Abstract
The aim of the present study was to investigate the seroprevalence of Hepatitis A virus antibodies in patients with clinical symptoms of viral hepatitis and molecular characterization of the detected isolates. The present study deals with the seroprevalence and the genetic diversity of HAV in 400 Tunisian patients presenting in dispensaries (160 patients) and in University Hospitals (240 patients) with hepatitis symptoms between 2006 and 2008. The patients with acute hepatitis were mainly from rural regions. However, the total number of patients was decreased over time. The collected samples were from patients with hepatitis symptoms occurring mainly during January–March (36.7, 26, and 35.5%) and September–December (39.4, 43.4, and 35.5%) during the three years of study, respectively. However, HAV infection was established for only 110 among 400 patients. The detected isolates were clustered within sub-genotype IA. The present study constituted another report of the continued surveillance of HAV infection in the region of Monastir and the molecular characterisation of the detected strains.
Keywords: Hepatitis A virus, Anti-HAV IgM, RT-PCR, Tunisia
Introduction
Hepatitis A virus (HAV) constitutes the principal etiological agent of acute hepatitis throughout the world, causing substantial morbidity in both developed and developing countries. The severity of hepatitis A is multifactorial, with age, gender, and drug toxicity [24]. HAV, member of the genus Hepatovirus of the Picornaviridae family, has been shown to be responsible for numerous disease outbreaks [5, 8] related to personal and sexual contacts [3, 6, 9]. In addition, several outbreaks were associated to contaminated food and water [9, 12], drug use [1], and other transmission pathways [33]. It is not possible to distinguish HAV strains by serotyping analysis, but seven genotypes can be differentiated with molecular methods: genotypes I, II, III and VII are associated with human infections whereas genotypes IV, V and VI cause infections in simians [29].
Genomic heterogeneity of HAV has been demonstrated based on different genome regions, including those encoding for the virion protein VP3 C-terminus [16], the VP1 N-terminus [26, 27] and the VP1/2A junction (168 bp) [4, 7, 27, 33], which is considered to be the most variable [33] and the most used for genotyping HAV.
Genotypes are distinguished by 15–25 % sequence diversity, whereas sub-genotypes (also referred to as subtypes) in each genotype differ in about 7.5 % of base positions [28]. Subtype IA appears to be responsible for the majority of hepatitis A cases worldwide, whereas subtype IB viruses have been found in the Mediterranean region. The others human genotypes are very infrequent [18, 28]. Comparison among nucleotide sequences allows genetically relating different strains during an outbreak and providing new insights into the molecular epidemiology of HAV.
The improvement in the environmental hygiene and the quality of drinking water in Tunisia has reduced hepatitis incidence and the risk of infection has been dropped significantly [17]. Although HAV incidence in Tunisia was studied [14], the surveillance of this infection need to be continued and the acquired data about this infection need to be updated and evaluated. Unfortunately, the surveillance of this infection was not done in a continued manner lack of facilities and sometimes practical reasons.
The aim of this study was to determine the seroprevalence of HAV in Tunisian patients and to complete the analysis by molecular characterization of HAV isolates.
Materials and Methods
Human Serum Samples
We analyzed 400 serum samples collected from the same number of patients (they were from 136 urban and 264 rural regions) between January 2006 and December 2008. The serum samples (220 males and 180 females) were from University Hospitals in Tunisia. Patient ranged in age from 1 to 60 years old with clinical symptoms of viral hepatitis such as jaundice (n = 250, 62.5 %), fever (n = 270, 67.5 %), fatigue (n = 380, 95 %), nausea (n = 240, 60 %), vomiting (n = 94, 23.5 %), and anorexia (n = 50, 12.5 %). The patients showed high levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST), and alkaline phosphatase (ALP) as compared to the normal levels. Based on their age, patients were categorized into three groups: <6 years, n = 134, 33.5 %; 6–15 years, n = 155, 38.75 %; >15 years, n = 111, 27.75 %. Two hundreds eighty-nine (72.25 %) out of the 400 serum samples were collected from patients under 15 years of age. Samples were used immediately or stored at −70 °C until use.
