Abstract
The fermentation of rice dietary fibre was measured by the cell yield, pH, optical density specific growth rate and biomass using nine co-cultures of four probiotics at 0, 6, 24 and 48 h incubation. The results from the fermentation of the soluble, insoluble and total dietary fibre (SDF, IDF, and TDF) of two rice varieties were compared. Overall there was no significant difference (p < 0.05) in the fermentation of the six different rice fibre fractions. However, co-cultures showed a preference for glucose as a fermentation substrate rather than the fibre fractions. Bifidobacteria species produced a higher cell count than the Lactobacillus species after 24 and 48 h of fermentation (p < 0.05). There was evidence of synergistic activity with increased growth observed when lactobacilli and Bifidobacterium were grown together. Growth was limited by the pH reaching 4.2–4.45. Specific growth rates of the co-cultures varied for different culture combinations. Combinations of the same species produced less biomass than combinations of mixed species. Bifidobacterium breve + Bifidobacterium longum + Lactobacillus rhamnosus was the most efficient. These findings showed that synergism between microorganisms in co-cultures affected the degree of fermentation of dietary fibre.
Keywords: Rice fibre, Fermentation, Growth parameters, Probiotics, Lactobacilli, bifidobacteria
Introduction
Microorganisms in the human gut have the potential to hydrolyse and ferment dietary fibre (Gibson and Roberfroid 1995). To understand this in detail, an in vitro fermentation system is more practical than expensive human and animal trials. Among the 400 species of bacteria in the human gut (Tannock 1995) Bifidobacterium and Lactobacillus genus are the most important (Gibson 1998; Mitsuoka 1990) in terms of digestion, absorption of nutrients, prevention of colonization of pathogens (Lankaputhra et al. 1996; Shin et al. 2000), and stimulation of immune responses (Yaeshima 1996).
Oligosaccharides are selectively utilized by all Bifidobacterium, some Lactobacillus and Bacteroides species (Bouhnik et al. 1997; Smiricky-Tjardes et al. 2003; Tzortzis et al. 2005). These microorganisms in the human gut work as a consortium (Tannock 1995). To understand the fermentation of dietary fibre by microorganisms, the following study used a consortium of probiotics.
Measuring culture viability in a fermentation medium is believed to be a good model to understand the probiotic activity in the human gut. The emphasis of this work was to determine the ability of co-cultures to achieve high viable cell populations using rice fibre fermentation. A concentration of approximately 107 cells ml−1 as the inoculum, is considered suitable (Gomes and Malcata 1999; Shortt 1999) to initiate the utilization of dietary fibre.
High cell growth and acidification rates help to reduce the fermentation time and boost the viability of the specific strain in the fermentation medium (Marklinder and Lönner 1992) and presumably in the human gut.
Physiological and taxonomical characteristics of Lactcobacillus and Bifidobacterium genus are important to select the best combinations for fermentation. Lactobacilli are fastidious microorganisms. Lactobacilli require fermentable carbohydrates, amino acids, vitamins of the B-complex, nucleic acids and minerals to achieve a high cell yield. Nutrient requirements are dependent on the strains of lactobacilli (Gomes and Malcata 1999). In contrast, Bifidobacterium have the ability to utilize a wide variety of nutrients thus they have a potential to adjust and compete for nutrients with other organisms in the environment (Crittenden et al. 2002). Therefore, substrate composition and nutritional requirements of the strain affect the overall performance of the microbial combination. Microbial growth also depends on extrinsic factors such as the pH, temperature and accumulation of metabolic end-products in the fermentation medium (Ganzle et al. 1998; Mercier et al. 1992).
The aim of present work was to study the fermentation of the rice dietary fibre by mixed cultures in order to understand the growth kinetics associated with this fermentation. The fermentation following the inoculation of rice fibre with combinations of microorganisms was measured using viable cell counts, biomass formation, acidification and the specific growth rate.
Materials and methods
Chemicals
Microbiological media de mann, Rogosa and Sharpe (MRS, Sigma, USA) and Reinforced Clostridial Medium (Sigma, USA) were used as the cultivation media for pure cultures of lactobacilli and bifidobacteria, respectively.
Rice varieties
Rice varieties from Sri Lanka were selected based on their milling grade to understand whether milling grade has an impact on the fermentation by co-cultures. Two rice varieties were chosen:—LD356 (RR1, red in colour, brown rice, dehulled) and AT353 (RR2, red in colour, unpolished, most of the germ having been removed).
Determination of soluble, insoluble and total dietary fibre
Rice samples were analyzed for soluble, insoluble and total dietary fibre (SDF, IDF and TDF) according to the AOAC method 991.43 (1995).
Filtration
In this study, the filtration was modified by using filter paper 541 (Whatman International LTD, Maidstone, UK as Celite proved difficult to be removed from the isolated fibre fractions (Titgemeyer et al. 1991).
Fermentation substrate
Dietary fibre fractions (TDF, IDF, SDF) obtained from the two rice samples were used as the substrates for the fermentation.
Bacterial strains
Bacterial strains Lactobacillus rhamnosus (ATCC 7469) (LR), Lactobacillus acidophilus (ATCC11975) (LA), Bifidobacterium breve (ATCC15700) (BB), and Bifidobacterium longum (ATCC15707) (BL), were obtained from the culture collection at the Institute of Environmental Science and Research Limited, New Zealand.
Co-cultures
Pure cultures of bacterial strains were combined in a 1:1 (v/v) ratio to prepare nine combinations in equal proportions. The combinations were LA+LR, BB+BL, BB+LA, BB+LR, BL+LA, BL+LR, BB+BL+LA, BB+BL+LR, and BB+BL+LA+LR,
Preparation of cell suspensions
Freeze dried cultures were rehydrated by inoculating Lactobacillus spp. in de Man, Rogosa, and Sharpe (MRS) medium and Bifidobacterium spp. in Reinforced Clostridial Medium, under strict anaerobic conditions. Anaerobic conditions were created by using an anaerobic chamber with the Gas pack (Oxoid Ltd., Hampshire, England) throughout the experiment. Lactobacillus spp. were incubated at 37 °C for 24 h and Bifidobacterium spp. were incubated at 37 °C for 72 h to obtain the complete growth curves of the organisms. For the fermentation trials, the bacteria were sub-cultured twice in 10 ml of the appropriate medium containing 10 g/l glucose as the carbon source. After incubation the bacterial cells were centrifuged, washed twice with physiological saline (0.85% NaCl solution), and resuspended in the basal medium (PYF solution) to remove excess carbon before the fermentation trials. The suspension was then diluted to 1:10 with the basal medium (Jaskari et al. 1998).
Preparation of growth medium
The basal medium, peptone/yeast extract/Fildes (PYF) solution, was used as the carbohydrate-free medium. PYF medium comprised 10 g Trypticase Peptone, 5 g yeast extract, 0.5 g L-cysteine hydrochloride, 40 mL digested horse blood, and 40 ml salt solution per 1 L. The salt solution contained 0.2 g CaCl2, 0.2 g MgSO4 ·7 H20, 1.0 g KH2 PO4, 1.0 g K2 HPO4, 10 g NaHCO3, and 2.0 g NaCl in 1 L deionized water (pH7.6) (Yoshimoto et al. 2005).
