Abstract
In animals, heterotrimeric guanine nucleotide–binding protein (G protein) signaling is initiated by G protein–coupled receptors (GPCRs), which activate G protein α subunits; however, the plant Arabidopsis thaliana lacks canonical GPCRs, and its G protein α subunit (AtGPA1) is self-activating. To investigate how AtGPA1 becomes activated, we determined its crystal structure. AtGPA1 is structurally similar to animal G protein α subunits, but our crystallographic and biophysical studies revealed that it had distinct properties. Notably, the helical domain of AtGPA1 displayed pronounced intrinsic disorder and a tendency to disengage from the Ras domain of the protein. Domain substitution experiments showed that the helical domain of AtGPA1 was necessary for self-activation and sufficient to confer self-activation to an animal G protein α subunit. These findings reveal the structural basis for a mechanism for G protein activation in Arabidopsis that is distinct from the well-established mechanism found in animals.
INTRODUCTION
Heterotrimeric guanine nucleotide–binding protein (G protein)–coupled receptors (GPCRs) convert extracellular signals to intracellular responses by activating the α subunit of the Gαβγ heterotrimer. GPCRs activate G proteins by promoting the release of guanosine diphosphate (GDP) from the G protein α subunit in favor of guanosine triphosphate (GTP). When bound to GTP, the α subunit undergoes conformational changes that result in the dissociation of the heterotrimer into a free α subunit and βγ dimer. Both can then stimulate or inhibit downstream effector enzymes. Signal propagation is halted after the α subunit hydrolyzes GTP and returns to the inactive, GDP-bound heterotrimeric state.
High-resolution, three-dimensional (3D) structures of animal G protein α subunits show that the guanine nucleotide is buried at the interface of two domains: (i) an all-α helical domain and (ii) a domain that resembles the small guanosine triphosphatase (GTPase) Ras (1). The Ras domain and domain linkers contain the residues that make contact with the magnesium ion (Mg2+), the GPCR, the regulator of G protein signaling (RGS) protein, the Gβ subunit, as well as residues that form the guanine nucleotide–binding pocket and catalyze the hydrolysis of GTP. Thus far, the only functions that have been ascribed to the helical domain are to interact with the guanine nucleotide exchange factor (GEF) p115RhoGEF and the GoLoco motif of RGS14 (2, 3), as well as to serve as a target of ubiquitination (4).
Some organisms, including Arabidopsis thaliana, lack canonical GPCRs and therefore rely on a distinct, unknown mechanism for G protein activation. Unlike animal G protein α subunits, the plant α subunit AtGPA1 has a rapid spontaneous rate of nucleotide exchange that is 50-fold faster than that of the next fastest α subunit (5). Thus, AtGPA1 does not require a GPCR or GEF to become activated, because the rate of nucleotide exchange is faster than that of GTP hydrolysis. Here, we used x-ray crystallography, molecular dynamics (MD) simulations, and biophysical and biochemical analyses to investigate the mechanism of activation of the AtGPA1. Our findings reveal a role for the α helical domain in regulating nucleotide exchange and provide a possible mechanism for GPCR-mediated nucleotide exchange.
RESULTS
AtGPA1 is activated in the absence of a GEF
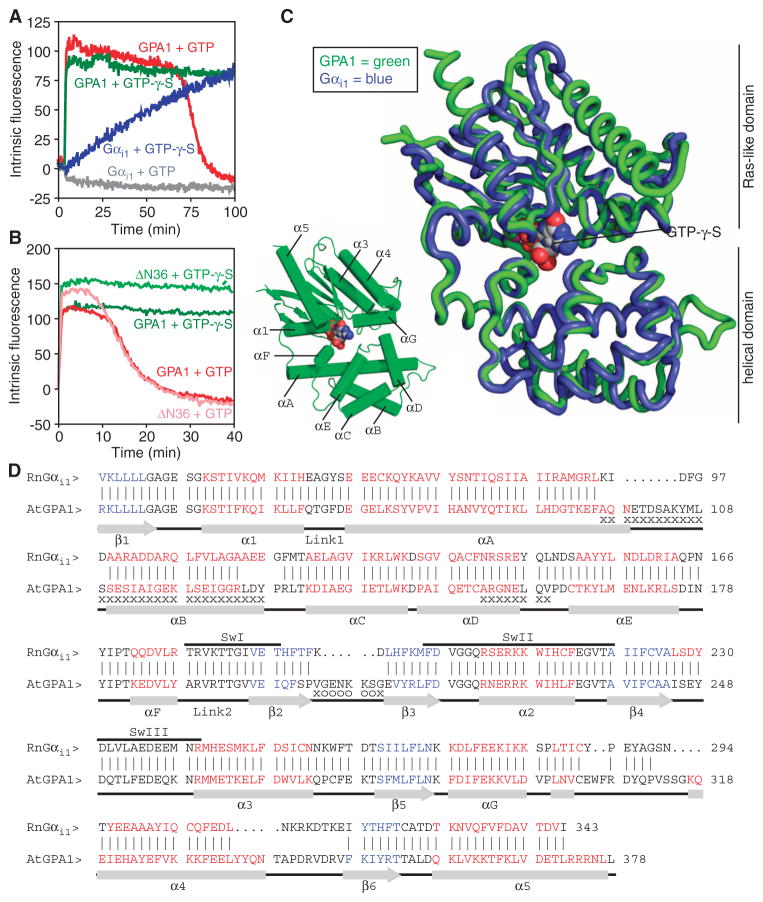
AtGPA1 has a fast rate of basal nucleotide exchange and therefore does not require a receptor for its activation. This property can be observed in a fluorescence assay in which GTP-bound α subunits have higher intrinsic fluorescence than do GDP-bound α subunits (6, 7). Here, we found that active AtGPA1 accumulated in the presence of GTP (until hydrolysis exhausted the GTP), because the rate of binding of GTP was faster than its hydrolysis (Fig. 1A). In contrast, Gαi1 remained inactive in the presence of GTP because its rate of GTP hydrolysis was faster than the rate of GTP binding (Fig. 1A). Both AtGPA1 and Gαi1 were activated by a nonhydrolyzable GTP analog [guanosine 5′-O-(3′-thiotriphosphate) (GTP-γ-S)], although Gαi1 was activated at a slower rate than AtGPA1. To shed light on this mechanism of G protein activation, we determined the crystal structure of the active form of AtGPA1, using data to a resolution of 2.3 Å (table S1). On the basis of past successes in crystallizing G protein α subunits, we deleted the N-terminal 36–amino acid residues of AtGPA1. The truncated protein (ΔN36) that was used for crystallization retained the same property of self-activation as the full-length protein (Fig. 1B).
