Abstract
Although a wide range of approaches have been developed to automatically assess the volume of brain regions from MRI, the reproducibility of these algorithms across different scanners and pulse sequences, their accuracy in different clinical populations and sensitivity to real changes in brain volume has not always been comprehensively examined. Firstly we present a comprehensive testing protocol which comprises 312 freely available MR images to assess the accuracy, reproducibility and sensitivity of automated brain segmentation techniques. Accuracy is assessed in infants, young adults and patients with Alzheimer’s disease in comparison to gold standard measures by expert observers using a manual technique based on Cavalieri’s principle. The protocol determines the reliability of segmentation between scanning sessions, different MRI pulse sequences and 1.5T and 3T field strengths and examines their sensitivity to small changes in volume using a large longitudinal dataset. Secondly we apply this testing protocol to a novel algorithm for segmenting the lateral ventricles and compare its performance to the widely used FSL FIRST and FreeSurfer methods. The testing protocol produced quantitative measures of accuracy, reliability and sensitivity of lateral ventricle volume estimates for each segmentation method. The novel algorithm showed high accuracy in all populations (intraclass correlation coefficient, ICC>0.95), good reproducibility between MRI pulse sequences (ICC>0.99) and was sensitive to age related changes in longitudinal data. FreeSurfer demonstrated high accuracy (ICC>0.95), good reproducibility (ICC>0.99) and sensitivity whilst FSL FIRST showed good accuracy in young adults and infants (ICC>0.90) and good reproducibility (ICC=0.98), but was unable to segment ventricular volume in patients with Alzheimer’s disease or healthy subjects with large ventricles. Using the same computer system, the novel algorithm and FSL FIRST processed a single MRI image in less than 10 minutes while FreeSurfer took approximately 7 hours. The testing protocol presented enables the accuracy, reproducibility and sensitivity of different algorithms to be compared. We also demonstrate that the novel segmentation algorithm and FreeSurfer are both effective in determining lateral ventricular volume and are well suited for multicentre and longitudinal MRI studies.
Keywords: segmentation, MRI reliability, sensitivity, lateral ventricles
Introduction
A range of automated segmentation algorithms are available for determining the volume of various local brain regions, including widely applied techniques such as FreeSurfer (Fischl et al., 2002), FSL FIRST (Patenaude et al., 2007), ANIMAL (Collins et al., 1999) and the LONI pipeline (Macdonald et al., 1994). Since their development these algorithms have been applied to neurological and psychiatric disorders such as Alzheimer’s disease (Cherubini et al., 2010), multiple sclerosis (Benedict et al., 2009) and schizophrenia (Kuperberg et al., 2003) and are also being used to investigate the developing brain in childhood and adolescence (Lenroot et al., 2007), However, early validation studies were limited to healthy young adults and did not report between session, pulse sequence or scanner reproducibility; measures of sensitivity to changes in regional brain volume were rarely presented. These issues are critically important for multi-centre and longitudinal studies, where segmentation algorithms should be sensitive to small changes in brain volume but insensitive to the use of different magnetic resonance imaging (MRI) scanners (reflecting differences in scanner hardware and software and performance differences between otherwise identical scanners). Another consideration is that some algorithms are not able to segment particular types of images, or require varying degrees of user intervention and therefore may become impractical for studies with large cohorts. These problems may explain why manual segmentation of brain regions is still commonplace in the literature (Doty et al., 2008; Dutt et al., 2009; Ettinger et al., 2007; Jack et al., 2008b) Recently, more rigorous studies have been published comparing segmentation algorithms in terms of accuracy (Babalola et al., 2009; Morey et al., 2009), test re-test reproducibility (Morey et al., 2010), sensitivity to changes in brain structure (Bergouignan et al., 2009), and the effect of MRI acquisition parameters on segmentation reproducibility in terms of global (de Boer et al., 2010; Shuter et al., 2008), subcortical and cortical volumes (Jovicich et al., 2009; Wonderlick et al., 2009). However to our knowledge, no publically available dataset exists that may be used to measure segmentation performance in terms of all the above parameters.
The aim of this paper is two-fold, a) to directly address this point by developing a comprehensive testing protocol to determine the accuracy, reproducibility and sensitivity of MRI neuroanatomical segmentation techniques using publically available data which can be used by other investigators and b) to apply the testing protocol to assess lateral ventricle segmentation using a new fully automated technique and to compare this with two popular freely available packages, FreeSurfer and FSL FIRST.
Specifically with respect to lateral ventricle segmentation:
The accuracy of the algorithms will be tested in healthy adults, patients with Alzheimer’s disease and infants, reflecting a wide range of brain morphology.
The reproducibility of the algorithms using the same participants will be tested between sessions, across pulse sequences and on data from a 1.5T and 3T MRI scanner; reflecting inter-session scanner variability, acquisition protocol variability and hardware variability.
The sensitivity of the algorithms to changes in ventricular volume will be tested on a longitudinal dataset where age related changes in brain morphology are expected to occur.
Our focus on the lateral ventricles is of clinical relevance and research interest because increased volume of this region has been implicated in a number of psychiatric and neurological disorders. Dilation of the lateral ventricles is one of the most consistent findings in both schizophrenia (Kempton et al., 2010; Wright et al., 2000) and bipolar disorder (Kempton et al., 2008). Although hippocampal volume reduction is the most prominent finding, ventricular volume increase is also a key sign of progression in Alzheimer’s disease (Zakzanis et al., 2003) and mild cognitive impairment (Carmichael et al., 2007).
Material and Methods
The segmentation testing protocol is described, followed by a description of a novel algorithm used to segment the lateral ventricles. Finally we demonstrate how the segmentation testing protocol is applied to assess the novel algorithm, FSL FIRST and FreeSurfer.
Segmentation Testing Protocol
Accuracy
Gold Standard
To establish the accuracy of the segmentation algorithms, manually determined lateral ventricle regions of interest (ROIs) were used as the ‘gold standard’ in each of the three groups described below (one independent rater for each group). The ROI analysis was conducted on the basis of sterological techniques and the Cavalieri principle implemented in PC-based software (MEASURE) which has been validated (Barta et al., 1997) and extensively used in ROI studies (Keller et al., 2009; McAlonan et al., 2002; McDonald et al., 2006). MEASURE superimposes a grid on the image and grid points falling within the lateral ventricles were manually marked by a trained rater. The region comprised the entire lateral ventricular system including the temporal horns. The lateral ventricles are bordered medially by the corpus callosum, septum pellucidum and interventricular foramen, anteriorly by the frontal cortex and posteriorly by the occipital cortex. Head tilt was corrected using manual reorientation in MEASURE in all brains before measurements to align images along the anterior commissure–posterior commissure (AC-PC) line and the interhemispheric fissure. A grid setting of 1×1×1 was used so that one grid point fell on each voxel in the image. The software allows the user to zoom in and out and view the image in 3 orthogonal planes. When using a high zoom setting trilinear interpolation is applied to the images, however the grid points are always displayed as one pixel. Raters were trained to use their judgement in classifying voxels which were affected by partial volume effects For each of the three groups below the lateral ventricles were analyzed by the rater on two occasions to obtain intra-rater reliability estimates and a random selection of 5 images from each group were analyzed by an independent rater to obtain inter-rater reliability estimates. Participants took part in this study in accordance with the Declaration of Helsinki and the procedures were approved by local ethics committees.
Young Adults
Seven young adults (mean ± SD, age = 23.8 ± 4.1 years) were scanned using a 1.5 Tesla GE Signa MRI scanner (General Electric, Milwaukee, WI). Images were acquired in the coronal plane using a three dimensional, T1 weighted, inversion-recovery prepared, steady state, spoiled gradient-echo pulse sequence (TR = 9.1 ms, TE = 2 ms, TI = 450 ms, flip angle = 20 degrees, slice thickness = 1.5 mm, matrix size = 256×256, voxel dimensions = 0.94×0.94×1.50 mm3, averages = 1, images available at http://sites.google.com/site/brainseg).