Serological Procedures
Sera were tested for hepatitis B surface antigen (HBsAg) and for IgM antibodies directed against the hepatitis B virus core protein (IgM antiHBc) using Hepanostika HBsAg Uni-form II and HBc IgM kits (both from Organon Teknika, Boxtel, Netherlands), respectively. In addition, sera were tested for hepatitis C virus using anti-HCV antibodies (UBI HCV EIA 4.0 kit, Organon). Samples were tested on the basis of their positivity for the anti-HAV IgM antibody using Abbott diagnosis kit (Abbott Laboratories, Rungies, France), according to the manufacturer’s instructions.
Nucleic Acid Extraction and Real-Time PCR
Viral RNA was extracted from 400 μl of each serum sample using QIAamp viral RNA kit (Qiagen, Paris, France), according to the manufacturer’s instructions. Viral RNA was eluted in 50 μl of RNase-free distilled water and was used immediately or stored at −80 °C until use. Positive anti-HAV IgM samples were analysed by Real-time PCR (RT-PCR) and by conventional reverse transcription PCR using several sets of primers in order to quantify the viral load and to determine the virus genotype. For RT-PCR, forward primer HAV1 (5′-TTT CCG GAG CCC CTC TTG-3′), as wild type reverse primers HAV2 (5′-AAA GGG AAA TTT AGC CTA TAG CC-3′) and HAV3 (5′-AAA GGG AAA ATT TAG CCT ATA GCC-3′), and HAV-probe (5′-FAM-ACT TGA TAC CTC ACC GCC GTT TGC CT-TAMRA-3′) were used to amplify 5′ non coding region (5′NCR), as described previously [7]. For conventional PCR, a set of primers were used to amplify the VP1/2A junction (168 pb) [15, 21, 31]. RT-PCRs were carried out using a Qiagen OneStep RT-PCR kit (Qiagen, hilden, germany) according to the manufacturer’s instructions.
Sequencing and Analysis of Viral Genome
PCR products of the VP1/2A junction were used for DNA sequencing using the commercial Terminator cycle sequencing ready reaction kit (ABI Prism BigDye, Applera Corporation, Forster City, CA). DNA sequencing was carried out using an automatic sequencer 373A DNA sequencing system (Applera Corporation, Forster City, CA). Sequences were aligned by using the ClustalW software [32], and optimized manually. Phylogenetic tree was constructed by using the computer program MEGA version 4.0 [30]. Statistical analyses were performed with SPSS software version 17. p values ≤ 0.05 were considered significant.
Results
Four hundred patients were enrolled in the present study. Most of patients with hepatitis symptoms were from rural regions (n = 264, 66 %). As shown in Table 1, the total number of patients showing hepatitis infection slightly decreased over times (2006, n = 147; 2007, n = 129; 2008, n = 124). In addition, the number of patients from urban regions was decreased (2006, n = 49; 2007, n = 47; 2008, n = 40), whereas the number of patients from rural regions was decreased in 2007 (n = 82 vs. n = 98) then it remains stable in 2008 (n = 84). The collected samples were from patients with hepatitis symptoms occurring mainly during January–March (36.7, 26, and 35.5 %) and September–December (39.4, 43.4, and 35.5 %) during the three years of study, respectively. All patients were tested negative for hepatitis B and hepatitis C viruses.