In vitro fermentation
Fermentations were conducted in sterile 100 ml bottles. Each bottle contained culture medium, substrate, and pure cultures. Culture medium (50 ml) and 1% (v/w,) substrate (mL/mg) (extracted TDF, SDF or IDF from each of the rice varieties separately) were added to each bottle and sealed for 24 h for complete hydration of the fibre. The bottles were maintained at 37 °C for 2 h prior to inoculation to initiate the fermentation as soon as inoculated and bottles of media were in the anaerobic jar with gas pack prior to inoculation. Broth medium (pH 7.6) was inoculated with 10% (v/v) of the bacterial suspension (107 colony forming units [cfu]/ml) and fermentation occurred under strict anaerobic conditions. All the bacteria were grown in duplicate fermentations in the appropriate basal medium. Aliquots, (5 ml for fibre analysis and 2 ml for SCFA analysis) were removed for analysis at 0, 6, 24 and 48 h after incubation (Yoshimoto et al. 2005; Jaskari et al. 1998).
Determination of pH, optical density and viable count
The pH of aliquots was determined using a pH probe, soon after removing the samples from the fermentation broth.
Bacterial growth was determined by measuring the optical density of samples (1.5 ml) at 600 nm. Samples were taken immediately following the incubation periods for optical density and for viable count. Two blanks were prepared to understand the effect of fibre suspension and the probiotic without the probiotic and fibre, respectively.
The viable counts of Bifidobacterium and Lactobacillus from BB+LA, BB+LR, BL+LA, and BL+LR were measured using selective media (de Mann Rogosa and Sharpe (MRS) (Sigma USA) and Reinforced Clostridial Medium (Sigma, USA) for Lactobacillus and Bifidobacterium, respectively) separately by the plate count method. The viable counts of Bifidobacterium and Lactobacillus of BB+BL, LA+LR, BB+BL+LA, BB+BL+LR, and BB+BL+LA+LR were measured using above selective media, but as a total count of Bifidobacterium and Lactobacillus individually.
Maximum specific growth rate
The maximum specific growth rate was calculated according to the following equation.
 |
- Nt
Viable count of time t
- N0
Viable count of time 0
- μ
specific growth rate (per hour)
Determination of the growth yield
The growth yield of organisms was calculated using the following formula.
 |
- A
net change in OD600 of culture grown on substrate X.
- B
net change in OD600 of culture grown without added substrate.
- C
net change in OD600 of culture grown on glucose. Biomass yield from growth on glucose was considered as 100% (Crittenden et al. 2002)
Statistical evaluation
The analyses were performed as two independent experiments and each experiment was done twice to have four observations of each parameter finally, and results were expressed as mean values with standard error. Data were analysed using the statistical analysis package of Microsoft Excel 2003. The differences between the experimental groups and the control containing glucose were evaluated using Student’s t-test for less numerous groups.
Differences between bacterial groups and counts/OD/cell yield/pH at 0, 6, 24 and 48 h fermentation for each substrate were tested for significance using paired t-tests assuming equal variances and considering two-tailed distribution. Differences were considered significant if p ≤ 0.05.
Results and discussion
Viable count
The viable cell count differed from one co-culture to another for different rice varieties (Table 1). The differences were significant (p < 0.05) between co-cultures of the same genus and co-cultures of different genus. However, all co-cultures were able to use both rice varieties as substrates. Co-cultures formed from more than two strains produced higher viable counts than the other combinations. Co-cultures produced maximum numbers of viable cells within 24 h incubation, suggesting that this is the optimal time for the fermentation of fibre.
Table 1.
Viable count of (Log10 (CFU)/mL) L. species and B. species in co-culture combinations on RR2
| Co-culture combinations | Fibre | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IDF | SDF | TDF | Glucose | |||||||||||||
| LA+LR | O h | 6 h | 24 h | 48 h | O h | 6 h | 24 h | 48 h | O h | 6 h | 24 h | 48 h | O h | 6 h | 24 h | 48 h |
| Lactobacillus speices | 9.4 ± 0.01 | 10.4 ± 0.01 | 10.4 ± 0.01 | 10.4 ± 0.01 | 9.4 ± 0.01 | 10.4 ± 0.01 | 10.4 ± 0.01 | 10.3 ± 0.01 | 9.4 ± 0.01 | 10.4 ± 0.01 | 10.4 ± 0.01 | 10.4 ± 0.01 | 9.4 ± 0.02 | 10.5 ± 0.02 | 10.4 ± 0.03 | 10.4 ± 0.02 |