Alzheimer’s Disease
Nine patients with Alzheimer’s disease (age = 77.4 ± 2.4 years, 6 females, mini mental state exam (MMSE) score = 23.7 ± 3.5, clinical dementia rating = 1.1 ± 0.4) were scanned using a 1.5 T General Electric Signa HDx MRI scanner (General Electric, Milwaukee, WI). One patient’s diagnosis was subsequently changed to depression with memory problems. Data acquisition was designed to be compatible with the Alzheimer Disease Neuroimaging Initiative (ADNI) (Jack et al., 2008a). Following a three plane localizer, a high resolution sagittal 3D MP-RAGE dataset was acquired (TR = 8.6 ms, TE = 3.8 ms, TI = 1000 ms, flip angle = 8 degrees, slice thickness = 1.2 mm, matrix size = 256×256, voxel dimensions = 0.938×0.938×1.2 mm3, averages = 1, images available from http://sites.google.com/site/brainseg).
Infants
The infant dataset was collected by an independent research group (Gousias et al., 2008) and is available at http://www.brain-development.org/. We selected a subset of 10 structural MRIs (subjects 2, 4, 8, 11, 14, 18, 21, 22, 26 and 27) from the full sample of 32 two-year old infants born prematurely (age = 24.8 ± 2.4 months, 16 females). Sagittal T1 weighted volumes were acquired from each subject (1.0T Phillips HPQ scanner, TR = 23ms, TE = 6ms, slice thickness = 1.6 mm, matrix size = 256×256, voxel dimensions = 1.04×1.04×1.6 mm3 resliced to 1.04×1.04×1.04 mm3).
Reproducibility
Test-retest reliability
To assess the reproducibility of the segmentation algorithms in the same subjects using the same MRI scanner and pulse sequence, we used the Open Access Series of Imaging Studies (OASIS, www.oasis-brains.org) database which includes structural MRI scans from 20 subjects (age = 23.4 ± 4.0 years, 8 females) who were scanned using the same pulse sequence (1.5T Siemens Vision scanner, TR = 9.7 ms, TE = 4 ms, TI = 20 ms, flip angle = 10 degrees, slice thickness = 1.25 mm, matrix size = 256×256, voxel dimensions = 1×1×1.25 mm3 resliced to 1×1×1 mm3, averages = 1) on 2 occasions within 90 days (Marcus et al., 2007).
Between scanner/ pulse sequence reproducibility
To determine the consistency of the segmentations when different MRI scanners and pulse sequences were used, 9 adults (age = 28 ± 8.5 years, 6 females) were each scanned using two MRI scanners (1.5T and 3.0T General Electric Signa HDx scanner) with 4 different pulse sequences in each scanner (8 images per subject in total, mean inter-scan interval between 1.5T and 3T scanner = 6.7 ± 4.2 days). The pulse sequences were all T1 weighted volumetric scans (see Table 1 for MRI sequence parameters, images available from http://sites.google.com/site/brainseg).
Table 1.
MRI sequences used for testing reproducibility between scanners and pulse sequences.
| Name | Pulse sequence |
Field strength (T) |
Orientation | TE (ms) |
TR (ms) |
TI (ms) |
Flip angle (°) |
Slice thickness (mm) |
In plane resolution (mm) |
|---|---|---|---|---|---|---|---|---|---|
| A1 | MP-RAGE* | 1.5 | Sagittal | 3.8 | 8.6 | 1000 | 8 | 1.20 | 0.938 |
| A2 | MP-RAGEx | 1.5 | Axial | 3.8 | 8.6 | 1000 | 8 | 1.20 | 0.938 |
| A3 | SPGR | 1.5 | Axial | 4.9 | 11.1 | 300 | 18 | 1.10 | 1.094 |
| A4 | SPGR | 1.5 | Coronal | 5.2 | 11.8 | 450 | 20 | 1.50 | 0.859 |
| B1 | MP-RAGE* | 3.0 | Sagittal | 2.8 | 6.6 | 900 | 8 | 1.20 | 1.016 |
| B2 | SPGR* | 3.0 | Sagittal | 2.8 | 7.0 | 650 | 8 | 1.20 | 1.016 |
| B3 | SPGR | 3.0 | Coronal | 2.8 | 7.1 | 450 | 20 | 1.10 | 1.094 |
| B4 | SPGR | 3.0 | Coronal | 3.1 | 8.1 | 500 | 20 | 1.50 | 0.781 |
ADNI compatible sequence (Jack et al., 2008a)
ADNI compatible sequence acquired in axial rather than sagittal orientation
For an extreme test of between pulse sequence reproducibility we obtained T2 weighted images collected for clinical reporting and T1 weighted scans from the same 15 young adults (age = 36.3 ± 13.4 years, 9 females). The images were acquired using the 1.5T scanner above with an axial T2 weighted sequence (TR = 3000 ms, TE = 97 ms, flip angle = 90 degrees, slice thickness = 3mm, matrix size = 256×256, voxel dimensions = 0.94×0.94×3 mm3, averages = 1) and sagittal T1 weighted scans (pulse sequence A1, Table 1, images available from http://sites.google.com/site/brainseg).
Sensitivity
Ventricular volume is known to increase with age in healthy adults from post-mortem (Hubbard and Anderson, 1981), CT (Schwartz et al., 1985) and MRI studies (Scahill et al., 2003). To examine the sensitivity of the algorithms to small changes in ventricular volume we used the longitudinal OASIS dataset (Marcus et al., 2009) which includes T1 weighted MR image pairs (1.5T Siemens Vision scanner, TR = 9.7 ms, TE = 4 ms, TI = 20 ms, flip angle = 10 degrees, slice thickness = 1.25 mm, matrix size = 256×256, voxel dimensions = 1×1×1.25 mm3, averages = 1) from the same healthy volunteers acquired at two time points (72 subjects, age at baseline scan = 75.4 ± 8.1 years, 50 females, mean inter-scan interval = 738 ± 249 days). The sensitivity of the algorithms was assessed by their ability to detect, i) a change in ventricular volume between the baseline and follow-up scan and ii) their ability to detect a correlation between the change in ventricular volume and the inter-scan interval.
Processing Speed
The average time taken for each algorithm to process one MR image was determined by processing 10 randomly chosen images from the OASIS dataset. All algorithms were run on a 2× Quad Core Xeon E5450 3.0GHz computer with 56Gb RAM using the CentOS 5.4 Linux operating system.
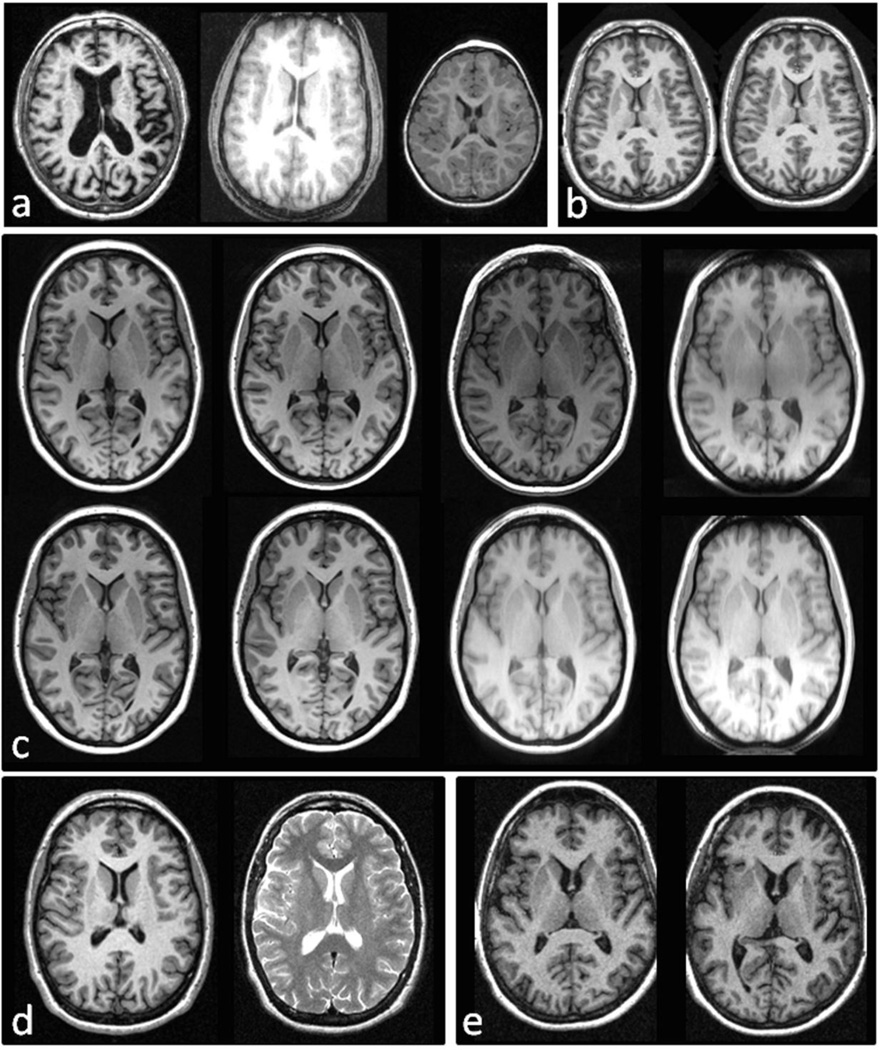
All of the data used in the testing protocol are publically available and are detailed in Figure 1 and Table 7.