Fig. 1.
The crystal structure of a self-activating G protein α subunit and its comparison to that of Gαi1. (A) Fluorescence-based measurement of the binding and hydrolysis of GTP. Purified AtGPA1 (GPA1) or Gαi1 (400 nM) was equilibrated at 23°C before GTP (1 μM) or nonhydrolyzable GTP-γ-S (1 μM) was added to the cuvette, and the change in intrinsic fluorescence (arbitrary units) of the G protein α subunit was monitored. Data are representative of three experiments. (B) Fluorescence-based measurement of the GTP-binding and hydrolysis properties of wild-type AtGPA1 and a truncated form of AtGPA1 that lacks the N-terminal 36–amino acid residues (ΔN36), as described for (A), but with 400 nM GTP or GTP-γ-S. (C) Superposition of the crystal structures of AtGPA1–GTP-γ-S (green) and Gαi1–GTP-γ-S (blue; PDB ID: 1GIA). (D) Structure-based sequence alignment of AtGPA1–GTP-γ-S and Rattus norvegicus Gαi1–GTP-γ-S. Vertical lines between sequences denote residues at equivalent locations in the two structures. Residues not modeled in any of the three molecules in the asymmetric unit of AtGPA1 are marked with “o.” Residues not modeled in at least two of the three molecules are marked with an “x.” Red residues constitute α helices, whereas blue residues constitute β strands. Abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
The structure of AtGPA1 is similar to that of other G protein α subunits
We found that the overall structure of AtGPA1 was highly similar to the previously reported structures of activated forms of Gαi1 and transducin. Specifically, the root mean square deviation (RMSD) between the backbones of the superimposed structures was 1.8 Å for 307 equivalent residues in Gαi1 and 1.8 Å for 315 equivalent residues in transducin (8, 9). Like other Gα proteins, AtGPA1 is organized into two domains (the helical and Ras domains) that surround the guanine nucleotide (Fig. 1, C and D). A detailed comparison of the structures of AtGPA1 and Gαi1 revealed a number of small differences between the proteins, including the existence of short, plant-specific loop inserts and variations in the lengths of secondary structure elements (Fig. 1, C and D). Moreover, when bound to AtGPA1, GTP-γ-S had 25.4 Å2 of solvent-accessible surface area, whereas only 5.0 Å2 was available when GTP-γ-S was bound to Gαi1 (fig. S1). This increase in the solvent accessibility of the AtGPA1-bound nucleotide can mostly be attributed to the absence of an interdomain interaction between Lys288 and Asp162 in AtGPA1, as is discussed later.
Guided by the crystal structure, we used site-directed mutagenesis to identify the residues responsible for the self-activation of AtGPA1. Initially, we focused on four areas of the Ras domain that affect nucleotide exchange in animal G protein α subunits: the guanine nucleotide–binding pocket (10), the switch regions (1), the domain linkers (11), and the α5 helix (12, 13). Mutagenesis of 16 sites that differ between AtGPA1 and Gαi1 revealed only modest changes (less than threefold) in the activation rate of AtGPA1 (table S2).
Crystallographic data indicate disorder in the helical domain of AtGPA1
In light of the mutagenesis data, we considered other regions of AtGPA1 that were not previously thought to be involved in nucleotide exchange. The electron density map of AtGPA1 showed substantial disorder in the helical domain. Of the three AtGPA1 monomers in the asymmetric unit, monomer A had the most complete electron density for the helical domain, which was likely due to the limitation of its motion by crystal packing contacts within the crystal lattice. Monomers B and C were missing substantial electron density, particularly in helices αA, αB, and αD (Fig. 1D, marked with x). Likewise, the average atomic displacement parameters for the helical domains of each AtGPA1 monomer were 1.7- to 2.1-fold higher than those of the respective Ras domains (table S3 and fig. S2). By contrast, the helical domains of animal G protein α subunits generally have average atomic displacement parameters that are similar to those of their Ras domains (table S3). Together, these data suggest that the helical domain of AtGPA1 is more mobile than the Ras domain, whereas in animal G protein α subunits, both domains generally have similar mobility.