Table 1.
Case distribution, results of serological tests and molecular characterization of HAV isolates
| 2006 | 2007 | 2008 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Month | No of sera | IgMc | RT-PCRb (%) |
Viral loada | No of sera | IgMc | RT-PCRb (%) |
Viral loada | No of sera | IgMc | RT-PCRb (%) |
Viral loada | |||
| Urban | Rural | Urban | Rural | Urban | Rural | ||||||||||
| January | 5 | 17 | 5/22 | 6/22 (27.2) | 3.1 ± 0.8 | 2 | 10 | 2/12 | 3/12 (25) | 2.3 ± 0.9 | 4 | 10 | 2/14 | 3/14 (42.8) | 3.2 ± 0.2 |
| February | 10 | 10 | 4/20 | 4/20 (20) | 4.2 ± 0.2 | 2 | 8 | 1/10 | 2/10 (20) | 2.1 ± 0.2 | 5 | 9 | 3/14 | 4/14 (28.5) | 1.7 ± 0.1 |
| March | 2 | 10 | 2/12 | 2/12 (16.6) | 3.2 ± 0.5 | 6 | 6 | 2/12 | 3/12 (25) | 3.1 ± 0.6 | 3 | 13 | 2/16 | 4/16 (25) | 3.9 ± 0.05 |
| April | 1 | 7 | 1/8 | 2/8 (25) | 2.8 ± 0.9 | 2 | 6 | 1/8 | 2/8 (25) | 3.3 ± 0.9 | 1 | 5 | 2/6 | 3/6 (50) | 3.1 ± 0.4 |
| May | 2 | 5 | 1/7 | 2/7 (28.5) | 3.1 ± 0.8 | 5 | 4 | 1/9 | 2/9 (22.2) | 2.5 ± 0.9 | 5 | 2 | 1/7 | 2/7 (28.5) | 2.1 ± 0.5 |
| June | 2 | 5 | 1/7 | 1/7 (14.2) | 1.9 ± 0.2 | 5 | 5 | 1/10 | 2/10 (20) | 3.0 ± 0.9 | 4 | 5 | 2/9 | 3/9 (33.3) | 2.0 ± 0.8 |
| July | 2 | 4 | 1/6 | 2/6 (33.3) | 1.9 ± 0.2 | 4 | 2 | 2/6 | 2/6 (33.3) | 2.5 ± 0.8 | 1 | 9 | 1/10 | 2/10 (20) | 1.8 ± 0.5 |
| August | 2 | 5 | 2/7 | 3/7 (42.8) | 2.5 ± 9.9 | 2 | 4 | 2/6 | 3/6 (50) | 2.7 ± 0.9 | 2 | 2 | 1/4 | 2/4 (50) | 3.6 ± 0.6 |
| September | 4 | 8 | 2/12 | 3/12 (25) | 3.2 ± 0.2 | 4 | 6 | 3/10 | 4/10 (40) | 4.3 ± 0.6 | 3 | 2 | 1/5 | 2/5 (40) | 4.0 ± 0.9 |
| October | 5 | 9 | 5/14 | 4/14 (28.5) | 4.1 ± 0.6 | 6 | 9 | 2/15 | 3/15 (20) | 2.5 ± 0.3 | 4 | 10 | 3/14 | 4/14 (28.5) | 2.3 ± 0.1 |
| November | 4 | 10 | 5/14 | 6/14 (42.8) | 2.6 ± 0.4 | 5 | 11 | 4/16 | 5/16 (31.2) | 2.6 ± 0.5 | 1 | 10 | 3/11 | 3/11 (27.2) | 2.0 ± 0.2 |
| December | 10 | 8 | 3/18 | 5/18 (27.7) | 2.8 ± 0.4 | 4 | 11 | 3/15 | 4/15 (26.6) | 1.3 ± 0.1 | 7 | 7 | 2/14 | 3/14 (42.8) | 2.4 ± 0.1 |
| Total (%) | 49 (33.3) | 98 (66.6) | 32/147 (21.7) | 40/147 (27.2) | 47 (36.4) | 82 (63.6) | 24/129 (18.6) | 35/129 (27.1) | 40 (32.3) | 84 (67.7) | 23/124 (18.5) | 35/124 (28.2) | |||
aMean viral load (Log10/ml)
bReal time PCR positives (RT-PCR) (%)
canti-HAV immunoglobulin class M positive (IgM)
Seroprevalence of HAV
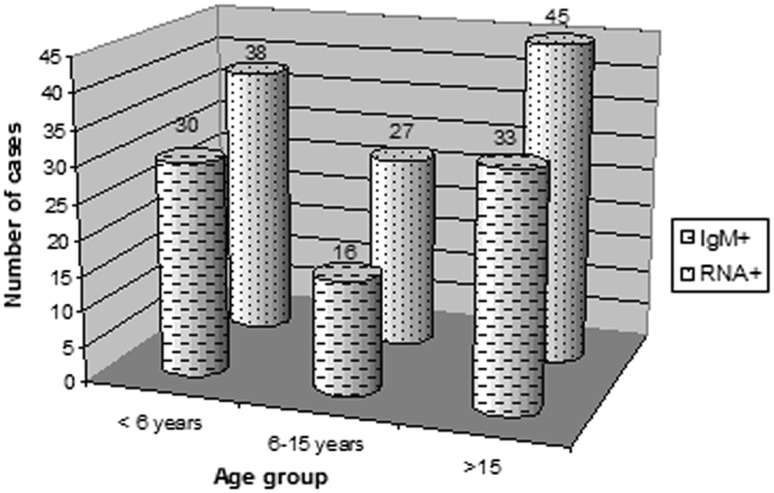
IgM antibodies against HAV were detected in 79 (19.75 %) of the 400 analysed samples. During the three years of the present study, 30 out of the 79 anti-HAV IgM positive samples were obtained from patients under 6 years (38 %); 16 anti-HAV IgM positive samples were obtained from patients aged between 6 and 15 years (20 %); 33 anti-HAV IgM positive samples were obtained from patients over 15 years old (42 %) (p = 0.009) (Fig. 1).