Figure 1.
MRI data used in the comprehensive testing protocol, a) Accuracy: Alzheimer’s disease, young adults, infants, b) Reproducibility: test-retest same scanner and pulse sequence; c) different scanner and pulse sequence, the same subject scanned with 8 sequences on a 1.5T scanner (top row) and 3T scanner (bottom row); d) T1 and T2 weighted images. e) Sensitivity: subject scanned as baseline and after 2 years.
Table 7.
Details of the publically available data used to test the segmentation algorithms
| Dataset | Number of subjects |
Number of images |
Mean age (years) |
Description All images are T1 weighted structural MR images unless otherwise stated |
Internet address |
|---|---|---|---|---|---|
| Young Adults | 7 | 7 | 23.8±4.1 | Healthy subjects | sites.google.com/site/brainseg |
| Alzheimer’s Disease | 9 | 9 | 77.4±2.3 | Patients with Alzheimer’s Disease | sites.google.com/site/brainseg |
| Infants | 10 | 10 | 2.1±0.2 | Infants born prematurely | sites.google.com/site/brainseg or www.brain-development.org |
| Lateral Ventricle ROIs on the above datasets | 26 | 26 | 34 (approx range 2–80) | Regions estimated by the Cavalieri principle. | sites.google.com/site/brainseg |
| Test-retest same scanner and pulse sequence | 20 | 40 | 23.4±4.0 | Healthy subjects scanned twice within 90 days | sites.google.com/site/brainseg or www.oasis-brains.org |
| Different scanner and pulse sequence | 9 | 72 | 28.0±8.5 | Healthy subjects scanned a total of 8 times using a 1.5T and 3T scanner with various T1 weighted sequences. | sites.google.com/site/brainseg |
| Reproducibility T1/T2 | 15 | 30 | 36.3±13.4 | Healthy subjects scanned with a T1 and T2 weighted acquisition | sites.google.com/site/brainseg |
| Sensitivity longitudinal dataset | 72 | 144 | 75.4±8.2 (baseline). Interscan interval 2.0±0.7 years | Healthy subjects scanned twice over a period of approximately 2 years | sites.google.com/site/brainseg or www.oasis-brains.org |
Lateral Ventricle Segmentation Algorithm ‘ALVIN’
Our novel algorithm for segmentation of the lateral ventricles, named ALVIN (Automatic Lateral Ventricle delIneatioN), uses ‘unified segmentation’ in SPM8 (Ashburner and Friston, 2005). Unified segmentation produces gray matter, white matter and cerebral spinal fluid (CSF) images from MRI data but does not segment subcortical structures. ALVIN works by applying a binary mask to spatially normalised cerebral spinal fluid (CSF) segmented images produced using unified segmentation. As the segmented images already demarcate the main boundaries of the lateral ventricles, the purpose of the mask was to exclude CSF outside the lateral ventricles, such as the third ventricle, superior cistern and sulcal CSF.
Creation of Binary Mask
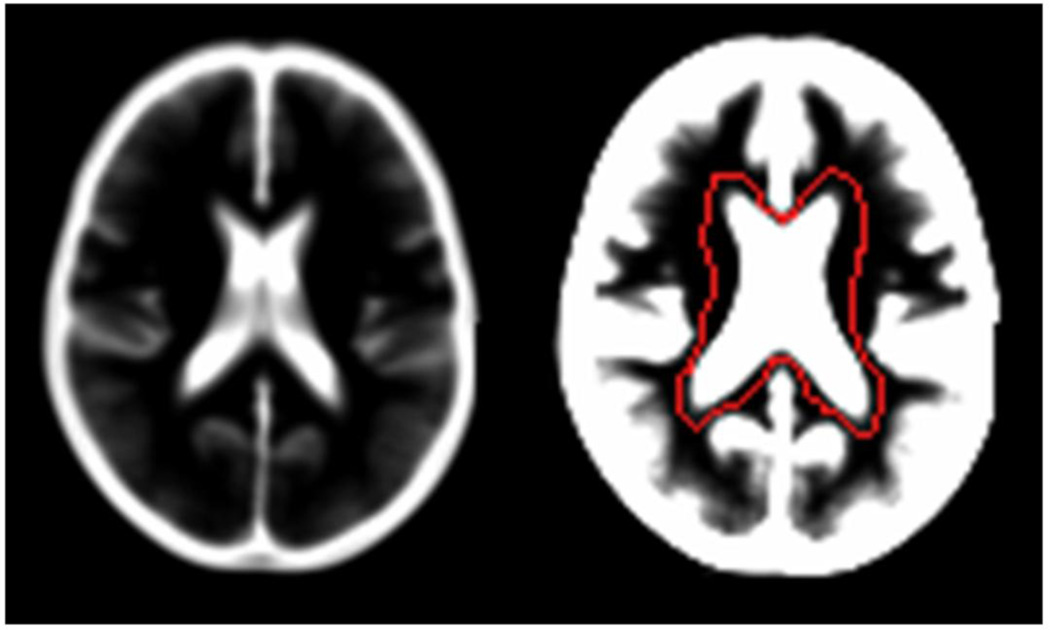
There is large inter-subject variability in the size and shape of the lateral ventricles even after spatial normalisation into Montreal Neurological Institute (MNI) space. Therefore it was important that the boundaries of the mask were made with reference to a large representative population. We used the healthy control sample from the cross-sectional OASIS, (www.oasis-brains.org) database which includes structural MRI scans from 316 healthy subjects aged 18 to 94 (Marcus et al., 2007). Images were averaged from 3 to 4 MP-RAGE scans (TR = 9.7 ms, TE = 4 ms, TI = 20 ms, flip angle = 10, slice thickness = 1.25 mm, matrix size = 256×256, voxel dimensions 1×1×1.25 mm3) obtained from the same subject on the same day. Modulated normalised CSF images were produced using unified segmentation in SPM8 with default options (Ashburner and Friston, 2005). Unified segmentation performs image registration, bias field correction, and tissue segmentation in one generative model. Images are spatially normalised into MNI space using affine transformations and non-linear basis functions; volume information at each voxel is conserved by multiplying tissue density values by the Jacobian determinant. We also applied a standard SPM algorithm (clean_gwc) which removes incorrectly segmented gray and white matter using an iterative conditional dilation and smoothing technique applied over combined gray and white matter maps. Of the 316 scans in the database 275 were successfully segmented by SPM. There was no significant difference in age or gender between subjects where segmentation had been successful compared to failed segmentations (both p>0.39). The face removal algorithm used by Marcus et al (2007) to ensure subject anonymity may have increased the segmentation failure rate, as priors used by SPM8 include facial features. A mean CSF image from the 275 segmented images was produced to enable the delineation of the lateral ventricle mask. The outlines of the mask was drawn using the ROI tool in MRIcro v1.40 (Rorden and Brett, 2000). To highlight all regions where CSF voxels existed in every subject, the mean CSF image was viewed using intensity window centre and width settings as 0.05 and 0.1 respectively (Figure 2). The mask boundaries were hand drawn to include the entire lateral ventricular system including the temporal horns. In a small number of regions/coordinates lateral ventricular CSF and non-ventricular CSF overlapped between subjects in normalised space (e.g. at the fornix boundary between the lateral ventricles and superior cistern, and in the occipital lobe between the posterior horn and parieto-occiptal sulcus). For these regions the image contrast was reduced and the mask boundary was marked within the CSF signal local minimum to ensure the ventricular CSF was included for the majority of subjects at these particular coordinates.