MD simulations reveal dynamic motion in the helical domain of AtGPA1
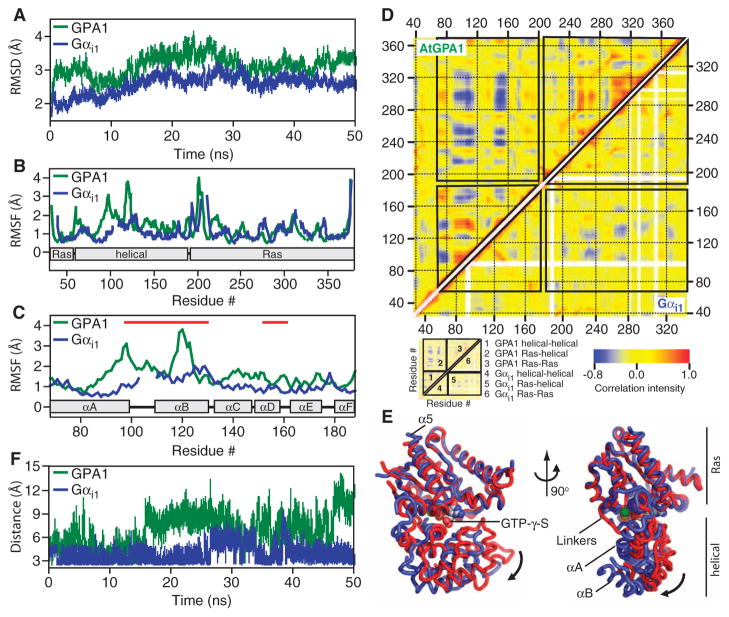
These analyses of AtGPA1 crystallographic data prompted us to assess the dynamic behavior of the helical domain of AtGPA1 with MD simulations. Our MD analysis indicated that AtGPA1 exhibited greater overall motion than did Gαi1 (Fig. 2A), with the largest fluctuations localized to its helical domain (Fig. 2, B and C, and Table 1). Within the helical domain, αA and αB exhibited the greatest fluctuations in the simulation (Fig. 2C and Table 1), consistent with the fragmented appearance of the electron density and the high average atomic displacement parameters for these two α helices in our AtGPA1 crystals (Fig. 1D, Table 1, and table S3). Moreover, covariance matrix plots showed strong anticorrelated movements between the helical domain and the Ras domain of AtGPA1 (Fig. 2D), indicating that these two domains frequently moved away from each other. We rendered one of the most frequent modes of motion from the AtGPA1 covariance matrix as a movie (movie S1), which illustrates the occurrence of a separation between the Ras domain and the helical domain in a manner that would enable nucleotide release. Within the Ras domain, only helix α5 moved relative to other secondary structural elements (movie S1 and Fig. 2E). In contrast, the helical domain displayed substantial intradomain movement, particularly with helices αA and αB (movie S1 and Fig. 2, D and E). Compared with AtGPA1, Gαi1 exhibited less anticorrelated movement between the helical domain and the Ras domain, consistent with it having a slower rate of nucleotide exchange (Fig. 2D).
Fig. 2.
All-atom MD simulations with AtGPA1 and Gαi1. All of the MD simulations were conducted with apo-Gαi1 (PDB# 1CIP, without GTP-γ-S) and apo-AtGPA1 (this report). (A) RMSDs from the starting crystal structures as a function of simulation time were calculated from the MD simulation. (B) RMSFs as a function of residue number. (C) RMSF for the G protein α-helical domains of AtGPA1 and Gαi1. Red lines mark residues that were not included in the crystallographic model for monomers B and C of AtGPA1 because of their poor electron density. (D) Covariance matrices for pairs of residues in apo-AtGPA1 and apo-Gαi1. The boxes in the main plot outline correlated movements between the Ras and helical domains as indicated in the schematic. Positively correlated movements are indicated in red, whereas negatively correlated movements are indicated in blue. (E) Structure and dynamics of AtGPA1 based on the most frequent modes of motion from movie S1. The crystal structure of AtGPA1 is depicted in red, and the ending structure for the third most frequent mode of motion is depicted in blue. (F) Distance between the side chains of Asp150 (D150) and Lys270 (K270) of Gαi1 and the comparable residues Asp162 (D162) and Lys288 (K288) of AtGPA1 during the 50-ns simulation.
Table 1.