Fig. 1.
Distribution of positive IgM anti-HAV and positive ARN cases detected in patients for three age groups: 1 to 5 years, 6 to 15 years, and >15 years. IgM+: Immunoglobulin class M positive (p = 0.009); RNA+: Viral RNA positive (p = 0.034)
Presence of HAV in Serum Samples
HAV genome was detected in 110 (27.5 %, 79 IgM positive and 31 IgM negative) out of 400 samples. All IgM positive samples were RT-PCR positive (p = 0.02). During January–March and September–December, HAV genome was detected in 75 % (80 % from rural areas), 68.5 % (85.7 % from rural areas), and 65.7 % (82.8 % from rural areas) of cases throughout the three years of the study, respectively. The highest viral loads during HAV viremia were detected in samples collected during January–March (mean; 3.5 Log10/ml ± 0.5, 2.5 Log10/ml ± 0.5, 2.9 Log10/ml ± 0.1) and September–December (mean; 3.17 Log10/ml ± 0.4, 2.7 Log10/ml ± 0.4, 2.7 Log10/ml ± 0.3) for the three years of the study, respectively (Table 1). 38 out of the 110 HAV RT-PCR positives were obtained from patients under 6 years (34.5 %); 27 HAV RT-PCR positives were obtained from patients aged between 6 and 15 years (24.5 %); 45 HAV RT-PCR positives were obtained from patients over 15 years old (41 %) (p = 0.034) (Fig. 1).
Sequence Analysis of HAV RNA Genomes
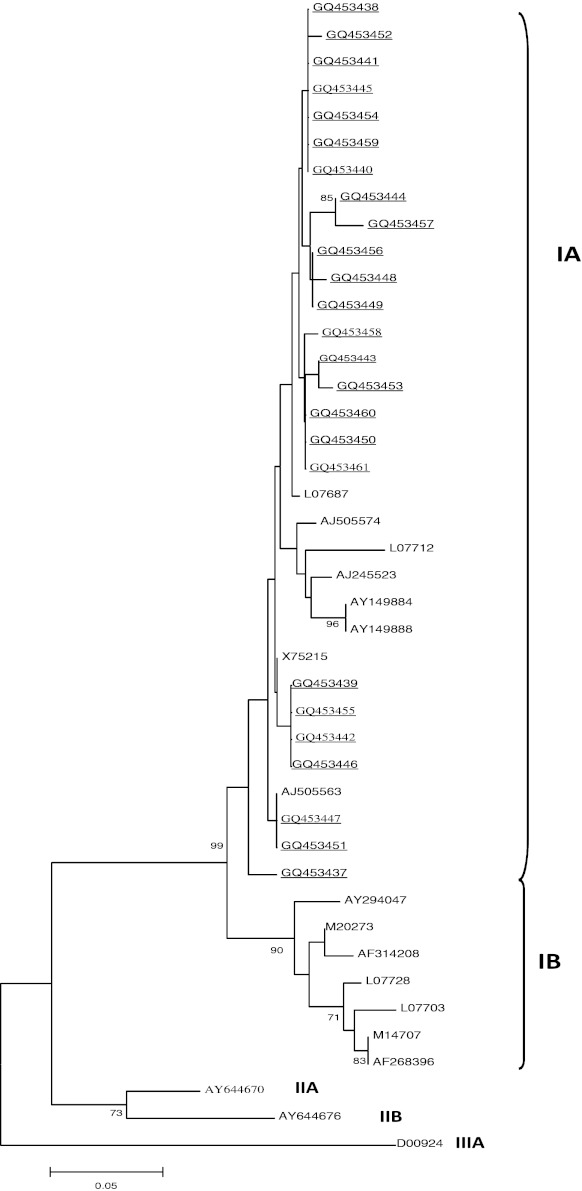
The HAV genome fragment, with a size of 168 nucleotides of the VP1/2A junction region, was amplified by RT-PCR. The amplified products were purified by phenol–chloroform extraction. Twenty five HAV isolates from sera were subjected to direct DNA sequencing. The nucleotide sequences of HAV were deposited in GenBank under the following accession numbers: GQ453437 (tun03-05), GQ453438 (tun06-05), GQ453439 (tun15-05), GQ453440 (tun95-06), GQ453441 (tun112-06), GQ453442 (tun113-06), GQ453443 (tun22-05), GQ453444 (tun29-05), GQ453445 (tun117-06), GQ453446 (tun37-05), GQ453447 (tun39-05), GQ453448 (tun123-06), GQ453449 (tun42-05), GQ453450 (tun59-05), GQ453451 (tun63-05), GQ453452 (tun67-05), GQ453453 (tun75-05), GQ453454 (tun76-05), GQ453455 (tun127-06), GQ453456 (tun80-05), GQ453457 (tun130-06), GQ453458 (tun145-06), GQ453459 (tun139-06), GQ453460 (tun48-06), and GQ453461 (tun155-06). Subsequently, the 168 pb products of the VP1/2A region were analyzed and compared with the reference strains L07687 (strain Arg90), AJ505574 (strain IT-SIB-01), L07712 (strain China81), AJ245523 (strain CU-26F), AY149884 (strain Sukhirin 7015), AY149888 (strain Yi-ngo 9250), AJ505563 (strain IT-CAP-00), AY294047 (strain IT-MAR-02), AF314208 (strain L-A-1), L07728 (strain Jor88), L07703 (strain Ag6014), AF268396 (strain HAF-203), M14707 (strain HM-175), X75215 (strain GMB), M20273 (strain MBB), AY644670 (SLF88), AY644676 (CF53/Berne), and D00924 (AGM27). Sequence analysis showed high similarity (99 %) among HAV strains isolated in serum samples. Comparative analysis classified the detected isolates within sub-genotype IA (Fig. 2).