| BB+BL | ||||||||||||||||
| Bifidobacterium species | 9.1 ± 0.02 | 10.5 ± 0.03 | 10.5 ± 0.02 | 10.4 ± 0.02 | 9.5 ± 0.01 | 10.5 ± 0.02 | 10.43 ± 0.03 | 10.5 ± 0.02 | 9.13 ± 0.03 | 10.5 ± 0.02 | 10.4 ± 0.02 | 10.4 ± 0.03 | 9.2 ± 0.02 | 10.4 ± 0.03 | 10.4 ± 0.02 | 10.4 ± 0.02 |
| BL+LR | ||||||||||||||||
| Bifidobacterium longum | 9.2 ± 0.02 | 9.4 ± 0.03 | 12.4 ± 0.03 | 10.4 ± 0.03 | 9.2 ± 0.02 | 9.4 ± 0.02 | 12.4 ± 0.02 | 10.4 ± 0.03 | 9.2 ± 0.03 | 9.5 ± 0.02 | 12.4 ± 0.01 | 10.4 ± 0.02 | 9.2 ± 0.02 | 9.4 ± 0.02 | 12.5 ± 0.02 | 10.4 ± 0.03 |
| Lactobacillus rhamnosus | 9.5 ± 0.02 | 10.4 ± 0.02 | 11.4 ± 0.02 | 8.4 ± 0.02 | 9.4 ± 0.02 | 10.4 ± 0.02 | 11.4 ± 0.02 | 9.4 ± 0.02 | 9.4 ± 0.03 | 10.4 ± 0.03 | 11.4 ± 0.01 | 10.5 ± 0.02 | 9.4 ± 0.02 | 10.5 ± 0.02 | 11.5 ± 0.03 | 9.4 ± 0.03 |
| BB+LR | ||||||||||||||||
| Bifidobacterium breve | 9.2 ± 0.02 | 9.4 ± 0.02 | 12.4 ± 0.03 | 10.4 ± 0.02 | 9.2 ± 0.03 | 9.4 ± 0.02 | 12.4 ± 0.02 | 10.4 ± 0.03 | 9.5 ± 0.01 | 9.4 ± 0.01 | 12.4 ± 0.02 | 10.4 ± 0.02 | 9.2 ± 0.03 | 9.4 ± 0.02 | 11.4 ± 0.02 | 10.4 ± 0.02 |
| Lactobacillus rhamnosus | 9.5 ± 0.02 | 10.4 ± 0.02 | 10.4 ± 0.02 | 8.4 ± 0.01 | 9.4 ± 0.02 | 10.5 ± 0.02 | 10.4 ± 0.01 | 9.4 ± 0.01 | 9.4 ± 0.02 | 10.4 ± 0.01 | 10.4 ± 0.02 | 8.4 ± 0.01 | 9.4 ± 0.02 | 10.5 ± 0.03 | 11.4 ± 0.01 | 9.4 ± 0.02 |
| BB+LA | ||||||||||||||||
| Bifidobacterium breve | 9.2 ± 0.02 | 9.4 ± 0.01 | 12.4 ± 0.02 | 10.4 ± 0.01 | 9.2±.03 | 9.5 ± 0.01 | 12.4 ± 0.02 | 10.4 ± 0.01 | 9.2 ± 0.02 | 9.4 ± 0.01 | 12.4 ± 0.02 | 10.4 ± 0.01 | 9.2 ± 0.03 | 9.4 ± 0.02 | 12.5 ± 0.01 | 10.4 ± 0.02 |
| Lactobacillus acidophilus | 9.4 ± 0.02 | 10.4 ± 0.02 | 10.4 ± 0.01 | 9.4 ± 0.02 | 9.4 ± 0.02 | 10.5 ± 0.01 | 10.4 ± 0.01 | 9.4 ± 0.02 | 9.4 ± 0.01 | 10.4 ± 0.02 | 10.4 ± 0.02 | 9.4 ± 0.02 | 9.4 ± 0.02 | 10.5 ± 0.01 | 12. 5 ± 0.01 | 9.4 ± 0.02 |
| BL+LA | ||||||||||||||||
| Bifidobacterium longum | 9.3 ± 0.03 | 9.4 ± 0.02 | 12.4 ± 0.02 | 11.4 ± 0.03 | 9.2 ± 0.02 | 9.4 ± 0.03 | 12.5 ± 0.02 | 11.4 ± 0.03 | 9.2 ± 0.02 | 9.5 ± 0.02 | 12.4 ± 0.02 | 11.4 ± 0.01 | 9.2 ± 0.01 | 9.4 ± 0.02 | 11.4 ± 0.02 | 11.4 ± 0.02 |
| Lactobacillus acidophilus | 9.4 ± 0.03 | 10.4 ± 0.01 | 11.4 ± 0.02 | 9.4 ± 0.03 | 9.4 ± 0.02 | 10.4 ± 0.01 | 11.5 ± 0.02 | 9.4 ± 0.01 | 9.4 ± 0.02 | 10.5 ± 0.02 | 11.5 ± 0.01 | 8.40 ± 0.02 | 9.4 ± 0.02 | 10.4 ± 0.02 | 10.4 ± 0.01 | 9.4 ± 0.03 |
| BB+BL+LR+LA | ||||||||||||||||
| Bifidobacterium species | 10.4 ± 0.01 | 10.4 ± 0.02 | 13.4 ± 0.02 | 12.4 ± 0.01 | 10.4 ± 0.02 | 10.4 ± 0.02 | 13.4 ± 0.01 | 12.4 ± 0.01 | 10.4 ± 0.02 | 10.4 ± 0.02 | 13.4 ± 0.02 | 12.4 ± 0.01 | 10.4 ± 0.03 | 10.4 ± 0.02 | 13.4 ± 0.01 | 12.4 ± 0.02 |
| Lactobacillus species | 10.2 ± 0.04 | 11.4 ± 0.02 | 11.4 ± 0.02 | 8.4 ± 0.01 | 10.3 ± 0.03 | 11.5 ± 0.02 | 11.4 ± 0.01 | 9.4 ± 0.03 | 10.3 ± 0.01 | 11.4 ± 0.02 | 11.5 ± 0.03 | 8.4 ± 0.01 | 10.2 ± 0.01 | 11.5 ± 0.03 | 13.4 ± 0.01 | 9.5 ± 0.01 |
| BB+BL+LA | ||||||||||||||||
| Bifidobacterium species | 10.4 ± 0.02 | 11.4 ± 0.03 | 14.4 ± 0.02 | 11.4 ± 0.01 | 10.4 ± 0.01 | 11.4 ± 0.02 | 14.4 ± 0.03 | 11.4 ± 0.02 | 10.5 ± 0.02 | 11.4 ± 0.02 | 14.4 ± 0.01 | 11.4 ± 0.01 | 10.4 ± 0.01 | 11.5 ± 0.01 | 14.4 ± 0.01 | 11.5 ± 0.0.01 |
| Lactobacillus species | 10.2 ± 0.01 | 11.4 ± 0.01 | 14.4 ± 0.02 | 8.4 ± 0.02 | 10.2 ± 0.01 | 11.4 ± 0.02 | 14.4 ± 0.03 | 9.4 ± 0.01 | 10.2 ± 0.01 | 11.4 ± 0.02 | 14.4 ± 0.01 | 8.4 ± 0.01 | 10.2 ± 0.02 | 11.5 ± 0.01 | 14.42 ± 0.01 | 9.43 ± 0.01 |
| BB+BL+LR | ||||||||||||||||
| Bifidobacterium species | 10.4 ± 0.01 | 12.4 ± 0.01 | 14.4 ± 0.01 | 11.5±.01 | 10.4 ± 0.01 | 12.4 ± 0.01 | 14.4 ± 0.01 | 11.4 ± 0.01 | 10.4 ± 0.01 | 12.4 ± 0.02 | 14.4 ± 0.02 | 11.5 ± 0.01 | 10.4 ± 0.02 | 12.5 ± 0.03 | 14.5 ± 0.01 | 11.4 ± 0.01 |
| Lactobacillus species | 10.2 ± 0.01 | 12.4 ± 0.02 | 14.4 ± 0.01 | 8.4 ± 0.01 | 10.2 ± 0.01 | 12.5 ± 0.02 | 14.4 ± 0.01 | 8.4 ± 0.01 | 10.2 ± 0.01 | 12.4 ± 0.01 | 14.4 ± 0.01 | 8.4 ± 0.01 | 10.2 ± 0.02 | 12.5 ± 0.03 | 14.5 ± 0.01 | 9.4 ± 0.02 |
Results are expressed as the mean value of two independent trials (n = 4). Standard error was between 0.01 and 0.03.Counting was done by growing Lactobacillus species and bifidobacterea species in selective media
pH
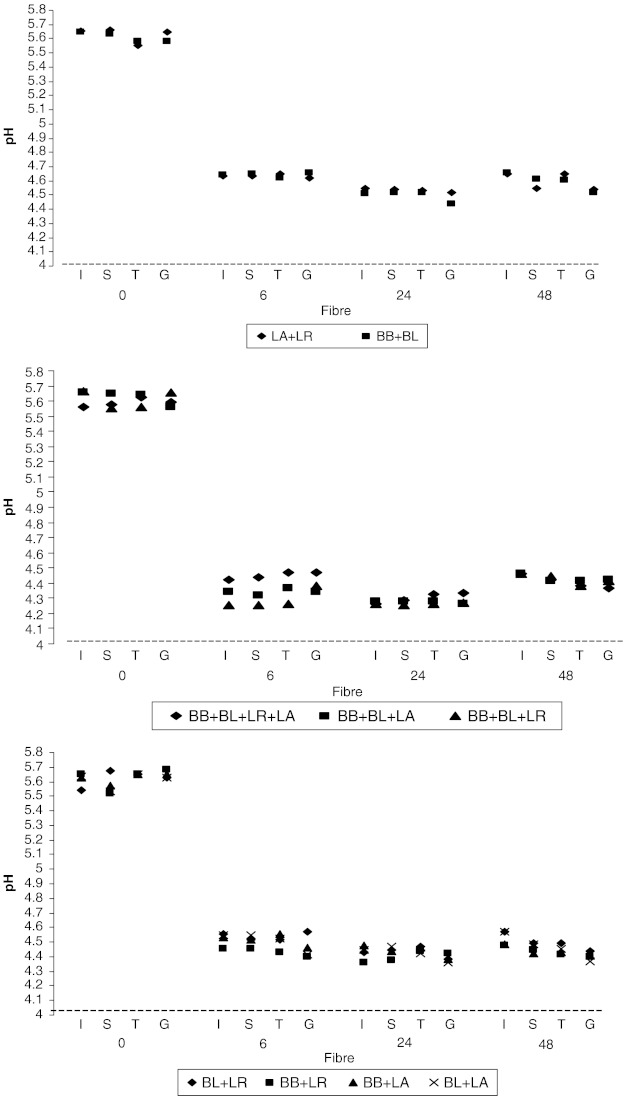
The pH of the fermentation medium varied with time (Fig. 1). The lowest pH was reported after 24 h incubation, indicating maximum metabolite formation. It was not possible to measure the acidification of each of the organisms making up the co-culture separately. However, this would be useful information to understand the contribution of each organism in the synergistic relationship.