Figure 2.
Mean image from 275 normalised and CSF segmented structural MRI scan from the OASIS healthy adult dataset (left). The same image with the intensity window=0.05 and width=0.1 showing lateral ventricle mask drawn in MRIcro v1.40 (right).
Determining the volume of the lateral ventricles using the binary mask
Unified segmentation in SPM8 was applied to each test image to produce a modulated CSF image which was multiplied by the binary mask giving a three-dimensional image of the lateral ventricles in MNI space. As the data was modulated, absolute volume of the lateral ventricles was calculated by summing the intensity over the entire normalised image.
Assessing ALVIN, FSL FIRST and FreeSurfer using the Segmentation Testing Protocol
The testing protocol was applied to the ALVIN algorithm described above, FSL (v4.1.7) FIRST and FreeSurfer (v4.5.0). Briefly the FSL FIRST algorithm performs subcortical volumetric and shape analysis using models constructed from manually segmented images (Patenaude, 2007). Initially the FIRST algorithm normalises the MR image into MNI space, after which the normalisation is checked manually. The spatial transformation is used to fit a subcortical mask to the image and a segmentation algorithm with a model of the left and right lateral ventricle is used to segment these structures. The algorithm requires the number of modes of variation as input, which is set to 40 for the lateral ventricles (as recommended by the authors of FSL FIRST). Finally a boundary correction algorithm which uses FSL’s segmentation tool, FAST is applied before the volume of the lateral ventricles is determined. The FreeSurfer package may be used to conduct subcortical segmentation and cortical surface parcellation. For the analysis used in this study the FreeSurfer pipeline (Fischl et al., 2002) performed intensity correction and skull stripping, followed by gray and white segmentation and segmentation of subcortical structures including the lateral ventricles using an atlas based approach. For each side of the brain FreeSurfer outputs two segmentations which are named ‘lateral ventricles’ and ‘inferior lateral ventricles’; the volumes of these regions were summed to obtain total lateral ventricle volume. The performance of the ALVIN algorithm was tested using SPM8, however to determine if the algorithm was compatible with SPM5 we also applied the entire testing protocol to ALVIN using both SPM versions. To determine spatial overlap in the segmentation produced by ALVIN and FSL FIRST and FreeSurfer, it was necessary to convert the segmented image produced by ALVIN from MNI space to native space. This was achieved by applying the inverse spatial normalisation parameters for each subject to the ALVIN binary mask of the lateral ventricles. The binary mask in native space was then used to mask a CSF segmented image in native space produced by the unified segmentation step. Finally the image of the lateral ventricles in native space was thresholded at 0.5 to produce a binary segmented image.
Statistical Analysis
To quantify accuracy and reproducibility we used the Intraclass Correlation Coefficient (ICC) measure (single measure, 2-way mixed consistency) (McGraw and Wong, 1996; Yaffee, 1998). For accuracy results, the ICC quantifies how well the automated segmentations agree with the gold standard measures, for reproducibility measures the ICC value quantifies the consistency of the segmentations. ICC values were calculated after the exclusion of failed segmentations, and were not calculated if more than 50% of segmentations failed. Spatial overlap of segmentations was assessed using the Dice coefficient (Crum et al., 2005). For the sensitivity analysis a paired t-test was used and the result was converted to a Z-score and Pearson’s r was used to determine the correlation between volume change and inter-scan interval. Statistical calculations were performed with SPSS 15.0 (SPSS Inc.) except for power calculations which were carried out using GPOWER 3.0 (Faul et al., 2007).
First Pass Failed Segmentations
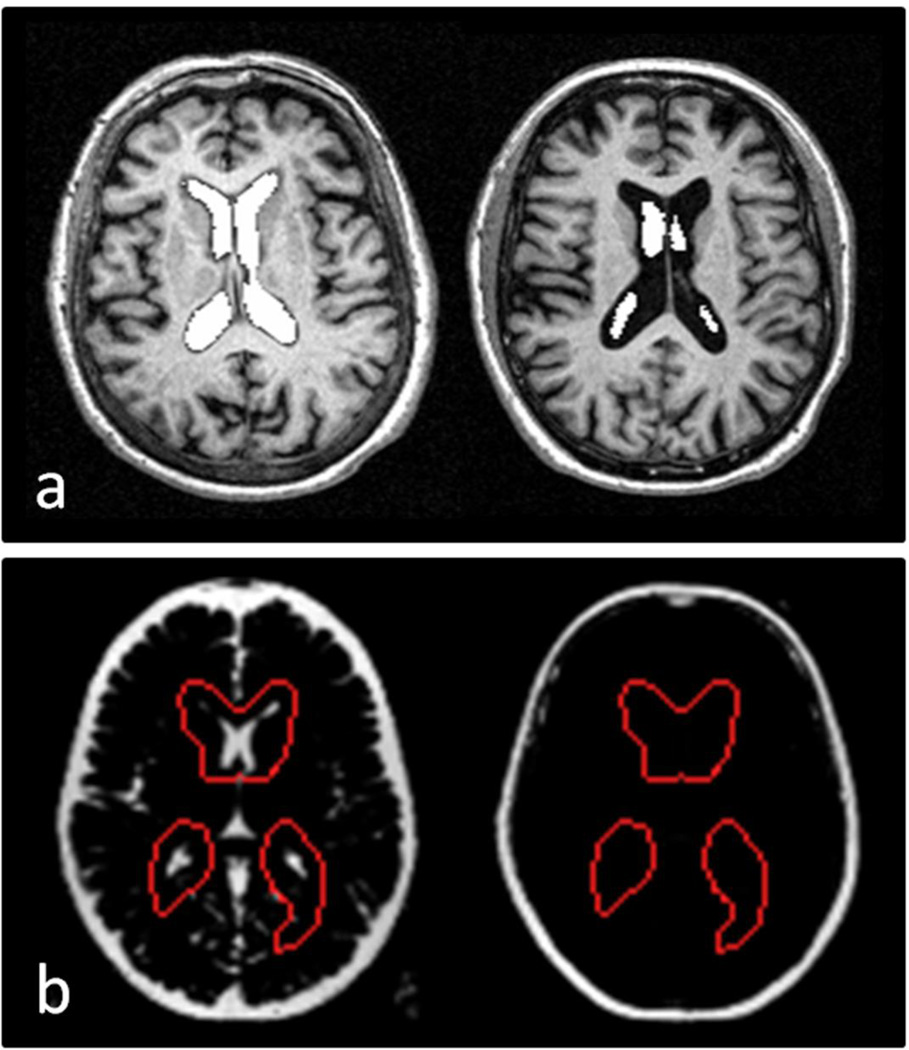
By visually inspecting each segmentation we recorded the number of cases where the algorithms failed to segment the lateral ventricles (see Figure 3 for examples). For consistency we did not attempt to adjust the default parameters in each algorithm and re-run the segmentation step or manually adjust the images.
Figure 3.
Images demonstrating successful (left) and failed segmentations (right) of the lateral ventricles. a) FSL FIRST b) ALVIN. FreeSurfer failures were recognised by the FreeSurfer software which terminated before a segmented image was produced.
Results
The performance of the 3 algorithms as assessed by the segmentation testing protocol is compared in Table 2 to 6.
Table 2.
Accuracy, reproducibility, sensitivity and speed of the 3 algorithms in segmenting the lateral ventricles.
| ALVIN | FSL FIRST | FreeSurfer | ||
|---|---|---|---|---|
| Accuracy | Young Adults (ICC) | 0.992 | 0.902 | 0.994 |
| Alzheimer’s Disease (ICC) | 0.973 | N/Aa | 0.998 | |
| Infants (ICC) | 0.953 | 0.901 | 0.959 | |
| Reproducibility | Test-retest, same scanner and pulse sequence (ICC) | 0.999 | 0.996 | 0.998 |
| Different scanner and pulse sequence (ICC) | 0.994 | 0.981 | 0.997 | |
| Reproducibility T1/T2 (ICC) | 0.982 | 0.925 | N/Aa | |
| Sensitivity | Paired t-test between baseline and follow-up scan (Z score) | 6.55, p<0.001 | N/Aa | 5.71, p<0.001 |
| Correlation of volume change and interscan interval (R value) | 0.52, p<0.001 | N/Aa | 0.60, p<0.001 | |
| Speed | Time to process one subject (minutes) | 8 | 7 | 420 |
Algorithms with a greater than 50% failure rate were not included in the ICC analysis
Table 6.