Fig. 2.
Phylogenetic tree of HAV strains and the 25 Tunisian HAV strains isolated from 2006 to 2008. The nucleotide sequences of VP1/2A were analyzed using UPGMA bootstrap test of phylogeny. The number at nodes indicates bootstrap percentages after 1,000 replicate sampling. The bar indicates genetic distance and values under 70% were hidden. Cut-off value for condensed tree is 70 %. HAV genotypes IA, IB, IIA, IIB, and IIIA were shown. HAV strains isolated in this study were underlined
Discussion
The decrease in number of patients suffering acute hepatitis was certainly in relation with the improvement of the socio-economic level and the hygiene conditions. However, the number of patients from rural regions was high comparing to urban regions. All IgM-positive samples in the present study were positive for HAV RNA, which provided information on the specificity of surveillance data and the absence of false-positive serologic results. However, the false-positive serologic results may occur as described previously for patients with persisting HAV IgM [34], cross-reactions in the test (e.g., in acute-phase infections with Epstein-Barr virus) [35], or nonspecific polyclonal activation of memory cells [36].
HAV RNA was detected in 31 IgM-positive patients. These samples were retested to exclude any false-positive HAV RNA possibly from RNA contamination of the clinical specimen. The same results obtained for each test suggest that HAV RNA can be detected in serum samples before IgM antibodies to HAV were detected as described previously [37]. These data demonstrated that HAV RNA testing could be more appropriate control for HAV infection. In our study, the age-distribution showed that HAV infection increased with age. Children under the age of 6 years are particularly effective transmitters of hepatitis A infection. This constitutes risk factor for HAV infection of adults. Thus, young children should be the primary focus of vaccination.
However one out of 10 patients arrives to the adult age without having been in contact with the HAV as reported previously [25], these individuals can make a firstly-infection therefore to an age sometimes advanced relatively with a severe clinical expression [19]. The current evidence that an important proportion of the population of young people is susceptible to HAV infection has a number of public health implications. These susceptible individuals will be at risk of acquiring HAV later in life, leading to a more severe disease. The prevalence of sub-group IA in the tested samples is in line with our previous reports [13], in which sub-genotype IA was the predominant type in acute HAV infection. This sub-genotype was predominant in other countries such as Brazil, Cuba, Costa Rica, Argentina, and Thailand [2, 10, 11, 20, 23]. Other reports described the prevalence of sub-genotype IB Turkey [22].