Fig. 1.
pH of the fermentation of rice fibre fractions. Results are expressed as the mean value of two trials ± std error (n = 4), I—Insoluble dietary fibre, S—Soluble dietary fibre, T—Total dietary fibre, G—Glucose
OD
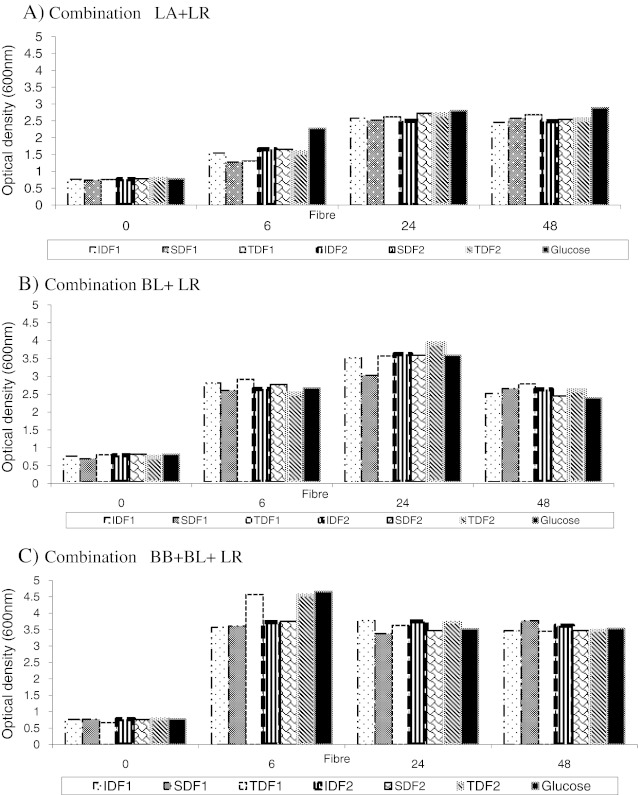
The optical density readings varied with different substrates (Fig. 2). Measuring the optical density is a convenient method to measure the growth curves of co-cultures on rice fibre. Viable counts of counterpart organisms demonstrated a rapid decline after the exponential phase, however, this was not observed in the results from the optical density readings for the mixed species co-cultures. This represents a change in the viable cell population but not the total cell content.
Fig. 2.
Growth curves for different co-culture combinations measured by optical density. Results are expressed as the mean value of two trials ± std error (n = 4)
Specific growth rate
The specific growth rates indicate the ability of the microorganisms to utilise the rice fibre substrates (Tables 2 and 3). The specific growth rate (CFU/mL/h) was established under the linearity assumption between the closest two time points in log phase using the formula,  (Gupthar et al. 2000). However, in this study time interval for most of the combinations were 6 and 18 h. Therefore, values of specific growth rate can be considered as the approximate values. The specific growth rates indicate the ability of the microorganisms to utilise the rice fibre substrates. Lactobacillus species and Bifidobacterium had different specific growth rates in different co-cultures. Even in the same co-culture, organisms demonstrated different specific growth rates indicating variation in the fermentation capacity of Bifidobacterium and the Lactobacillus species. However, the microorganisms in the following co-culture—BB+BL+LR—produced similar specific growth rates.
(Gupthar et al. 2000). However, in this study time interval for most of the combinations were 6 and 18 h. Therefore, values of specific growth rate can be considered as the approximate values. The specific growth rates indicate the ability of the microorganisms to utilise the rice fibre substrates. Lactobacillus species and Bifidobacterium had different specific growth rates in different co-cultures. Even in the same co-culture, organisms demonstrated different specific growth rates indicating variation in the fermentation capacity of Bifidobacterium and the Lactobacillus species. However, the microorganisms in the following co-culture—BB+BL+LR—produced similar specific growth rates.
Table 2.