Quantification of helical domain dynamics. The Ras domain of AtGPA1 comprises amino acid residues 37 to 63 and 197 to 372, whereas that of Gαi1 comprises residues 34 to 58 and 185 to 343. The helical domain of AtGPA1 contains residues 68 to 188, whereas that of Gαi1 contains residues 62 to 176. The αA-αB helices contain residues 68 to 125 of AtGPA1 and residues 62 to 117 of Gαi1. The RMSF calculations are from the data shown in Fig. 2B. The B values were calculated from crystallographic data collected for monomer A of AtGPA1 and Gαi1.
| Domain | RMSF (Å)
|
B value (Å2)
|
||
|---|---|---|---|---|
| GPA1 | Gαi1 | GPA1 | Gαi1 | |
| Ras | 1.34 | 1.08 | 27.9 | 20.4 |
| Helical | 1.83 | 1.10 | 48.4 | 19.0 |
| αA-αB | 2.02 | 1.13 | 52.5 | 21.4 |
Disruption of interdomain interactions affects the nucleotide exchange rates of animal G protein α subunits (14–17). Likewise, distance plots obtained through our MD simulations showed that residues that form interdomain interactions in the crystal structures of animal G protein α subunits were consistently within the distance required to form hydrogen bonds in Gαi1, but were rarely in hydrogen bond distance in AtGPA1 (Fig. 2F). Together, these analyses suggest that the helical domain of AtGPA1 is more dynamic than that of Gαi1 and that the motion in AtGPA1 is dominated by movements consistent with the presumed structural requirements of nucleotide exchange (18).
The helical domain of the G protein α subunit influences protein stability
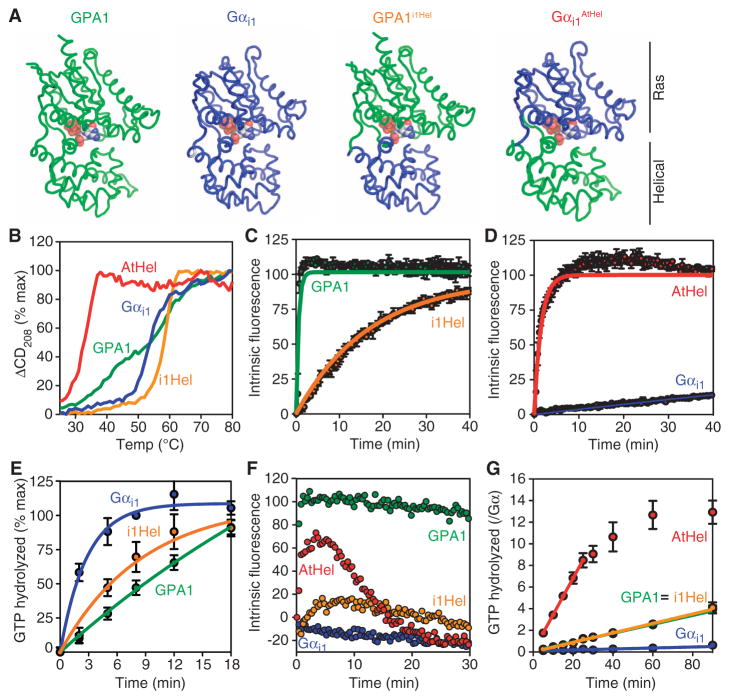
The crystal structure and MD simulations suggested that the Arabidopsis G protein α subunit was distinct from other G protein α subunits in that its helical domain was dynamic and frequently dissociated from the Ras domain. To test the hypothesis that structural features in the helical domain of AtGPA1 were responsible for self-activation, we replaced the helical domain of the plant α subunit with that of Gαi1. We refer to the resulting chimera as AtGPA1i1Hel (Fig. 3A). We also generated the reciprocal chimera by replacing the helical domain of Gαi1 with that of AtGPA1, a chimera that we refer to as Gαi1AtHel (Fig. 3A). First, we investigated protein stability with circular dichroism (CD) spectroscopy by monitoring changes in secondary structure as a function of temperature. Consistent with the comparatively weak electron density and dynamic motion in the helical domain of AtGPA1 (Fig. 2, B to F, and table S3), this domain conferred reduced stability to the chimeric α subunit (Fig. 3B). In contrast, the helical domain of Gαi1 conferred increased stability to AtGPA1i1Hel. These experiments showed that the nature of the helical domain strongly influenced the unfolding properties of the G protein α subunit.
Fig. 3.
Kinetics and unfolding properties of G protein α subunit chimeras. (A) Cartoon representations of the chimeras used in this study. (B) Temperature-induced unfolding of the indicated G protein α subunits as monitored by CD spectroscopy at 208 nm. (C and D) GTP-γ-S binding rates were measured from intrinsic fluorescence changes as described in Fig. 1 with GTP-γ-S (2 μM). (E) Single-turnover GTP hydrolysis. Purified His-tagged G protein α subunits (900 nM) were loaded with [γ-32P]GTP before the reaction was started by addition of Mg2+. Hydrolyzed 32PO4 was extracted with charcoal and quantified. (F) Fluorescence-based GTP binding and hydrolysis were measured as described in Fig. 1 with GTP (400 nM). (G) Steady-state GTP hydrolysis. Purified G protein α subunits (400 nM) were incubated with [γ-32P]GTP (10 μM) for the indicated times before hydrolyzed 32PO4 was extracted with charcoal and quantified. For all of the panels, data are representative of at least two experiments. Error bars indicate SEM.