We are aware that this study has some limitations. The exclusion of patients attending private hospitals meant that it was not possible to better evaluate the incidence rate of HAV infection. This exclusion criterion was justifiable for operational reasons. However, HAV still represents a major public health problem and permanent surveillance could prove to be a valuable strategy in the study of prevalence and of the incidence of various pathogens, especially when there is a lack of sufficient clinical data. Recommendation of vaccination depends on the HAV endemicity of the region. HAV vaccination was recommended in intermediate endemicity areas. Vaccination interrupts virus transmission very efficiently and can lead to a substantial reduction in the incidence of HAV infection in the entire population.
Acknowledgments
The authors are grateful to all technician staff of the Laboratory of Transmissible Diseases and Biological Active Substances (Faculty of Pharmacy, Monastir, Tunisia) and Department of Microbiology (Faculty of Pharmacy, Nantes, France) for their help and support.
Footnotes
Hakima Gharbi-Khelifi and Nabil Ben Salem Abid are contributed equally to this work.
Nucleotide sequence data reported are available in the GenBank databases under the accession numbers: GQ453437, GQ453438, GQ453439, GQ453440, GQ453441, GQ453442, GQ453443, GQ453444, GQ453445, GQ453446, GQ453447, GQ453448, GQ453449, GQ453450, GQ453451, GQ453452, GQ453453, GQ453454, GQ453455, GQ453456, GQ453457, GQ453458, GQ453459, GQ453460, and GQ453461.
References
- 1.Almasio PL, Amoroso P. HAV infection in chronic liver disease: a rationale for vaccination. Vaccine. 2003;21(19–20):2238–2241. doi: 10.1016/S0264-410X(03)00139-7. [DOI] [PubMed] [Google Scholar]
- 2.Arauz-Ruiz P, Sundqvist L, Garcia Z, Taylor L, Visona K, Norder H, Magnius LO. Presumed common source outbreaks of hepatitis A in an endemic area confirmed by limited sequencing within the VP1 region. J Med Virol. 2001;65(3):449–456. doi: 10.1002/jmv.2056. [DOI] [PubMed] [Google Scholar]
- 3.Bell BP, Shapiro CN, Alter MJ, Moyer LA, Judson FN, Mottram K, Fleenor M, Ryder PL, Margolis HS. The diverse patterns of hepatitis A epidemiology in the United States-implications for vaccination strategies. J Infect Dis. 1998;178(6):1579–1584. doi: 10.1086/314518. [DOI] [PubMed] [Google Scholar]
- 4.Bruisten SM, van Steenbergen JE, Pijl AS, Niesters HGM, van Doornum GJJ, Coutinho RA. Molecular epidemiology of hepatitis A virus in Amsterdam, the Netherlands. J Med Virol. 2001;63:88–95. doi: 10.1002/1096-9071(20000201)63:2<88::AID-JMV1001>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 5.Byun KS, Kim JH, Song K-J, Baek LJ, Song J-W, Park SH, Kwon OS, Yeon JE, Kim JS, Bak YT, Lee CH. Molecular studies: hepatitis A virus and hepatectomy. Molecular epidemiology of hepatitis A virus in Korea. J Gastroenterol Hepatol. 2001;16:519–524. doi: 10.1046/j.1440-1746.2001.02481.x. [DOI] [PubMed] [Google Scholar]
- 6.Cooksley WG. What did we learn from the Shanghai hepatitis A epidemic? J Viral Hepat. 2000;7(Suppl 1):1–3. doi: 10.1046/j.1365-2893.2000.00021.x. [DOI] [PubMed] [Google Scholar]
- 7.Costa-Mattioli M, Monpoeho S, Nicand E, Aleman MH, Billaudel S, Ferre V. Quantification and duration of viraemia during hepatitis A infection as determined by real-time RT-PCR. J Viral Hepat. 2002;9(2):101–106. doi: 10.1046/j.1365-2893.2002.00336.x. [DOI] [PubMed] [Google Scholar]
- 8.Costa-Mattioli M, Monpoeho S, Schvoerer C, Besse B, Aleman MH, Billaudel S, Cristina J, Ferré V. Genetic analysis of hepatitis A virus outbreak in France confirms the co-circulation of subgenotypes Ia, Ib and reveals a new genetic lineage. J Med Virol. 2001;65(2):233–240. doi: 10.1002/jmv.2025. [DOI] [PubMed] [Google Scholar]