Mean (SD) lateral ventricle volume in ml of paired images in the reliability and sensitivity datasets
| Alvin | FSL FIRST | FreeSurfer | ||||
|---|---|---|---|---|---|---|
| Scan 1 | Scan2 | Scan1 | Scan2 | Scan1 | Scan2 | |
| Test-retest: baseline, follow-up | 14.71 (10.54) | 14.73 (10.42) | 13.33 (4.74) | 13.42 (4.89) | 12.97 (10.96) | 12.91 (10.88) |
| Different scanner: 1.5T, 3T | 17.18 (9.53) | 17.56 (9.56) | 14.74 (4.67) | 14.76 (5.00) | 20.93 (12.39) | 21.33 (12.25) |
| Reproducibility: T1,T2 | 15.26 (6.39) | 20.33 (6.11) | 14.77 (4.65) | 17.85 (4.44) | N/Aa | N/Aa |
| Sensitivity: baseline, follow-up | 42.67 (20.05) | 45.38 (21.50) | N/Aa | N/Aa | 35.12 (18.87) | 37.67 (20.47) |
Algorithms with a greater than 50% failure rate were not included in this table
Accuracy
The intra-rater agreement (same rater), in terms of intra-class correlation coefficients (ICCs) for gold standard manual segmentation of the lateral ventricles was 0.994 for young adults, 0.999 in patients with Alzheimer’s disease and 0.973 in infants. Inter-rater reliability (independent raters) for adults, patients with Alzheimer’s disease and infants was 0.995, 0.991 and 0.993 respectively. All three algorithms demonstrated high accuracy compared to manual gold standard segmentation (Table 2). In terms of segmentation failures, the Alzheimer’s disease images were the most problematic for all 3 segmentation algorithms, particularly FSL FIRST which was unable to segment any of the images (Table 3). Overall FreeSurfer demonstrated the highest segmentation accuracy. Segmentation consistency was good between the three algorithms (Table 4). ICC and Dice coefficient measures both indicated that ALVIN and FreeSurfer most closely agreed, except for the young adults dataset where the latter measure suggested a closer agreement between ALVIN and FSL FIRST. In terms of absolute volume measures (Table 5) ALVIN reported a consistently higher volume than the other techniques.
Table 3.
First pass segmentation failures for each algorithm and dataset. Examples of segmentation failures are shown in Figure 3.
| ALVIN | FSL FIRST | FreeSurfer | |
|---|---|---|---|
| Young Adults | 0% | 14% | 0% |
| Alzheimer’s Disease | 11% | 100% | 11% |
| Infants | 0% | 0% | 0% |
| Test-retest, same scanner and pulse sequence | 0% | 5% | 0% |
| Different scanner and pulse sequence | 0% | 13% | 0% |
| Reproducibility T1/T2 | 0% | 7% | 50% |
| baseline and follow-up scan | 7% | 63% | 10% |
| All images | 3.5% | 36.2% | 9.6% |
Table 4.
Paired comparisons of segmentation by ALVIN, FSL FIRST and FreeSurfer for the young adult, Alzheimer’s disease and infant validation dataset. The consistency of the segmentations is measured using the intraclass correlation coefficient and Dice coefficient.
| ALVIN vs FSL FIRST |
ALVIN vs FreeSurfer | FSL vs FreeSurfer | ||
|---|---|---|---|---|
| Young Adults | ICC | 0.835 | 0.988 | 0.941 |
| Dice Coefficient (SD) | 0.869 (0.052) | 0.807 (0.052) | 0.757 (0.057) | |
| Alzheimer’s Disease | ICC | N/Aa | 0.987 | N/Aa |
| Dice Coefficient (SD) | N/Aa | 0.901 (0.015) | N/Aa | |
| Infants | ICC | 0.869 | 0.990 | 0.900 |
| Dice Coefficient (SD) | 0.753 (0.187) | 0.772 (0.094) | 0.698 (0.181) | |
Algorithms with a greater than 50% failure rate were not included in this table
Table 5.
Mean (SD) lateral ventricle volume in ml for manual segmentation and the automated algorithms for the young adult, Alzheimer’s disease and infant validation datasets.
| Manual | Alvin | FSL FIRST | FreeSurfer | |
|---|---|---|---|---|
| Young Adults | 13.32 (8.00) | 20.17 (8.11) | 15.00 (3.00) | 15.20 (7.74) |
| Alzheimer’s Disease | 54.93 (26.24) | 57.20 (22.96) | N/Aa | 55.83 (25.82) |
| Infants | 7.10 (4.42) | 13.75 (5.26) | 10.99 (3.43) | 10.04 (4.87) |
Algorithms with a greater than 50% failure rate were not included in this table
Reproducibility
ALVIN showed the highest test-retest and T1/T2 reproducibility (Table 2). FSL FIRST showed good reproducibility, but suffered from a reasonably high failure rate on the inter-scanner/ pulse-sequence dataset. FreeSurfer demonstrated good reproducibility in the test-retest dataset and the highest inter-scanner/pulse-sequence reproducibility, but was unable to process any of the clinical T2 weighted images. Absolute volume estimates (Table 6) also showed highly consistent values between paired scans, and demonstrated that ventricular volume estimates were higher from T2 weighted images.
Sensitivity
ALVIN and FreeSurfer were able to detect a change in ventricular volume between baseline and follow-up scan, estimating an increase in volume of 2.7 ml and 2.5 ml respectively over the 2 year period. Both algorithms were also able to detect the expected correlation between the volume increase and interscan interval. FSL FIRST had a failure rate of 63% which precluded a sensitivity analysis. A power analysis suggests that ALVIN would require a sample size of 11 subjects, and FreeSurfer a sample size of 13 subjects to detect a significant change in lateral ventricle volume between the baseline and follow-up scan (two tailed, alpha=0.05, power=0.8). In terms of detecting a correlation between change in ventricular volume and interscan interval, ALVIN and FreeSurfer would require a sample size of 26 and 19 subjects, respectively (two tailed, alpha=0.05, power=0.8).
First Pass Segmentation failures
Segmentation failures were conspicuous during visual inspection and were characteristic for each algorithm. ALVIN failures most commonly occurred at the SPM unified segmentation stage where the scalp was incorrectly classified as CSF (example shown in Figure 3b). FSL FIRST failures occurred at the main segmentation stage and were revealed when the segmented lateral ventricles were overlaid on the MRI scan; as shown (Figure 3a) only small fragments of the lateral ventricles were segmented. FreeSurfer segmentation errors occurred at the normalisation or skull stripping stage and were recognised by the algorithm which terminated the procedure. ALVIN demonstrated the lowest segmentation failure rate of 3.2% over all images followed by FreeSurfer with a failure rate of 9.6% and FSL FIRST with a failure rate of 36.2%.
Processing Speed
Manual segmentations took approximately 80 minutes per subject. ALVIN and FSL FIRST were both faster than manual segmentation taking 8 and 7 minutes respectively. FreeSurfer was an order of magnitude slower than the other algorithms taking approximately 7 hours, although during this time the software segmented a number of subcortical structures, as it was not possible to segment the lateral ventricles only (Table 2).
ALVIN backward compatibility
Lateral ventricles volumes obtained using ALVIN in SPM8 agreed well with those produced using SPM5 (ICC>0.999 over all images in the testing protocol) suggesting the ALVIN algorithm worked effectively with both versions of SPM.
The ALVIN algorithm which takes MRI images in native space as inputs and outputs ventricular volumes, is freely available as an SPM extension and may be downloaded from sites.google.com/site/brainseg. The images used in the testing protocol are freely available and may be downloaded from the websites listed in Table 7.
Discussion
We have developed a testing protocol for assessing the accuracy, reproducibility and sensitivity of segmentation algorithms based on publically available data and validated a conceptually simple technique for automatically extracting the lateral ventricles. The availability of the testing protocol will enable other researchers to validate future segmentation algorithms.