The helical domain of AtGPA1 is necessary and sufficient for self-activation
To determine the role of the helical domain in nucleotide exchange, we measured the activation rates of the wild-type and chimeric G protein α subunits. In a fluorescence-based experiment with GTP-γ-S, we found that the nucleotide exchange rate of AtGPA1 was about 400-fold faster than that of Gαi1 (Fig. 3, C and D). In contrast, the rate of basal nucleotide exchange of AtGPA1i1Hel was 35-fold slower than that of wild-type AtGPA1 (Fig. 3C and Table 2), indicating that the helical domain of AtGPA1 was necessary for its rapid nucleotide exchange. The rate of basal nucleotide exchange of Gαi1AtHel was 168-fold faster than that of wild-type Gαi1 (Fig. 3D and Table 2) and nearly equivalent to that of AtGPA1. Because the intrinsic fluorescence-based assay is an indirect measure of nucleotide exchange, we corroborated these data by directly measuring the binding of the GTP-γ-35S radionucleotide to the G protein α subunit and the release of a nonhydrolyzable GTP analog (MANT-GMPPNP) (fig. S3). Together, these data showed that the helical domain of AtGPA1 was necessary and sufficient for rapid nucleotide exchange.
Table 2.
Summary of GTP binding and hydrolysis measurements. GTP binding was measured by fluorescence as described in Fig. 3, C and D. Single-turnover GTP hydrolysis was measured as described in Fig. 3E. For Gαi1AtHel, the rate of GTP hydrolysis was inferred from the steady-state hydrolysis experiment shown in Fig. 3G. Steady-state turnover was determined from the experiment in Fig. 3G. The rate-limiting step was determined by comparing the rates of GTP binding and hydrolysis. The percentage of bound GTP was approximated by the following equation: GTP binding rate/(GTP binding rate + GTP hydrolysis rate). Note that GTP/GDP ratio, GDIs (guanine nucleotide dissociation inhibitors), and GAPs (GTPase-activating proteins) will affect the percentage of GTP bound to a G protein α subunit in vivo. Rates were measured at 24°C and are reported as min−1.
| GPA1 | Gαi1 | GPA1iHel | Gαi1AtHel | |
|---|---|---|---|---|
| GTP-γ-S binding | 1.7 (0.02) | 0.0038 (0.0003) | 0.059 (0.001) | 0.64 (0.03) |
| GTP hydrolysis | 0.032 (0.02) | 0.32 (0.07) | 0.15 (0.03) | 0.13* (0.01) |
| Steady-state turnover | 0.041 (0.002) | 0.0045 (0.0005) | 0.044 (0.003) | 0.13 (0.01) |
| Rate-limiting step | Hydrolysis | Binding | Binding | Hydrolysis |
| % GTP bound | 98 | 1 | 28 | 83 |
Rate could not be measured in a single-turnover assay and is inferred from steady-state turnover.
AtGPA1 activates itself because the rate of nucleotide exchange (GTP binding) is faster than that of GTP hydrolysis (5). To determine whether the rate of binding of GTP to the chimeric G protein α subunits was faster than their rate of GTP hydrolysis, we measured GTP hydrolysis in a single-turnover experiment. The rate of binding of GTP to AtGPA1i1Hel (0.059 min−1) was slower than that of GTP hydrolysis (0.15 min−1) (Fig. 3E). These and other data (Table 2) suggested that replacing the helical domain of AtGPA1 with that of Gαi1 was sufficient to disrupt the self-activation property of the plant G protein. To test this hypothesis directly, we measured G protein activation by fluorescence in the presence of fully hydrolyzable GTP. As expected, activated AtGPA1 accumulated in the presence of GTP because its rate of nucleotide exchange was faster than that of GTP hydrolysis. Activated Gαi1 did not accumulate because its rate of nucleotide exchange was slower than that of GTP hydrolysis (Fig. 3F). Unlike wild-type Gαi1, activated Gαi1AtHel was detectable in the presence of GTP (Fig. 3F). Conversely, AtGPA1i1Hel formed far less activated G protein than did wild-type AtGPA1 (Fig. 3F). The fast exchange property of AtGPA1 and Gαi1AtHel was further manifested as rapid steady-state cycling between the GDP- and the GTP-bound forms (Fig. 3G and Table 2). In contrast, the cycling of Gαi1 and AtGPA1i1Hel was limited by slow nucleotide exchange (Fig. 3G and Table 2). Together, our biochemical and biophysical data indicate that the nature of the helical domain dictates protein stability and the rate-limiting step for G protein activation.
DISCUSSION
The distinct function of the helical domain of AtGPA1 suggests that the plant and animal kingdoms use distinct mechanisms of signal activation. Whereas animal G protein α subunits must be activated by an exchange factor, the Arabidopsis α subunit is self-activated as a result of the properties of its helical domain. Our data suggest that the stability of the helical domain and the interdomain interactions observed in animal G protein α subunits provide a means of regulating G protein activity that is not found in Arabidopsis. In contrast to AtGPA1, animal G protein α subunits have more stable interdomain and intradomain interactions, which likely stabilize the helical domain and limit basal nucleotide exchange.