Specific growth rates of different co-culture combinations between 0 and 6 h of fermentation
| Time points | h | ||||||
|---|---|---|---|---|---|---|---|
| 0–6 | 0–6 | 0–6 | 0–6 | 0–6 | 0–6 | 0–6 | |
| Co culture combinations | |||||||
| LA+LR | IDF1 | SDF1 | TDF1 | IDF2 | SDF2 | TDF2 | Glucose |
| Lactobacillus species | 0.40 ± 0.01 | 0.38 ± 0.01 | 0.39 ± 0.01 | 0.38 ± 0.01 | 0.38 ± 0.01 | – | 0.41 ± 0.02 |
| BB+BL | |||||||
| Bifidobacterium species | 0.46 ± 0.02 | 0.47 ± 0.02 | 0.48 ± 0.01 | 0.48 ± 0.01 | 0.44 ± 0.01 | – | 0.49 ± 0.02 |
| BL+LR | |||||||
| Bifidobacterium longum | 0.04 ± 0.01 | 0.07 ± 0.01 | 0.08 ± 0.01 | 0.08 ± 0.02 | 0.08 ± 0.01 | 0.38 ± 0.01 | 0.07 ± 0.01 |
| Lactobacillus rhamnosus | 0.44 ± 0.01 | 0.45 ± 0.02 | 0.42 ± 0.01 | 0.40 ± 0.01 | 0.38 ± 0.01 | 0.09 ± 0.01 | 0.37 ± 0.01 |
| BB+LR | |||||||
| Bifidobacterium breve | 0.06 ± 0.02 | 0.08 ± 0.01 | 0.08 ± 0.01 | 0.08 ± 0.01 | 0.08 ± 0.01 | 0.38 ± 0.01 | 0.09 ± 0.03 |
| Lactobacillus rhamnosus | 0.39 ± 0.01 | 0.40 ± 0.01 | 0.41 ± 0.01 | 0.41 ± 0.02 | 0.41 ± 0.01 | – | 0.41 ± 0.01 |
| BB+LA | |||||||
| Bifidobacterium breve | 0.07 ± 0.01 | 0.09 ± 0.01 | 0.10 ± 0.01 | 0.08 ± 0.01 | 0.09 ± 0.01 | 0.38 ± 0.01 | 0.08 ± 0.01 |
| Lactobacillus acidophilus | 0.38 ± 0.02 | 0.39 ± 0.01 | 0.39 ± 0.01 | 0.40 ± 0.02 | 0.41 ± 0.01 | – | 0.40 ± 0.01 |
| BL+LA | |||||||
| Bifidobacterium longum | 0.05 ± 0.01 | 0.07 ± 0.01 | 0.06 ± 0.01 | 0.07 ± 0.01 | 0.07 ± 0.03 | 0.38 ± 0.01 | 0.08 ± 0.01 |
| Lactobacillus acidophilus | 0.39 ± 0.01 | 0.42 ± 0.02 | 0.41 ± 0.01 | 0.41 ± 0.01 | 0.39 ± 0.01 | 0.09 ± 0.01 | 0.39 ± 0.01 |
| BB+BL+LR+LA | |||||||
| Bifidobacterium species | 0.02 ± 0.02 | 0.02 ± 0.01 | 0.01 ± 0.01 | 0.01 ± 0.02 | – | 0.39 ± 0.01 | – |
| Lactobacillus species | 0.48 ± 0.01 | 0.48 ± 0.02 | 0.46 ± 0.02 | 0.47 ± 0.01 | 0.46 ± 0.01 | 0.02 ± 0.01 | 0.48 ± 0.01 |
| BB+BL+LA | |||||||
| Bifidobacterium species | 0.39 ± 0.02 | 0.39 ± 0.01 | 0.39 ± 0.01 | 0.38 ± 0.01 | 0.38 ± 0.021 | 0.01 ± 0.01 | 0.40 ± 0.01 |
| Lactobacillus species | 0.47 ± 0.01 | 0.50 ± 0.01 | 0.48 ± 0.01 | 0.46 ± 0.01 | 0.48 ± 0.01 | 0.39 ± 0.02 | 0.470.01 |
| BB+BL+LR | |||||||
| Bifidobacterium species | 0.79 ± 0.01 | 0.77 ± 0.02 | 0.77 ± 0.01 | 0.77 ± 0.01 | 0.77 ± 0.01 | 0.19 ± 0.01 | 0.79 ± 0.01 |
| Lactobacillus species | 0.84 ± 0.01 | 0.84 ± 0.01 | 0.85 ± 0.01 | 0.85 ± 0.02 | 0.88 ± 0.01 | 0.19 ± 0.01 | 0.87 ± 0.01 |
Results are expressed as the mean value of two independent trials (n = 4)
Table 3.
Specific growth rates of different co-culture combinations between 6 and 24 h of fermentation
| Time points | h | ||||||
|---|---|---|---|---|---|---|---|
| 6–24 | 6–24 | 6–24 | 6–24 | 6–24 | 6–24 | 6–24 | |
| Co culture combinations | |||||||
| LA+LR | IDF1 | SDF1 | TDF1 | IDF2 | SDF2 | TDF2 | Glucose |
| Lactobacillus species | – | – | – | – | – | – | – |
| BB+BL | |||||||
| Bifidobacterium species | – | – | – | – | – | – | – |
| BL+LR | |||||||
| Bifidobacterium longum | 0.26 ± 0.01 | 0.25 ± 0.02 | 0.26 ± 0.01 | 0.39 ± 0.01 | 0.39 ± 0.01 | 0.38 ± 0.02 | 0.39 ± 0.01 |
| Lactobacillus rhamnosus | – | – | – | 0.13 ± 0.01 | 0.13 ± 0.01 | 0.13 ± 0.01 | 0.14 ± 0.01 |
| BB+LR | |||||||
| Bifidobacterium breve | 0.39 ± 0.02 | 0.39 ± 0.02 | 0.38 ± 0.01 | 0.38 ± 0.01 | 0.38 ± 0.01 | 0.38 ± 0.01 | 0.26 ± 0.01 |
| Lactobacillus rhamnosus | 0.13 ± 0.01 | 0.13 ± 0.01 | 0.13 ± 0.01 | – | – | – | 0.13 ± 0.01 |
| BB+LA | |||||||
| Bifidobacterium breve | 0.26 ± 0.01 | 0.26 ± 0.01 | 0.26 ± 0.01 | 0.38 ± 0.01 | 0.38 ± 0.01 | 0.38 ± 0.01 | 0.39 ± 0.01 |
| Lactobacillus acidophilus | – | 0.01 ± 0.01 | – | – | – | – | 0.26 ± 0.01 |
| BL+LA | |||||||
| Bifidobacterium longum | 0.39 ± 0.01 | 0.38 ± 0.01 | 0.38 ± 0.01 | 0.39 ± 0.01 | 0.39 ± 0.01 | 0.38 ± 0.01 | 0.26 ± 0.01 |
| Lactobacillus acidophilus | 0.01 ± 0.01 | – | – | 0.13 ± 0.01 | 0.13 ± 0.02 | 0.13 ± 0.02 | – |
| BB+BL+LR+LA | |||||||
| Bifidobacterium species | 0.26 ± 0.01 | 0.26 ± 0.01 | 0.26 ± 0.01 | 0.39 ± 0.01 | 0.39 ± 0.01 | 0.39 ± 0.01 | 0.38 ± 0.01 |
| Lactobacillus species | – | – | – | 0.02 ± 0.001 | 0.02 ± 0.001 | 0.02 ± 0.001 | 0.25 ± 0.01 |
| BB+BL+LA | |||||||
| Bifidobacterium species | 0.13 ± 0.01 | 0.13 ± 0.01 | 0.13 ± 0.01 | 0.39 ± 0.01 | 0.38 ± 0.01 | 0.39 ± 0.01 | 0.38 ± 0.01 |
| Lactobacillus species | 0.13 ± 0.01 | 0.13 ± 0.02 | 0.13 ± 0.02 | 0.38 ± 0.02 | 0.39 ± 0.01 | 0.39 ± 0.01 | 0.38 ± 0.01 |
| BB+BL+LR | |||||||
| Bifidobacterium species | 0.13 ± 0.01 | 0.13 ± 0.01 | 0.130.01 | 0.26 ± 0.01 | 0.26 ± 0.02 | 0.26 ± 0.02 | 0.26 ± 0.01 |
| Lactobacillus species | 0.13 ± 0.01 | 0.13 ± 0.02 | 0.13 ± 0.01 | 0.26 ± 0.02 | 0.25 ± 0.02 | 0.25 ± 0.01 | 0.26 ± 0.01 |
Results are expressed as the mean value of two independent trials (n = 4)
Biomass
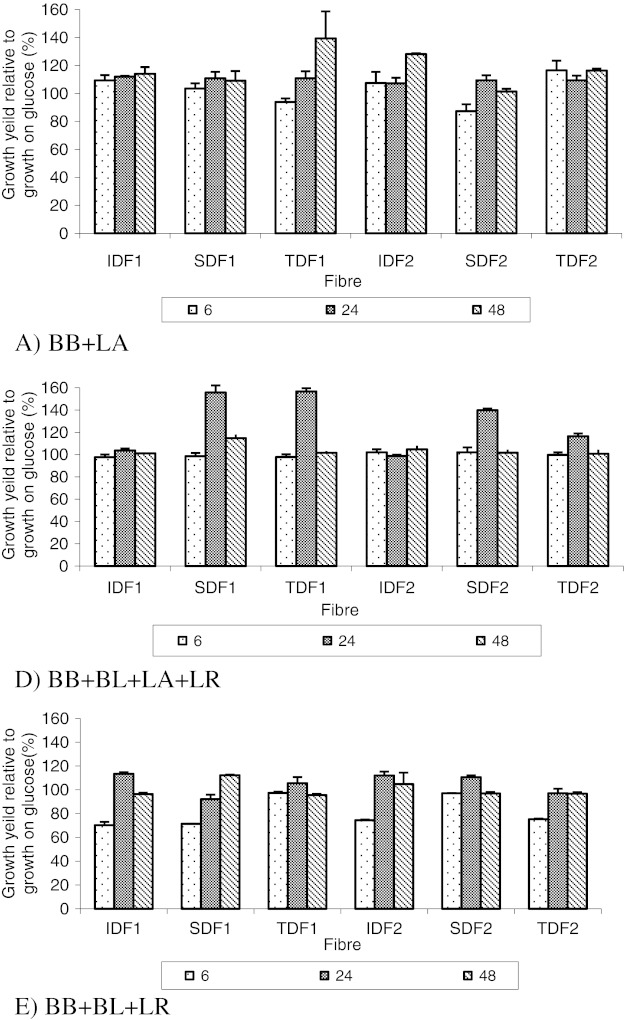
Some of the co-cultures produced maximum biomass at 24 h and some at 48 h incubation (Fig. 3). Since the co-cultures obtained the maximum viable count at 24 h, those producing the highest biomass yield at 24 h suggest that the maximum metabolite formation is also occurring at this time. The combinations with the highest biomass at 48 h, suggests metabolite formation occurs for longer and there is a number of non viable cells at this later stage of fermentation since organisms are in death phase.
Fig. 3.
Biomass yield of co-culture combinations at 6, 24 and 48 h incubation. Results are expressed as the mean value of two trials ± std error (n = 4)
