledgments
The authors are thankful to the Director, CSIR-National Botanical Research Institute, Lucknow for research facilities and Council of Scientific and Industrial Research, New Delhi, India for fellowship to D. Pratap and S. Kumar.
References
- 1.Boccard F, Baulcombe DC. Mutational analysis of cis-acting sequences and gene function in RNA3 of Cucumber mosaic virus. Virology. 1993;193:563–78. [DOI] [PubMed]
- 2.Bonnet J, Fraile A, Sacristan S, Malpica JM, Garcia-Arenal F. Role of recombination in the evolution of natural population of Cucumber mosaic virus, a tripartite RNA plant virus. Virology. 2005;332:359–368. doi: 10.1016/j.virol.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 3.Brigneti G, Voinnet WX, Li H, Ding SW, Baulcombe DC. Viral pathogenecity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 1998;17:6739–6746. doi: 10.1093/emboj/17.22.6739. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Francki RIB, Mossop DW, Hatta T. Cucumber mosaic virus. In: Harrison BD, Mutant AF, editors. CMI/AAB Descriptions of Plant Viruses, vol. 213. 1979. p. 6.
- 5.Gallitelli D, Di Franco A, Vovlas C, Kaper JM. Infezioni miste del virus del mosaico del cetriolo (CMV) e di potyvirus in colture ortive di Puglia e Basilicata. Informa Fitopatol. 1988;12:57–64. [Google Scholar]
- 6.Kiranmai G, Sreenivasulu P, Nayudu MV. Characterization of cucumber mosaic cucumovirus isolates naturally infecting three solanaceous vegetables crops in Andhra Pradesh. Indian Phytopathol. 1997;50:437–438. [Google Scholar]
- 7.Kiranmai G, Sreenivasulu P, Nayudu MV. Epidemiology of cucumber mosaic cucumovirus isolates naturally infecting three solanaceous vegetable crops around Tirupati. Indian Phytopathol. 1998;51:315–318. [Google Scholar]
- 8.Koundal V, Haq QMR, Praveen S. Characterization, genetic diversity, and evolutionary link of Cucumber mosaic virus strain New Delhi from India. Biochem Genet. 2011;49:25–38. doi: 10.1007/s10528-010-9382-8. [DOI] [PubMed] [Google Scholar]
- 9.Kumar S. Molecular characterization of Cucumovirus causing severe mosaic and ring spot disease in chrysanthemum and development of their management strategies. Ph.D. thesis, Lucknow: University of Lucknow; 2008.
- 10.McGarvey P, Tousignant M, Geletka L, Cellini F, Kaper JM. The complete sequence of Cucumber mosaic virus from Ixora that is deficient in the replication of satellite RNAs. J Gen Virol. 1995;76:2257–2270. doi: 10.1099/0022-1317-76-9-2257. [DOI] [PubMed] [Google Scholar]
- 11.Morgenstern B. DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics. 1999;15:211–218. doi: 10.1093/bioinformatics/15.3.211. [DOI] [PubMed] [Google Scholar]
- 12.Ng JCK, Liu S, Perry KL. Cucumber mosaic virus mutants with altered physical properties and defective in aphid vector transmission. Virology. 2000;276:395–403. doi: 10.1006/viro.2000.0569. [DOI] [PubMed] [Google Scholar]
- 13.Palukaitis P, Garcia-Arenal F. Cucumoviruses. Adv Virus Res. 2003;62:241–323. doi: 10.1016/S0065-3527(03)62005-1. [DOI] [PubMed] [Google Scholar]
- 14.Pawlowski K, Kunze R, Vries SD, Bisseling T. Isolation of total, poly (A) and polysomal RNA from plant tissues. Plant Mol Bio Man. 1994;D5:1–13. [Google Scholar]
- 15.Perry KL, Zhang L, Palukaitis P. Amino acid changes in the coat protein of Cucumber mosaic virus differentially affect transmission by the aphids Myzus persicae and Aphis gossypii. Virology. 1998;242:204–210. doi: 10.1006/viro.1998.8991. [DOI] [PubMed] [Google Scholar]
- 16.Pratap D, Kumar S, Raj SK. First molecular identification of a Cucumber mosaic virus isolate causing shoestring symptoms on tomato in India. Aust Plant Dis Notes. 2008;3:57–58. doi: 10.1071/DN08023. [DOI] [Google Scholar]
- 17.Raj SK, Srivastava A, Chandra G, Singh BP. Natural occurrence of Cucumber mosaic virus on Datura innoxia L. in India. Bull OEPP/EPPO Bull. 1999;29:455–457. doi: 10.1111/j.1365-2338.1999.tb01418.x. [DOI] [Google Scholar]
- 18.Raj SK, Srivastava A, Chandra G, Singh BP. Characterization of Cucumber mosaic virus isolate infecting Gladiolus cultivars and comparative evaluation of serological and molecular methods for sensitive diagnosis. Cur Sci. 2002;83:1132–1136. [Google Scholar]
- 19.Renart J, Sandoval IV. Western blots. Methods Enzymol. 1984;104:455–460. doi: 10.1016/S0076-6879(84)04114-8. [DOI] [PubMed] [Google Scholar]
- 20.Roossinck MJ, Zhang L, Hellward K. Rearrangements in the 5′ nontranslated region and phylogenetic analyses of Cucumber mosaic virus RNA3 indicate radial evolution of three subgroups. J Virol. 1999;73:6752–6758. doi: 10.1128/jvi.73.8.6752-6758.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samad A, Raj SK, Srivastava A, Chandra G, Ajaykumar PV, Zaim M, Singh BP. Characterization of a Cucumber mosaic virus isolate infecting Egyptian henbane (Hyoscyamus muticus L.) in India. Acta Virol. 2000;44:131–136. [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor: Cold Spring Harbor Laboratory, Press; 1989.
- 23.Smith TJ, Chase E, Schmidt T, Perry K. The structure of Cucumber mosaic virus and comparison to Cowpea chlorotic mottle virus. J Virol. 2000;74:7578–7586. doi: 10.1128/JVI.74.16.7578-7586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srivastava A, Chandra G, Raj SK. Molecular characterization of a strain of Cucumber mosaic virus based on coat protein and movement protein genes. Acta Virol. 2004;48:229–239. [PubMed] [Google Scholar]
- 25.Sudhakar N, Prasad DN, Mohan N, Murugesan K. First report of Cucumber mosaic virus subgroup II infecting Lycopersicon esculantum in India. Plant Dis. 2006;90:1457. doi: 10.1094/PD-90-1457B. [DOI] [PubMed] [Google Scholar]
- 26.Swapna Geetanjali A, Kumar R, Srivastava PS, Mandal B. Biological and molecular characterization of two distinct tomato strains of Cucumber mosaic virus based on complete RNA3 genome and subgroup specific diagnosis. Indian J Virol. 2011;22:117–126. doi: 10.1007/s13337-011-0051-2. [DOI] [PMC free article] [PubMed] [Google Scholar]