Current models of receptor-catalyzed exchange suggest that the receptor uses the α5 helix of the G protein α subunit (and possibly the Gβγ dimer) to expel the guanine nucleotide from between the two domains (18). Our simulation data indicate that animal G protein α subunits retain some ability to exchange nucleotides by a method similar to that of AtGPA1. Whether receptors catalyze nucleotide exchange by increasing this type of motion or by a distinct mechanism remains to be determined. Perhaps receptor-induced movement of the α5 helix propagates to the helical domain. Nonetheless, our findings reveal a mechanism for receptor-independent G protein activation and may yield distinct mechanisms of G protein activation by receptors.
MATERIALS AND METHODS
Purification of Gα proteins
AtGPA1 or human Gαi1 with an N-terminal 6×His (histidine) tag cleavable with tobacco etch virus (TEV) protease was expressed from the pPROEx-Htb plasmid (Invitrogen) in Escherichia coli Codon Plus (RIPL) BL21 cells (Stratagene). Proteins were purified essentially as described previously (19). Briefly, cells were lysed by sonication in N20 buffer [25 mM tris-HCl (pH 7.4), 100 mM NaCl, 5% (v/v) glycerol, 20 mM NaF, 30 μM AlCl3, 20 mM imidazole, 50 μM GDP, 1 mM dithiothreitol (DTT), protease inhibitor cocktail (Roche), and 1 mM phenylmethylsulfonyl fluoride (PMSF)]. After clarification by ultracentrifugation, the supernatant was applied to Ni Sepharose Fast Flow resin (GE Healthcare) and gently rocked for 90 min at 4°C. The nickel resin was washed in N20 buffer before elution in N250 buffer (N20 buffer containing 230 mM imidazole). Elution fractions with G protein α subunits were pooled, concentrated, subjected to size exclusion chromatography with an S200 column (GE Healthcare), and resolved in SB [25 mM tris-HCl (pH 7.4), 100 mM NaCl, 5% (v/v) glycerol, 1 mM DTT, and 50 μM GDP]. Peak fractions were pooled and concentrated, and protein concentrations were determined by measuring absorbance at 280 nm. Complementary DNAs (cDNAs) encoding chimeric proteins were generated by polymerase chain reaction (PCR) with the domain-swapping protocol from Stratagene. For crystallization, the region encoding the N-terminal 36–amino acid residues of AtGPA1 was deleted by PCR mutagenesis. The purification of AtGPA1ΔN36 included two additional steps after the first round of nickel affinity chromatography: cleavage of the 6×His tag with TEV followed by a second round of nickel affinity chromatography. The flow-through from the second round of nickel affinity chromatography was then subjected to size exclusion chromatography as described earlier. AtGPA1-bound GDP was replaced with GTP-γ-S by buffer exchange during the protein concentration step.
Crystallization and x-ray diffraction data collection
Crystals of AtGPA1 were grown at 4°C by the hanging-drop vapor-diffusion method. Drops contained 1.5 μl of protein solution [AtGPA1 (20 mg/ml) in 25 mM tris-HCl (pH 7.4), 5% (v/v) glycerol, 150 mM NaCl, 1 mM DTT, and 500 μM GTP-γ-S] and were equilibrated against 1.5 μl of 0.3 M magnesium sulfate, 0.1 M sodium cacodylate (pH 6.0), and 21% (w/v) PEG 8000 (polyethylene glycol, molecular weight 8000). Initially obtained crystals were used for macroseeding. Crystals reached their final rod-shaped form within 14 days after macroseeding and were cryoprotected in the mother liquor with 8% (v/v) glycerol. Our AtGPA1 crystals exhibited the symmetry of space group P212121 (with cell dimensions of a = 67 Å, b = 119 Å, and c = 162 Å), contained three molecules in the asymmetric unit (solvent content, 53%), and diffracted x-rays to a maximum resolution of about 2.2 Å. Diffraction data were collected at 100 K at a wavelength of 1.000 Å with the Advanced Photon Source 23ID beamline. Data were processed with HKL2000 (20). Diffraction spots from our crystals generally had a streaky appearance at higher resolution, and the data were characterized by mild anisotropy. Data collection statistics are presented in table S1.
Structure determination and refinement
The structure of AtGPA1 was determined by molecular replacement with the program Phaser (21) using the structure of rat Gαi1 [Protein Data Bank (PDB) ID: 1GIA] as the search model. Refinement was performed with the program Refmac5 (22) from the CCP4 suite (CCP4, 1994), consisting of conjugate-gradient minimization and refinement of individual atomic displacement and TLS (translation, libration, screw rotation) parameters, interspersed with manual revisions of the model with the program Coot (23). The current model contained three AtGPA1 monomers (labeled A, B, and C, respectively) in the asymmetric unit. They were characterized by different degrees of disorder. Molecule A was almost entirely visible in the electron density map with the exception of the N-terminal residues 30 to 31, the C-terminal residues 381 to 383, and the small loop containing residues 205 to 210. Molecules B and C contained additional larger segments that could not be located in the electron density map. For B, these included amino acid residues 97 to 128 and 153 to 160, whereas for C these segments included amino acid residues 65 to 72, 91 to 133, 145 to 157, and 204 to 212. Some helical fragments of molecules B and C were accompanied by very weak, but characteristic, electron densities. These helices were identified by comparing the respective electron densities to the equivalent regions in molecule A, where they were well defined. The anisotropy in the diffraction data was reflected in the overall anisotropic scale factors B11 = 0.02, B22 = 1.38, and B33 = −1.40, as defined by Refmac5. Of all of the residues in the model, 98.5% fell into the “favored” region in the Ramachandran plot, with the remaining 1.5% falling into the “generally allowed” region, as determined by MolProbity (24). Solvent accessibilities of the GTP-γ-S ligands were determined with the AreaIMol program from the CCP4 suite. RMSDs from rat Gαi1 and bovine transducin were calculated with the DALI server (25). Cys152 and Cys163 of AtGPA1 appeared to be chemically modified, consistent with previous reports for G protein α subunits crystallized from cacodylate (26).