The aim of this research was to observe and compare the growth characteristics of combinations of pure cultures of L. acidophilus, L. rhamnosus, B. breve and B. longum in unformulated dietary rice fibre. Composition of raw material, specific growth rate of the combinations, final cell count, biomass and acidification rate were the parameters used to evaluate the fermentation process of dietary fibre. This is the first report to compare the in vitro fermentation of rice fibre by co-cultures of probiotics.
In this study, high cell counts of the starter cultures ensured the high initial levels of cell counts in the fermentation medium: 108–109 cfu/mL. The addition of fibre substrate to fermentation medium in general had an observable (p < 0.05) effect on the culture growth and the viability, especially at 24 h incubation, for all the microbial combinations (Table 1). This indicates the ability of the co-cultures to use rice fibre as a fermentable substrate. Previous work with oat fibre using some species of the genus Lactobacillus produced a 2.8 log increase in the cell count from a similar inoculum (Angelov et al. 2005), less than the increase observed in the current research. This may be due to the different substrate and/or different fermentation capabilities of the strains used in each experiment.
Cell counts during fermentation in the current study were substantially species and time dependant. Bifidobacterium species produced a higher cell count (p < 0.05) than Lactobacillus species, especially after 24 and 48 h incubation for all the substrates (Table 1). Fibre had a greater effect on the cell numbers of Bifidobacterium than the Bactobacillus species. Previous work with species of genus Bifidobacterium has shown that bifidobacteria can ferment a variety of carbohydrates (Bezkorovainy 1999) and that bifidobacteria vary in their fermentation profiles on different monosaccharides (Beverly and George 1991). This particular ability of Bifidobacterium will help these organisms to out compete other organisms in the human gut when dietary fibre is provided in the diet.
Microorganisms belong to different genera, showed slow growth when the pH was 4.2–4.45 after 24 h incubation (Fig. 1). These experiments were conducted in a synthetic media without pH control. These uncontrolled conditions in the fermentation medium would have been resulted the accumulation of the metabolic products such as short chain fatty acids. These organic acids can inhibit the microbial growth (Passos et al. 1993). However, previous studies have found that the contribution of metabolites (organic acids) in limiting the growth of microorganisms is less than the effect of pH (Giraud et al. 1991). Thus the higher cell population of Bifidobacterium species compared with the Bactobacillus species in combinations can be attributed to the higher growth of bifiobacteria in the fermentation medium before the pH value drops to 4.2–4.45. Previous studies have found that the pH value that limits the growth of Lactobacillus species (L. acidophilus) is dependent on the cereal used in the fermentation (Lönner and Preve-Åkesson 1988). Less acidification in the medium may allow an increase the exponential phase of the microbial combinations and allow the formation of more metabolites in the fermentation medium. It is still not clear whether the limit of microbial growth with these microbial combinations is due to the acidification of the medium or deficiency in the nutrients.
However, there were differences in the growth curves (OD-optical density) for the fermentation of the different fibre fractions—IDF, SDF, and for TDF (Fig. 2). The growth curves for co-cultures of species from same genus were different from the mixed co-cultures (different genus) and among the mixed cultures the binary combinations had a different pattern than those consisting of more than two species, demonstrating synergism between the Lactobacillus and Bifidobacterium species (Fig. 2). This indicates different synergistic relationships between the different microbial combinations during fermentation. This relationship is considered as synergistic since higher numbers of viable cells were produced during log phase of growth using co-cultures compared with the individual organisms (un published data). The variations of cell numbers between the different co-cultures were statistically significant (p < 0.05). Previous work has found evidence of symbiosis between L. acidophilus and B. lactis when grown in milk (Gomes et al. 1998). Lactobacilli have a higher proteolytic activity and can supply Bifidobacterium with peptides and amino acids (Gomes et al. 1998) which they need to produce a higher yield. Bifidobacterium species can also be inhibited by a fast growing Lactobacillus strain (Gomes and Malcata 1999). However, this study did not observe higher growth of Lactobacillus species compared with the Bifidobacterium species. This indicates the Lactobacillus species tested in this trial are not fast growing organisms when fibre is used as the fermentation substrate. However, among the Lactobacillus species, L. acidophilus showed slower growth than the L. rhamnosus (Tables 1, 2 and 3). The slower increase in cell numbers of L. acidophilus could be explained by the absence of the nutrients in the medium (Gomes and Malcata 1999). Previous research has demonstrated that this organism has a high nutrient requirement and has poor growth in media lacking supplements such as yeast extract and peptone (Gomes and Malcata 1999).