- 9.Cotter SM, Sansom S, Long T, Koch E, Kellerman S, Smith F, Averhoff F, Bell BP. Outbreak of hepatitis A among men who have sex with men: implications for hepatitis A vaccination strategies. J Infect Dis. 2003;187(8):1235–1240. doi: 10.1086/374057. [DOI] [PubMed] [Google Scholar]
- 10.de Paula VS, Lu L, Niel C, Gaspar AM, Robertson BH. Genetic analysis of hepatitis A virus isolates from Brazil. J Med Virol. 2004;73(3):378–383. doi: 10.1002/jmv.20101. [DOI] [PubMed] [Google Scholar]
- 11.Diaz BI, Sariol CA, Normann A, Rodriguez L, Flehmig B. Genetic relatedness of Cuban HAV wild-type isolates. J Med Virol. 2001;64(2):96–103. doi: 10.1002/jmv.1023. [DOI] [PubMed] [Google Scholar]
- 12.Franco E, Giambi C, Ialacci R, Coppola RC, Zanetti AR. Risk groups for hepatitis A virus infection. Vaccine. 2003;21(19–20):2224–2233. doi: 10.1016/S0264-410X(03)00137-3. [DOI] [PubMed] [Google Scholar]
- 13.Gharbi-Khelifi H, Ferre V, Sdiri K, Berthome M, Fki L, Harrath R, Billaudel S, Aouni M. Hepatitis A in Tunisia: phylogenetic analysis of hepatitis A virus from 2001 to 2004. J Virol Methods. 2006;138(1–2):109–116. doi: 10.1016/j.jviromet.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Gharbi-Khelifi H, Sdiri K, Ferre V, Harrath R, Berthome M, Billaudel S, Aouni M. A 1-year study of the epidemiology of hepatitis A virus in Tunisia. Clin Microbiol Infect. 2007;13(1):25–32. doi: 10.1111/j.1469-0691.2006.01588.x. [DOI] [PubMed] [Google Scholar]
- 15.Hussain Z, Das BC, Husain SA, Asim M, Chattopadhyay S, Malik A, Poovorawan Y, Theamboonlers A, Kar P. Hepatitis A viral genotypes and clinical relevance: clinical and molecular characterization of hepatitis A virus isolates from northern India. Hepatol Res. 2005;32(1):16–24. doi: 10.1016/j.hepres.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 16.Jansen RW, Siegl G, Lemon SM. Molecular epidemiology of human hepatitis A virus defined by an antigen-capture polymerase chain reaction method. Proc Natl Acad Sci USA. 1990;87:2867–2871. doi: 10.1073/pnas.87.8.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Letaief A, Kaabia N, Gaha R, Bousaadia A, Lazrag F, Trabelsi H, Ghannem H, Jemni L. Age-specific seroprevalence of hepatitis a among school children in central Tunisia. Am J Trop Med Hyg. 2005;73(1):40–43. [PubMed] [Google Scholar]
- 18.Lu L, Ching KZ, de Paula VS, Nakano T, Siegl G, Weitz M, Robertson BH. Characterization of the complete genomic sequence of genotype II hepatitis A virus (CF53/Berne isolate) J Gen Virol. 2004;85(10):2943–2952. doi: 10.1099/vir.0.80304-0. [DOI] [PubMed] [Google Scholar]
- 19.Mathur P, Arora NK. Epidemiological transition of hepatitis A in India: issues for vaccination in developing countries. Indian J Med Res. 2008;128(6):699–704. [PubMed] [Google Scholar]
- 20.Mbayed VA, Sookoian S, Alfonso V, Campos RH. Genetic characterization of hepatitis A virus isolates from Buenos Aires, Argentina. J Med Virol. 2002;68(2):168–174. doi: 10.1002/jmv.10194. [DOI] [PubMed] [Google Scholar]
- 21.Najarian R, Caput D, Gee W, Potter SJ, Renard A, Merryweather J, Van Nest G, Dina D. Primary structure and gene organization of human hepatitis A virus. Proc Nat Acad Sci USA. 1985;82(9):2627–2631. doi: 10.1073/pnas.82.9.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Normann A, Badur S, Onel D, Kilic A, Sidal M, Larouze B, Massari V, Müller J, Flehmig B. Acute hepatitis A virus infection in Turkey. J Med Virol. 2008;80(5):785–790. doi: 10.1002/jmv.21137. [DOI] [PubMed] [Google Scholar]