Segmentation Testing Protocol
We primarily used intraclass correlation coefficients (ICC) to measure reproducibility and accuracy. Although segmentation may be assessed with metrics which measure the overlap of regions (Fischl et al., 2002) this was problematic with our data because the gold standard measures were made on the basis of the Cavalieri principle, and the software used did not produce volumetric image files representing manual segmentations. However we were able quantify segmentation overlap between the automated algorithms using the Dice coefficient. The ICC measure of agreement is a widely used statistic, spanning genetics (Gibert et al., 1998), functional neuroimaging (Caceres et al., 2009) and clinical rating scales (Nuechterlein et al., 2008), and is also the standard measure for assessing intra-rater and inter-rater reliability on manually drawn ROIs in structural MRI studies (DeLisi et al., 1997; Doty et al., 2008; McClure et al., 2006) and has previously been used to determine the reliability of FreeSurfer (Wonderlick et al., 2009) and FSL FIRST (Morey et al., 2010). We used an ICC which measures consistency rather than absolute agreement, thus high ICCs reported in this paper suggest the segmentation algorithms would give very similar statistical results when comparing two groups of subjects. However as each algorithm is likely to give a systematic difference in volume (Table 5) it would not be possible to combine data produced by different algorithms in a single study.
The validation dataset within the testing protocol is comparable to the Internet Brain Segmentation Repository (http://www.cma.mgh.harvard.edu/ibsr/) a dataset which includes 18 MRI scans with manual segmentations of 43 individual structures. Our dataset includes manual segmentation of the lateral ventricles only, but includes infants and patients with Alzheimer’s disease to reflect a wider range of brain morphology. A related online resource, BrainWeb (Cocosco et al., 1997) (http://mouldy.bic.mni.mcgill.ca/brainweb/) allows the user to enter customizable MRI sequence parameters to produce a simulated MRI image of the brain and others have highlighted the importance of simulation for segmentation (Simmons et al., 1996). The BrainWeb tool has been used to validate a number of segmentation algorithms (Amato et al., 2003; Chao et al., 2009). Our dataset of 9 individuals scanned with 8 sequences on at 1.5T and 3T could also be used to verify that segmentation algorithms are reproducible when applied to images using a range of MRI parameters.
To assess sensitivity we examined the impact of aging on lateral ventricle volume, as this is reasonably robust effect (Scahill et al., 2003). However not knowing the real change in ventricular volume is problematic in assessing the sensitivity of these algorithms. A different approach is to use simulated data, such as Camara et al. (2008) who used a deformation model to mimic atrophy in Alzheimer’s disease to assess algorithms which measure brain atrophy. The advantage of simulated data is that the investigator precisely knows the location and magnitude of the changes that have occurred, however such an approach relies on the simulation accurately mirroring the effects of pathology on brain structure which may not always be possible and does not reflect small differences that might be caused by, for example, changes in head positioning, hydration and RF coil performance over time. Within our testing protocol it would have been preferable to use a larger group of validation images.
In this study our strategy was to use subgroups which reflected a wide range in brain morphology rather than one large healthy adult group. Our hope is that other investigators will add to the pool of publically available manually segmented images allowing future algorithms to be validated against a larger number of healthy adults and patients with other neurological and psychiatric disorders. A valid criticism of a study which compares an investigator’s own algorithm against others is that there may be biases in selecting the testing protocol. However in this study we have used data that is already publicly available where possible and made our own additional data and software freely available so that other researchers may verify the methods we have used.
Performance of ALVIN, FSL FIRST and FreeSurfer
Using the testing protocol we have validated ALVIN, our segmentation algorithm in adults, patients with Alzheimer’s disease, and infants, and shown it to be reliable between MRI scanners and pulse sequences and sensitive to small changes in ventricular volume. In developing this technique we have built on existing neuroimaging software and datasets; ALVIN relies on the unified segmentation methodology developed by Ashburner and Friston (2005) and the ventricular mask which was based on the representative OASIS MRI dataset (Marcus et al., 2007). The algorithm was comparable to FreeSurfer in terms of accuracy, reproducibility and sensitivity. ALVIN produced volume estimates which were higher than manual segmentation values and the other automated techniques. This was most marked in the infant dataset although the other two automated techniques also gave higher values than manual segmentation. Inspection of the infant dataset segmented with ALVIN revealed that in some cases small parts of the superior cistern and parieto-occiptal sulcus were classified as ventricular CSF due to their relative position in normalised MNI space. Unfortunately altering the ventricular mask to improve segmentation in infants would adversely impact segmentation in older age groups due to increased size of the ventricles with aging. A possible improvement would be for ALVIN to automatically select different ventricular masks based on brain structure, or to use a more accurate spatial normalisation procedure to closely match ventricular size and shape to a standard template. Differences in absolute volume estimates between the manual and automated methods are also likely to arise from partial volume effects. Both manual and automated methods use intensity information when classifying voxels, however small differences in the threshold applied may lead to different volume estimates particularly in structures with a large surface area to volume ratio. Clinical and research questions are usually concerned with volume differences, either between patients and controls, or baseline and follow-up scans where reproducibility may be more important than absolute volume. As highlighted previously it is not possible to combine data from different algorithms in a single study if the algorithms exhibit systematic differences. ICC values demonstrated that ALVIN closely agreed with manual measures in terms of the relative distribution of volumes in a group and was also sensitive to longitudinal changes in volume. In terms of processing speed, if a user required ventricular volumes, ALVIN was 50 times faster than FreeSurfer. However an important limitation of ALVIN is that it is only able to segment a single region while both FSL FIRST and FreeSurfer are able to segment a number of cortical and subcortical regions
In terms of previous reproducibility studies, Morey et al (2010) reported test-retest ICC values of 0.993–0.999 for the lateral ventricles segmented using FreeSurfer which compared well with our value of 0.998, and reported ICC values of 0.977–0.998 for FSL FIRST which agreed with the value of 0.996 reported in this study. Our results also concur with Jovicich et al (2009) who report that different T1 weighted images had only a relatively small effect on segmented lateral ventricle volume compared to inter-subject variability using FreeSurfer.
The FreeSurfer algorithm was valid in all groups, while the FSL FIRST algorithm was valid in young adults and infants and both techniques demonstrated good reproducibility. The poor performance of FSL FIRST in patients with Alzheimer’s disease was surprising, especially as the training dataset used to develop the algorithm included patients with Alzheimer’s disease (Patenaude, 2007). Inspection of the FIRST segmented images revealed that in some cases only small sections of the ventricles were identified, leading to erroneous volume estimates. In addition the ventricular model within FIRST did not appear to include the temporal horn which contributed to a lower accuracy estimate. Examination of the data showed that FIRST had particular problems with large ventricles and was not able to segment any images with ventricles larger than 35ml. Thus FIRST failure rates were particularly high in the Alzheimer dataset and the OASIS longitudinal dataset used in the sensitivity analysis (Table 3) which included participants with a mean age of 75. The poor performance of FSL FIRST ventricular segmentation is unusual for FSL structural MRI processing tools. Indeed a recent publication has shown that FSL FIRST accurately segments other subcortical structures (Patenaude et al., 2011) and in terms of our own studies we have previously demonstrated that FSL SIENA was sensitive enough to detect small changes in brain morphology from acute dehydration (Kempton et al., 2009).
The ability of ALVIN and FSL FIRST to segment the lateral ventricles from clinical T2 weighted images is useful, as it demonstrates the techniques may be used with lower resolution data, allowing the algorithms to be applied to older images or studies where acquisition time is required be kept to a minimum.
ALVIN and FreeSurfer are well suited to multicentre and/or longitudinal studies due to their relatively high inter-scanner reproducibility and sensitivity to changes in ventricular volume. For multicentre projects the different scanners would still need to be modelled at the statistical analysis stage, however by using these algorithms, the inter-scanner variance would be efficiently accounted for, increasing sensitivity to small changes in ventricular size.