Measurement of GTP binding by intrinsic fluorescence
Purified G protein α subunit (400 nM) was equilibrated in a cuvette with 1 ml of TEMNG buffer [25 mM tris-HCl (pH 8), 1 mM EDTA, 5 mM MgCl2, 100 mM NaCl, and 5% (v/v) glycerol]. GTP (400 nM or 2 μM) or nonhydrolyzable GTP-γ-S (400 nM or 2 μM) was added to the cuvette, and the activation and inactivation rates of AtGPA1 were monitored by measuring the change in the intrinsic fluorescence of AtGPA1 (excitation at 284 nm, emission at 340 nm) that resulted from structural rearrangement of the Switch II tryptophan residue (7). Measurements were made with a Perkin Elmer Luminescence Spectrometer and analyzed with FL WinLab software. The rate of binding of GTP-γ-S was determined by fitting a one-phase exponential association curve with GraphPad Prism software.
Measurement of steady-state and single-turnover GTP hydrolysis
For steady-state measurements, we mixed purified Gα protein (800 nM) in HEL buffer [50 mM Hepes (pH 7.0), 1 mM EDTA, 0.1% Lubrol, and 1 mM DTT] with an equal volume of [γ-32P]GTP buffer (HEL buffer containing 10 mM MgCl2, 2 μM GTP, and [γ-32P]GTP at about 5000 to 10,000 cpm/pmol) to start the hydrolysis reaction. At a given time point, the reaction was stopped by quenching duplicate 100-μl aliquots in ice-cold HPO4 buffer (pH 2.0) containing 5% (w/v) charcoal. After quenching and charcoal extraction, which denatures proteins and removes organic compounds, the amount of 32PO4 hydrolyzed was quantified by Cherenkov radiation counting of supernatants. For single-turnover reactions, purified α subunit (900 nM) was preloaded with a mixture of GTP (3 μM) and [γ-32P] GTP for 10 to 30 min at room temperature before the reaction was moved to ice for 5 min. The hydrolysis reaction was then started by the addition of MgCl2 (10 mM) and GTP-γ-S (100 μM). Reactions were quenched, extracted, and processed as described earlier. Single-turnover GTP hydrolysis was not successfully measured for Gαi1AtHel, likely because of the instability of this protein (see Fig. 3B) and the inefficiency of GTP binding in the absence of Mg2+.
Measurement of protein stability by CD spectroscopy
Purified α subunit [0.2 mg/ml in 10 mM PO4 buffer (pH 7.5), with 10 μM GDP and 2 mM MgCl2] was melted over a temperature range of 15°C to 80°C at a ramp speed of 1°C/min, whereas CD was monitored at 208 nm. Values reported are the percentage maximum change in the CD signal for one experiment and are representative of three experiments. Measurements were collected with a Chirascan plus CD spectrometer and processed with Chirascan software. The unfolding processes for all of the proteins tested were irreversible in that they lead to protein precipitation. Thus, it was not possible to perform a detailed thermodynamic analysis of the unfolding processes.
MD simulations and analysis
MD simulations with the Amber 10.0 software suite were run on apo-AtGPA1 and apo-Gαi1 from rat (PDB ID: 1CIP) essentially as described previously (27, 28). Apoenzymes were generated by removing nucleotide ligands and modeled water molecules from the crystal structures. Modeller software (CCP4 Suite) was used to rebuild residues missing from the crystal structure of AtGPA1 (Fig. 1D, marked with “o”). Hydrogen atoms were added, the electrostatic charge of the system was neutralized with counterions, and proteins were solvated with 15,000 to 21,000 TIP3P water molecules with the XLEAP module of Amber 10.0. All MD simulations were conducted for 50 ns with a 2-fs time step with the force field described by Duan et al. (29). Analysis was performed with the PTRAJ module from Amber 10.0. All-atom, mass-weighted RMSDs were calculated for each trajectory with the crystal structure as a reference. We used an averaged structure as a reference to determine root mean square fluctuations (RMSFs). Normalized pairwise correlation coefficients between pairs of Cα atoms from two residues were computed as described previously (30). These values range from −1 to +1 and reflect anticorrelated movements (a negative value corresponds to movement in opposing directions) or correlated movements (a positive value indicates movement in the same direction). We rendered one of the most frequent modes of motion from a quasiharmonic analysis completed with PTRAJ as a movie (movie S1). This mode represents the third most frequent mode of motion for AtGPA1 and is similar to the most frequent mode of motion for Gαi1. Although the guanine nucleotide was not included in the MD simulation, it is pictured in the movie for reference.