- 23.Poovorawan Y, Theamboonlers A, Chongsrisawat V, Jantaradsamee P, Chutsirimongkol S, Tangkijvanich P. Clinical features and molecular characterization of hepatitis A virus outbreak in a child care center in Thailand. J Clin Virol. 2005;32(1):24–28. doi: 10.1016/j.jcv.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 24.Rezende G, Roque-Afonso AM, Samuel D, Gigou M, Nicand E, Ferre V, Dussaix E, Bismuth H, Féray C. Viral and clinical factors associated with the fulminant course of hepatitis A infection. Hepatology. 2003;38(3):613–618. doi: 10.1053/jhep.2003.50366. [DOI] [PubMed] [Google Scholar]
- 25.Rezig D, Ouneissa R, Mhiri L, Mejri S, Haddad-Boubaker S, Ben Alaya N, Triki H. Seroprevalences of hepatitis A and E infections in Tunisia. Pathol Biol (Paris) 2008;56(3):148–153. doi: 10.1016/j.patbio.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 26.Robertson BH, Averhoff F, Cromeans TL, Han X-H, Khoprasert B, Nainan OV, Rosenberg J, Paikoff L, DeBess E, Shapiro CN, Margolis HS. Genetic relatedness of hepatitis A virus isolates during a community-wide outbreak. J Med Virol. 2000;62:144–150. doi: 10.1002/1096-9071(200010)62:2<144::AID-JMV4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 27.Robertson BH, Khanna B, Nainan OV, Margolis HS. Epidemiologic patterns of wild-type hepatitis A virus determined by genetic variation. J Infect Dis. 1991;163:286–292. doi: 10.1093/infdis/163.2.286. [DOI] [PubMed] [Google Scholar]
- 28.Robertson BH, Jansen RW, Khanna B, Totsuka A, Nainan OV, Siegl G, Widell A, Margolis HS, Isomura S, Ito K, Ishizu T, Moritsugu Y, Lemon SM. Genetic relatedness of hepatitis A virus strains recovered from different geographical regions. J Gen Virol. 1992;73(6):1365–1377. doi: 10.1099/0022-1317-73-6-1365. [DOI] [PubMed] [Google Scholar]
- 29.Sanchez G, Populaire S, Butot S, Putallaz T, Joosten H. Detection and differentiation of human hepatitis A strains by commercial quantitative real-time RT-PCR tests. J Virol Methods. 2006;132(1–2):160–165. doi: 10.1016/j.jviromet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 30.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 31.Theamboonlers A, Jantaradsamee P, Chatchatee P, Chongsrisawat V, Mokmula M, Poovorawan Y. Molecular characterization of hepatitis-A-virus infections, in the context of two outbreaks in southern Thailand. Ann Trop Med Parasitol. 2002;96(7):727–734. doi: 10.1179/000349802125001898. [DOI] [PubMed] [Google Scholar]
- 32.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang JY, Lee SD, Tsai YT, Lo KJ, Chiang BN. Fulminant hepatitis A in chronic HBV carrier. Dig Dis Sci. 1986;31(1):109–111. doi: 10.1007/BF01347921. [DOI] [PubMed] [Google Scholar]
- 34.Kao HW, Ashcavai M, Redeker AG. The persistence of hepatitis A IgM antibody after acute clinical hepatitis A. Hepatology. 1984;4:933–936. doi: 10.1002/hep.1840040525. [DOI] [PubMed] [Google Scholar]
- 35.Fikar CR, McKee C. False positivity of IgM antibody to Epstein-Barr viral capsid antigen during acute hepatitis A infection. Pediatr Infect Dis J. 1994;13:413–414. doi: 10.1097/00006454-199405000-00016. [DOI] [PubMed] [Google Scholar]
- 36.Roque-Afonso AM, Grangeot-Keros L, Roquebert B, Desbois D, Poveda JD, Mackiewicz V, Dussaix E. Diagnostic relevance of immunoglobulin G avidity for hepatitis A virus. J Clin Microbiol. 2004;42:5121–5124. doi: 10.1128/JCM.42.11.5121-5124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen JI, Feinstone S, Purcell RH. Hepatitis A virus infection in a chimpanzee: duration of viremia and detection of virus in saliva and throat swabs. J Infect Dis. 1989;5:887–890. doi: 10.1093/infdis/160.5.887. [DOI] [PubMed] [Google Scholar]