Acknowledgements
The authors acknowledge financial support from the National Institute for Health Research (NIHR) Specialist Biomedical Research Centre for Mental Health award to the South London and Maudsley NHS Foundation Trust and the Institute of Psychiatry, King’s College London. W.R. Crum acknowledges support from the King’s College London Centre of Excellence in Medical Engineering funded by the Wellcome Trust and EPSRC (WT 088641/Z/09/Z). We are grateful to the Open Access Structural Imaging Series for the use of this data and include their following grant numbers: P50 AG05681, P01 AG03991, R01 AG021910, P20 MH071616, U24 RR021382. Ulrich Ettinger acknowledges support from the Deutsche Forschungsgemeinschaft (ET 31/2-1).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amato U, Larobina M, Antoniadis A, Alfano B. Segmentation of magnetic resonance brain images through discriminant analysis. J Neurosci Methods. 2003;131:65–74. doi: 10.1016/s0165-0270(03)00237-1. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Babalola KO, Patenaude B, Aljabar P, Schnabel J, Kennedy D, Crum W, Smith S, Cootes T, Jenkinson M, Rueckert D. An evaluation of four automatic methods of segmenting the subcortical structures in the brain. Neuroimage. 2009;47:1435–1447. doi: 10.1016/j.neuroimage.2009.05.029. [DOI] [PubMed] [Google Scholar]
- Barta PE, Dhingra L, Royall R, Schwartz E. Improving stereological estimates for the volume of structures identified in three-dimensional arrays of spatial data. J Neurosci Methods. 1997;75:111–118. doi: 10.1016/s0165-0270(97)00049-6. [DOI] [PubMed] [Google Scholar]
- Benedict RH, Ramasamy D, Munschauer F, Weinstock-Guttman B, Zivadinov R. Memory impairment in multiple sclerosis: correlation with deep grey matter and mesial temporal atrophy. J Neurol Neurosurg Psychiatry. 2009;80:201–206. doi: 10.1136/jnnp.2008.148403. [DOI] [PubMed] [Google Scholar]
- Bergouignan L, Chupin M, Czechowska Y, Kinkingnehun S, Lemogne C, Le Bastard G, Lepage M, Garnero L, Colliot O, Fossati P. Can voxel based morphometry, manual segmentation and automated segmentation equally detect hippocampal volume differences in acute depression? Neuroimage. 2009;45:29–37. doi: 10.1016/j.neuroimage.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Caceres A, Hall DL, Zelaya FO, Williams SC, Mehta MA. Measuring fMRI reliability with the intra-class correlation coefficient. Neuroimage. 2009;45:758–768. doi: 10.1016/j.neuroimage.2008.12.035. [DOI] [PubMed] [Google Scholar]
- Camara O, Schnabel JA, Ridgway GR, Crum WR, Douiri A, Scahill RI, Hill DL, Fox NC. Accuracy assessment of global and local atrophy measurement techniques with realistic simulated longitudinal Alzheimer's disease images. Neuroimage. 2008;42:696–709. doi: 10.1016/j.neuroimage.2008.04.259. [DOI] [PubMed] [Google Scholar]
- Carmichael OT, Kuller LH, Lopez OL, Thompson PM, Dutton RA, Lu A, Lee SE, Lee JY, Aizenstein HJ, Meltzer CC, Liu Y, Toga AW, Becker JT. Ventricular volume and dementia progression in the Cardiovascular Health Study. Neurobiol Aging. 2007;28:389–397. doi: 10.1016/j.neurobiolaging.2006.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao WH, Chen YY, Lin SH, Shih YY, Tsang S. Automatic segmentation of magnetic resonance images using a decision tree with spatial information. Comput Med Imaging Graph. 2009;33:111–121. doi: 10.1016/j.compmedimag.2008.10.008. [DOI] [PubMed] [Google Scholar]
- Cherubini A, Spoletini I, Peran P, Luccichenti G, Di Paola M, Sancesario G, Gianni W, Giubilei F, Bossu P, Sabatini U, Caltagirone C, Spalletta G. A multimodal MRI investigation of the subventricular zone in mild cognitive impairment and Alzheimer's disease patients. Neurosci Lett. 2010;469:214–218. doi: 10.1016/j.neulet.2009.11.077. [DOI] [PubMed] [Google Scholar]
- Cocosco CA, Kollokian V, Kwan RKS, Evans AC. BrainWeb: Online Interface to a 3D MRI Simulated Brain Database. Neuroimage. 1997;5:S425. [Google Scholar]
- Collins DL, Zijdenbos AP, Baare WFC, Evans AC. Information Processing in Medical Imaging. Heidelberg: Springer Berlin; 1999. ANIMAL+INSECT: Improved Cortical Structure Segmentation; pp. 210–223. [Google Scholar]
- Crum WR, Camara O, Rueckert D, Bhatia KK, Jenkinson M, Hill DL. Generalised overlap measures for assessment of pairwise and groupwise image registration and segmentation. Med Image Comput Comput Assist Interv. 2005;8:99–106. doi: 10.1007/11566465_13. [DOI] [PubMed] [Google Scholar]
- de Boer R, Vrooman HA, Ikram MA, Vernooij MW, Breteler MM, van der Lugt A, Niessen WJ. Accuracy and reproducibility study of automatic MRI brain tissue segmentation methods. Neuroimage. 2010;51:1047–1056. doi: 10.1016/j.neuroimage.2010.03.012. [DOI] [PubMed] [Google Scholar]
- DeLisi LE, Sakuma M, Tew W, Kushner M, Hoff AL, Grimson R. Schizophrenia as a chronic active brain process: a study of progressive brain structural change subsequent to the onset of schizophrenia. Psychiatry Res. 1997;74:129–140. doi: 10.1016/s0925-4927(97)00012-7. [DOI] [PubMed] [Google Scholar]
- Doty TJ, Payne ME, Steffens DC, Beyer JL, Krishnan KR, LaBar KS. Agedependent reduction of amygdala volume in bipolar disorder. Psychiatry Res. 2008;163:84–94. doi: 10.1016/j.pscychresns.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutt A, McDonald C, Dempster E, Prata D, Shaikh M, Williams I, Schulze K, Marshall N, Walshe M, Allin M, Collier D, Murray R, Bramon E. The effect of COMT, BDNF, 5-HTT, NRG1 and DTNBP1 genes on hippocampal and lateral ventricular volume in psychosis. Psychol Med. 2009:1–15. doi: 10.1017/S0033291709990316. [DOI] [PubMed] [Google Scholar]
- Ettinger U, Picchioni M, Landau S, Matsumoto K, van Haren NE, Marshall N, Hall MH, Schulze K, Toulopoulou T, Davies N, Ribchester T, McGuire PK, Murray RM. Magnetic resonance imaging of the thalamus and adhesio interthalamica in twins with schizophrenia. Arch Gen Psychiatry. 2007;64:401–409. doi: 10.1001/archpsyc.64.4.401. [DOI] [PubMed] [Google Scholar]
- Faul F, Erdfelder E, Lang AG, Buchner A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods. 2007;39:175–191. doi: 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Gibert P, Moreteau B, Moreteau J, David JR. Genetic variability of quantitative traits in Drosophila melanogaster (fruit fly) natural populations: analysis of wild-living flies and of several laboratory generations. Heredity. 1998;80:326–335. [Google Scholar]
- Gousias IS, Rueckert D, Heckemann RA, Dyet LE, Boardman JP, Edwards AD, Hammers A. Automatic segmentation of brain MRIs of 2-year-olds into 83 regions of interest. Neuroimage. 2008;40:672–684. doi: 10.1016/j.neuroimage.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Hubbard BM, Anderson JM. Age, senile dementia and ventricular enlargement. J Neurol Neurosurg Psychiatry. 1981;44:631–635. doi: 10.1136/jnnp.44.7.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, J LW, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DL, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008a;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Lowe VJ, Senjem ML, Weigand SD, Kemp BJ, Shiung MM, Knopman DS, Boeve BF, Klunk WE, Mathis CA, Petersen RC. 11C PiB and structural MRI provide complementary information in imaging of Alzheimer's disease and amnestic mild cognitive impairment. Brain. 2008b;131:665–680. doi: 10.1093/brain/awm336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicich J, Czanner S, Han X, Salat D, van der Kouwe A, Quinn B, Pacheco J, Albert M, Killiany R, Blacker D, Maguire P, Rosas D, Makris N, Gollub R, Dale A, Dickerson BC, Fischl B. MRI-derived measurements of human subcortical, ventricular and intracranial brain volumes: Reliability effects of scan sessions, acquisition sequences, data analyses, scanner upgrade, scanner vendors and field strengths. Neuroimage. 