The combinations of species from different genus showed a very short stationary phase and a long exponential phase except for the LA+LR and BB+BL combinations. A long exponential phase indicates greater utilization of fibre by the microorganisms. Due to the fast depletion of the fibre after the log phase, the microorganisms had a very short stationary phase before the populations started to decline. Maximal cell counts of 1010–1012/mL were obtained in 24 h and this is likely to be similar in the human gut since total transit time of fibre in human gut has been estimated as 24 to72 h (Wrick et al. 1983). Co-cultures of BB+BL and LA+LR did not enter the decline phase like the other combinations. Their population was small compared with other combinations, thus nutrition limitation might not have affected them as rapidly as other combinations.
In this study, the specific growth rates were expected to be similar for the same organisms in binary and more than binary combinations for the same fibre fraction. Interestingly the same organism in different co-cultures exhibited different specific growth rates for the same substrates and the specific growth rate was also different between the different co-cultures. The variations in chemical structure and the quantity of sugars of fibre fractions may contribute to variations in the specific growth rates. Specific growth rates may change when the substrate concentration decreases with the incubation time. In this study, substrates used by the microorganisms as the energy source, would have been used for more than just growth (eg; reproduction, metabolite formation etc.). Therefore, the growth rate of the microorganisms may be slower than the expected. On the other hand, the energy requirement of the microorganisms making up the co-culture may differ from combination to combination. Therefore, organisms may exhibit different specific growth rates for the same substrate when organisms are present in different combinations. Metabolic waste that accumulates in the medium may inhibit the cells and may affect the specific growth rate. The capacity of microorganisms to compete for nutrients with the other microorganisms in the combination might also affect the specific growth rate (Tables 2 and 3).
This combination of factors may explain the different specific growth rates for the fermentation of the same substrate when microorganisms are in different co-cultures. Therefore, it is impossible to know whether Lactobacillus or Bifidobacterium species had a higher specific growth rate on fibre when they are in combinations compared with individual species. However, combinations of the same species demonstrated a higher specific growth rate than other combinations with the lowest specific growth rate reported from the combination of BB+BL+LR.
Most of the combinations of microorganisms showed significantly (p < 0.05) higher growth on glucose compared with the fibre (Fig. 3). This indicates the preference of co-cultures is more towards glucose than the fibre fractions. However, microorganisms in co-cultures of the same genus showed an equal preference for glucose and for fibre while co-cultures of more than one genus showed a higher preference for glucose than fibre. This result disagrees with previous work showing a higher growth by Bifidobacterium on dietary fibre (fructo-oligosaccharides) than on glucose (Gibson and Wang 1994; Wang and Gibson 1993). Co-cultures of different species showed a preference for glucose and for TDF in terms of the viable count and optical density measurements. It was interesting to note that all potential prebiotics (IDF, SDF and TDF) produced similar fermentations with the co-cultures used, irrespective of the rice variety. Generally the difference in cell count for co-culture fermentations of fibre substrates (IDF, SDF, and TDF) at each time point was not significant (p > 0.05). The combined cultures utilized the fibre fractions equally well. We conclude, that the milling grade of these rice varieties has a minimal effect in the composition of sugar content in the fibre fractions, because, in this study we used the same quantity of fibre as IDF, SDF, TDF from two different rice varieties. These fractions were 99% pure. It is true that, initial fibre content of these rice varieties varied (Fernando et al. 2008).
Among the combinations evaluated, those of the species from same genus demonstrated less biomass formation than the other combinations during the fermentation (p < 0.05). There was no significant difference in the biomass yield from the different fibre substrates (p > 0.05). The highest pH drop after 24th h incubation indicates that the physiological state of cells at 24 h favours the formation of acids rather than biomass.
The combination of BB+BL+LR showed the greatest stimulation of growth by the presence of the potential prebiotics TDF, SDF and IDF, and Glucose in terms of viable count, optical density and pH. With an average count of 13.6 log CFU/mL for this combination, this indicates a good synergistic relationship between these probiotic microorganisms, capable of using the rice fibre fractions. LR or LR in co-culture with BB or BL or BB+BL, produced high counts (12–14 log CFU/mL on average) but only in the presence of glucose.
Conclusion
Co-cultures grew on fibre from both rice varieties, reaching high cell yields. This demonstrates synergistic relationships between these bacterial combinations. This study found that there was no higher preference for any particular dietary fibre fraction. However, most co-cultures showed a preference for glucose rather than fibre as a substrate. This study observed an increase in the cell count of Bifidobacterium and Lactobacillus species with combinations from a different genus rather than combinations from the same genus. This suggests a certain degree of symbiosis between the members of each genus. The microorganisms making up the combinations reached the death phase after a very short stationary phase, between 24 to 48 h, except in fermentations consisting of organisms from the same genus. The pH drop in all fermentations occurred rapidly, reaching the lowest level after 6 h fermentation. Co-cultures made up of more than one genus reached a lower pH than those made up of organisms from the same genus, adding further evidence for the synergistic association in these co-cultures. The specific growth rate differed among the co-cultures on all the fibre substrates indicating the differences in the ability to utilize fibre. BB+BL+LR was the combination showing the most growth on all substrates tested (TDF, SDF, IDF and Glucose).The results from this work suggest that fermentation of dietary fibre from rice could be enhanced by the addition of probiotics bacteria in specific combinations.
Acknowledgement
This work was supported by the Asian Development Board. We would like to thank Technicians (Magi) of Nutrition laboratory of Massey University, for their assistance with this work.