2009;46:177–192. doi: 10.1016/j.neuroimage.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller SS, Roberts N, Hopkins W. A comparative magnetic resonance imaging study of the anatomy, variability, and asymmetry of Broca's area in the human and chimpanzee brain. J Neurosci. 2009;29:14607–14616. doi: 10.1523/JNEUROSCI.2892-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempton MJ, Ettinger U, Schmechtig A, Winter EM, Smith L, McMorris T, Wilkinson ID, Williams SC, Smith MS. Effects of acute dehydration on brain morphology in healthy humans. Hum Brain Mapp. 2009;30:291–298. doi: 10.1002/hbm.20500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempton MJ, Geddes JR, Ettinger U, Williams SC, Grasby PM. Meta-analysis, database, and meta-regression of 98 structural imaging studies in bipolar disorder. Arch Gen Psychiatry. 2008;65:1017–1032. doi: 10.1001/archpsyc.65.9.1017. [DOI] [PubMed] [Google Scholar]
- Kempton MJ, Stahl D, Williams SC, Delisi LE. Progressive lateral ventricular enlargement in schizophrenia: A meta-analysis of longitudinal MRI studies. Schizophr Res. 2010;120:54–62. doi: 10.1016/j.schres.2010.03.036. [DOI] [PubMed] [Google Scholar]
- Kuperberg GR, Broome MR, McGuire PK, David AS, Eddy M, Ozawa F, Goff D, West WC, Williams SC, van der Kouwe AJ, Salat DH, Dale AM, Fischl B. Regionally localized thinning of the cerebral cortex in schizophrenia. Arch Gen Psychiatry. 2003;60:878–888. doi: 10.1001/archpsyc.60.9.878. [DOI] [PubMed] [Google Scholar]
- Lenroot RK, Gogtay N, Greenstein DK, Wells EM, Wallace GL, Clasen LS, Blumenthal JD, Lerch J, Zijdenbos AP, Evans AC, Thompson PM, Giedd JN. Sexual dimorphism of brain developmental trajectories during childhood and adolescence. Neuroimage. 2007;36:1065–1073. doi: 10.1016/j.neuroimage.2007.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald D, Avis D, Evans AC. Multiple surface identification and matching in magnetic resonance images. Proceedings of SPIE. 1994;2359:160–169. [Google Scholar]
- Marcus DS, Fotenos AF, Csernansky JG, Morris JC, Buckner RL. Open Access Series of Imaging Studies: Longitudinal MRI Data in Nondemented and Demented Older Adults. J Cogn Neurosci. 2009 doi: 10.1162/jocn.2009.21407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus DS, Wang TH, Parker J, Csernansky JG, Morris JC, Buckner RL. Open Access Series of Imaging Studies (OASIS): cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. J Cogn Neurosci. 2007;19:1498–1507. doi: 10.1162/jocn.2007.19.9.1498. [DOI] [PubMed] [Google Scholar]
- McAlonan GM, Daly E, Kumari V, Critchley HD, van Amelsvoort T, Suckling J, Simmons A, Sigmundsson T, Greenwood K, Russell A, Schmitz N, Happe F, Howlin P, Murphy DG. Brain anatomy and sensorimotor gating in Asperger's syndrome. Brain. 2002;125:1594–1606. doi: 10.1093/brain/awf150. [DOI] [PubMed] [Google Scholar]
- McClure RK, Phillips I, Jazayerli R, Barnett A, Coppola R, Weinberger DR. Regional change in brain morphometry in schizophrenia associated with antipsychotic treatment. Psychiatry Res. 2006;148:121–132. doi: 10.1016/j.pscychresns.2006.04.008. [DOI] [PubMed] [Google Scholar]
- McDonald C, Marshall N, Sham PC, Bullmore ET, Schulze K, Chapple B, Bramon E, Filbey F, Quraishi S, Walshe M, Murray RM. Regional brain morphometry in patients with schizophrenia or bipolar disorder and their unaffected relatives. Am J Psychiatry. 2006;163:478–487. doi: 10.1176/appi.ajp.163.3.478. [DOI] [PubMed] [Google Scholar]
- McGraw KO, Wong SP. Forming inferences about some intraclass correlation coefficients. Psychological Methods. 1996;1:30–46. [Google Scholar]
- Morey RA, Petty CM, Xu Y, Hayes JP, Wagner HR, 2nd, Lewis DV, LaBar KS, Styner M, McCarthy G. A comparison of automated segmentation and manual tracing for quantifying hippocampal and amygdala volumes. Neuroimage. 2009;45:855–866. doi: 10.1016/j.neuroimage.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey RA, Selgrade ES, Wagner HR, 2nd, Huettel SA, Wang L, McCarthy G. Scan-rescan reliability of subcortical brain volumes derived from automated segmentation. Hum Brain Mapp. 2010;31:1751–1762. doi: 10.1002/hbm.20973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuechterlein KH, Green MF, Kern RS, Baade LE, Barch DM, Cohen JD, Essock S, Fenton WS, Frese FJ, 3rd, Gold JM, Goldberg T, Heaton RK, Keefe RS, Kraemer H, Mesholam-Gately R, Seidman LJ, Stover E, Weinberger DR, Young AS, Zalcman S, Marder SR. The MATRICS Consensus Cognitive Battery, part 1: test selection, reliability, and validity. Am J Psychiatry. 2008;165:203–213. doi: 10.1176/appi.ajp.2007.07010042. [DOI] [PubMed] [Google Scholar]
- Patenaude B. D.Phil Thesis. University of Oxford; 2007. Bayesian Statistical Models of Shape and Appearance for Subcortical Brain Segmentation. [Google Scholar]
- Patenaude B, Smith S, Kennedy D, Jenkinson M. FIRST - FMRIB's integrated registration and segmentation tool. Human Brain Mapping Conference. 2007 [Google Scholar]
- Patenaude B, Smith SM, Kennedy DN, Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage. 2011;56:907–922. doi: 10.1016/j.neuroimage.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorden C, Brett M. Stereotaxic display of brain lesions. Behav Neurol. 2000;12:191–200. doi: 10.1155/2000/421719. [DOI] [PubMed] [Google Scholar]
- Scahill RI, Frost C, Jenkins R, Whitwell JL, Rossor MN, Fox NC. A longitudinal study of brain volume changes in normal aging using serial registered magnetic resonance imaging. Arch Neurol. 2003;60:989–994. doi: 10.1001/archneur.60.7.989. [DOI] [PubMed] [Google Scholar]
- Schwartz M, Creasey H, Grady CL, DeLeo JM, Frederickson HA, Cutler NR, Rapoport SI. Computed tomographic analysis of brain morphometrics in 30 healthy men, aged 21 to 81 years. Ann Neurol. 1985;17:146–157. doi: 10.1002/ana.410170208. [DOI] [PubMed] [Google Scholar]
- Shuter B, Yeh IB, Graham S, Au C, Wang SC. Reproducibility of brain tissue volumes in longitudinal studies: effects of changes in signal-to-noise ratio and scanner software. Neuroimage. 2008;41:371–379. doi: 10.1016/j.neuroimage.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Simmons A, Arridge SR, Barker GJ, Williams SC. Simulation of MRI cluster plots and application to neurological segmentation. Magn Reson Imaging. 1996;14:73–92. doi: 10.1016/0730-725x(95)02040-z. [DOI] [PubMed] [Google Scholar]
- Wonderlick JS, Ziegler DA, Hosseini-Varnamkhasti P, Locascio JJ, Bakkour A, van der Kouwe A, Triantafyllou C, Corkin S, Dickerson BC. Reliability of MRIderived cortical and subcortical morphometric measures: effects of pulse sequence, voxel geometry, and parallel imaging. Neuroimage. 2009;44:1324–1333. doi: 10.1016/j.neuroimage.2008.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright IC, Rabe-Hesketh S, Woodruff PW, David AS, Murray RM, Bullmore ET. Meta-analysis of regional brain volumes in schizophrenia. Am J Psychiatry. 2000;157:16–25. doi: 10.1176/ajp.157.1.16. [DOI] [PubMed] [Google Scholar]
- Yaffee RA. Enhancement of Reliability Analysis: Application of Intraclass Correlations with SPSS/Windows v.8. 1998 [Google Scholar]
- Zakzanis KK, Graham SJ, Campbell Z. A meta-analysis of structural and functional brain imaging in dementia of the Alzheimer's type: a neuroimaging profile. Neuropsychol Rev. 2003;13:1–18. doi: 10.1023/a:1022318921994. [DOI] [PubMed] [Google Scholar]