References
- AOAC Method 991.43. (1995). Total, insoluble and soluble dietary fibre in food—enzymatic-gravimetric method, MES-TRIS buffer. In: Official methods of analysis, 16th edition, AOAC International, Gaithersburg, MD
- Angelov A, Gotcheva V, Hristozova T, Gargova S. Application of pure and mixed probiotic lactic acid bacteria and yeast cultures for oat fermentation. J Sci Food Agric. 2005;85:2134–2141. doi: 10.1002/jsfa.2223. [DOI] [Google Scholar]
- Beverly AD, George TM. Comparison of carbohydrate substrate preferences in eight species of Bifidobacteria. FEMS Microbiol Lett. 1991;84:151–156. doi: 10.1111/j.1574-6968.1991.tb04588.x. [DOI] [PubMed] [Google Scholar]
- Bezkorovainy A. Biochemistry and physiology of Bifidobacteria. In: Miller-Catchpole R, Bezkorovainy A, editors. Nutrition and metabolism of Bifidobacteria. Boca Raton: CRC; 1999. pp. 93–129. [Google Scholar]
- Bouhnik Y, Flourie B, D’Agay-Abensour L, Pochart P, Gramet G, Durand M, Rambaud J-C. Administration of transgalacto-oligosaccharides increases fecal Bifidobacteria and modifies colonic fermentation metabolism in healthy humans. J Nutr. 1997;127:444–448. doi: 10.1093/jn/127.3.444. [DOI] [PubMed] [Google Scholar]
- Crittenden R, Karppinen S, Ojanen S, Tenkanen M, Fagerstrom R, Matto J, Saarela M, Mattila-Sandholm T, Poutanen K. In vitro fermentation of cereal dietary fibre carbohydrates by probiotic and intestinal bacteria. J Sci Food Agric. 2002;82:781–789. doi: 10.1002/jsfa.1095. [DOI] [Google Scholar]
- Fernando WMADB, Ranaweera KKDS, Bamunuarachchi A, Brennan CS. The influence of rice fibre fractions on the in vitro fermentation production of short chain fatty acids using human faecal micro flora. Int J Food Sci Technol. 2008;43:2237–2244. doi: 10.1111/j.1365-2621.2008.01861.x. [DOI] [Google Scholar]
- Ganzle MG, Ehmann M, Hammes WP. Modeling of growth of Lactobacillus sanfranciscensis and candida milleri in response to process parameters of sourdough fermentation. Appl Environ Microbiol. 1998;64:2616–2623. doi: 10.1128/aem.64.7.2616-2623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson GR. Dietary modulation of human gut microflora using prebiotics. Br J Nutr. 1998;80:S209–S212. [PubMed] [Google Scholar]
- Gibson GR, Roberfroid MB. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Appl Microbiol. 1995;125:1401–1412. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- Gibson GR, Wang X. Regulatory effects of Bifidobacteria on the growth of other colonic bacteria. J Appl Bacteriol. 1994;77:412–420. doi: 10.1111/j.1365-2672.1994.tb03443.x. [DOI] [PubMed] [Google Scholar]
- Giraud E, Lelong B, Raimbault M. Influence of pH and initial lactate concentration on the growth of Lactobacillus plantarum. Appl Microbiol Biotechnol. 1991;36:96–99. doi: 10.1007/BF00164706. [DOI] [Google Scholar]
- Gomes AMP, Malcata FX. Bifidobacterium spp. and Lactobacillus acidophilus: biological, biochemical, technological and therapeutical properties relevant for use as probiotics. Trends Food Sci Technol. 1999;10:139–157. doi: 10.1016/S0924-2244(99)00033-3. [DOI] [Google Scholar]
- Gomes AMP, Malcata FX, Klaver FAM. Growth enhancement of Bifidobacterium lactis bo and Lactobacillus acidophilus ki by milk hydrolyzates. J Dairy Sci. 1998;81:2817–2825. doi: 10.3168/jds.S0022-0302(98)75840-0. [DOI] [PubMed] [Google Scholar]
- Gupthat AS, Bhattacharya S, Basu TK. Evaluation of the maximum specific growth rate of yeast indicating non-linear growth trends in batch culture. World J Microbiol Biotechnol. 2000;16:613–616. doi: 10.1023/A:1008980317225. [DOI] [Google Scholar]
- Jaskari J, Kontula P, Siitonen A, Jousimies-Somer H, Mattila-Sandholm T, Poutanen K. Oat β-glucan and xylan hydrolysates as selective substrates for Bifidobacterium and Lactobacillus strains. Appl Microbiol Biotechnol. 1998;49:175–181. doi: 10.1007/s002530051155. [DOI] [PubMed] [Google Scholar]
- Lankaputhra WEV, Shah NP, Britz M. Survival of Bifidobacteria during refrigerated storage in the presence of acid and hydrogen peroxide. Milchwissenschaft. 1996;51:65–70. [Google Scholar]
- Lönner C, Preve-Åkesson K. Acidification properties of lactic acid bacteria in rye sour doughs. Food Microbiol. 1988;5:43–58. doi: 10.1016/0740-0020(88)90007-X. [DOI] [Google Scholar]
- Marklinder I, Lönner C. Fermentation properties of intestinal strains of Lactobacillus, of a sour dough and of a yoghurt starter culture in an oat-based nutritive solution. Food Microbiol. 1992;9:197–205. doi: 10.1016/0740-0020(92)80047-8. [DOI] [Google Scholar]
- Mercier P, Yerushalmi L, Rouleau D, Dochain D. Kinetics of lactic acid fermentation on glucose and corn by Lactobacillus amylophilus. J Chem Technol Biotechnol. 1992;55:111–121. doi: 10.1002/jctb.280550204. [DOI] [Google Scholar]
- Mitsuoka T. Bifidobacteria and their role in human health. J Ind Microbiol Biotechnol. 1990;6:263–267. [Google Scholar]
- Passos FV, Fleming HP, Ollis DF, Hassan HM, Felder RM. Modelling the specific growth rate of Lactobacillus plantarum in cucumber extract. Appl Microbiol Biotechnol. 1993;40:143–150. doi: 10.1007/BF00170443. [DOI] [Google Scholar]
- Shin HS, Lee JH, Pestka JJ, Ustunol Z. Growth and viability of commercial Bifidobacterium spp. in skim milk containing oligosaccharides and inulin. J Food Sci. 2000;65:884–887. doi: 10.1111/j.1365-2621.2000.tb13605.x. [DOI] [Google Scholar]
- Shortt C. The probiotic century: historical and current perspectives. Trends Food Sci Technol. 1999;10:411–417. doi: 10.1016/S0924-2244(00)00035-2. [DOI] [Google Scholar]
- Smiricky-Tjardes MR, Grieshop CM, Flickinger EA, Bauer LL, GCJr F. Dietary galactooligosaccharides affect ileal and total-tract nutrient digestibility, ileal and fecal bacterial concentrations, and ileal fermentative characteristics of growing pigs. J Anim Sci. 2003;81:2535–2545. doi: 10.2527/2003.81102535x. [DOI] [PubMed] [Google Scholar]
- Titgemeyer EC, Bourquin LD, Fahey GC, Jr, Garleb KA. Fermentability of various fibre sources by human fecal bacteria in vitro. Am J Clin Nutr. 1991;53:1418–1424. doi: 10.1093/ajcn/53.6.1418. [DOI] [PubMed] [Google Scholar]
- Tannock GW. Role of probiotics. In: Gibson GR, Macfarlane GT, editors. Human colonic bacteria: role in nutrition, physiology and pathology. London: CRC; 1995. pp. 257–271. [Google Scholar]
- Tzortzis G, Goulas AK, Gee JM, Gibson GR. A novelgalactooligosaccharide mixture increases the Bifidobacterial population numbers in a continuous in vitro fermentation system and in the proximal colonic contents of pigs in vivo. J Nutr. 2005;135:1726–1731. doi: 10.1093/jn/135.7.1726. [DOI] [PubMed] [Google Scholar]
- Wang X, Gibson GR. Effects of the in vitro fermentation of oligofructose and inulin by bacteria growing in the human large intestine. J Appl Microbiol. 1993;75:373–380. doi: 10.1111/j.1365-2672.1993.tb02790.x. [DOI] [PubMed] [Google Scholar]
- Wrick KL, Robertson JB, Vansoest PJ, Lewis BA, Rivers JM, Roe DA, Hackler LR. The influence of dietary fiber source on human intestinal transit and stool output. J Nutr. 1983;113:1464–1479. doi: 10.1093/jn/113.8.1464. [DOI] [PubMed] [Google Scholar]
- Yaeshima T. Benefits of Bifidobacteria to human health. Bull IDF. 1996;313:36–42. [Google Scholar]
- Yoshimoto M, Yamakawa O, Tanoue H. Potential chemopreventive properties and varietal difference of dietary fiber from sweet potato (Ipomoea batatas L.) root. JARQ. 2005;39:37–43. [Google Scholar]