Supplementary Material
Fig. S1. Solvent accessibility of guanine nucleotides in plant and animal G protein α subunits.
Fig. S2. B value analysis of AtGPA1.
Fig. S3. Nucleotide exchange by G protein α subunits.
Fig. S4. CD spectra of GDP-bound G protein α subunits.
Table S1. Data collection and refinement statistics for AtGPA1.
Table S2. GTP-binding rates for wild-type and mutant G protein α subunits.
Table S3. B value analysis of crystal structures of G protein α subunits.
Movie S1. Movie depicting domain motion in AtGPA1 generated from MD simulations.
Acknowledgments
Funding: This work was supported by grants from the National Institutes of General Medical Sciences (NIGMS) (R01GM065989), Department of Energy (DE-FG02-05er15671), and National Science Foundation (MCB-0723515 and MCB-0718202) to A.M.J. and NIGMS (R01GM080739) to H.G.D. J.C.J. was supported by a Ruth L. Kirschstein postdoctoral award.
We thank J. Vanhooke for valuable help throughout the crystallization process, B. Wallace and M. Miley for collecting diffraction data, A. Tripathy (University of North Carolina Macromolecular Interactions Facility) for instruction in CD spectroscopy, and J. Sondek for sharing equipment and insightful comments on the manuscript.
Footnotes
Author contributions: J.C.J. managed the project, made reagents, designed the experiments, acquired and analyzed the data, and wrote the manuscript; J.W.D. made reagents, acquired and analyzed the data, and crystallized the protein; M.M. solved the crystal structure, analyzed the data, and wrote the manuscript; B.R.S.T. designed the experiments, acquired and analyzed the data, and wrote the manuscript; H.G.D. and A.M.J. analyzed the data, managed the project, and wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Accession numbers: Structure coordinates for AtGPA1 are deposited in the Protein Data Bank (www.pdb.org) as 2XTZ.
REFERENCES AND NOTES
- 1.Sprang SR. G protein mechanisms: Insights from structural analysis. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 2.Kimple RJ, Kimple ME, Betts L, Sondek J, Siderovski DP. Structural determinants for GoLoco-induced inhibition of nucleotide release by Gα subunits. Nature. 2002;416:878–881. doi: 10.1038/416878a. [DOI] [PubMed] [Google Scholar]
- 3.Chen Z, Singer WD, Sternweis PC, Sprang SR. Structure of the p115RhoGEF rgRGS domain-Gα13/i1 chimera complex suggests convergent evolution of a GTPase activator. Nat Struct Mol Biol. 2005;12:191–197. doi: 10.1038/nsmb888. [DOI] [PubMed] [Google Scholar]
- 4.Marotti LA, Jr, Newitt R, Wang Y, Aebersold R, Dohlman HG. Direct identification of a G protein ubiquitination site by mass spectrometry. Biochemistry. 2002;41:5067–5074. doi: 10.1021/bi015940q. [DOI] [PubMed] [Google Scholar]
- 5.Johnston CA, Taylor JP, Gao Y, Kimple AJ, Grigston JC, Chen JG, Siderovski DP, Jones AM, Willard FS. GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proc Natl Acad Sci USA. 2007;104:17317–17322. doi: 10.1073/pnas.0704751104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higashijima T, Ferguson KM, Sternweis PC, Ross EM, Smigel MD, Gilman AG. The effect of activating ligands on the intrinsic fluorescence of guanine nucleotide-binding regulatory proteins. J Biol Chem. 1987;262:752–756. [PubMed] [Google Scholar]
- 7.Higashijima T, Ferguson KM, Smigel MD, Gilman AG. The effect of GTP and Mg2+ on the GTPase activity and the fluorescent properties of Go. J Biol Chem. 1987;262:757–761. [PubMed] [Google Scholar]
- 8.Coleman DE, Berghuis AM, Lee E, Linder ME, Gilman AG, Sprang SR. Structures of active conformations of Giα1 and the mechanism of GTP hydrolysis. Science. 1994;265:1405–1412. doi: 10.1126/science.8073283. [DOI] [PubMed] [Google Scholar]
- 9.Noel JP, Hamm HE, Sigler PB. The 2.2 Å crystal structure of transducin-α complexed with GTPγS. Nature. 1993;366:654–663. doi: 10.1038/366654a0. [DOI] [PubMed] [Google Scholar]
- 10.Iiri T, Herzmark P, Nakamoto JM, van Dop C, Bourne HR. Rapid GDP release from Gsα in patients with gain and loss of endocrine function. Nature. 1994;371:164–168. doi: 10.1038/371164a0. [DOI] [PubMed] [Google Scholar]
- 11.Majumdar S, Ramachandran S, Cerione RA. Perturbing the linker regions of the α-subunit of transducin: A new class of constitutively active GTP-binding proteins. J Biol Chem. 2004;279:40137–40145. doi: 10.1074/jbc.M405420200. [DOI] [PubMed] [Google Scholar]
- 12.Oldham WM, Van Eps N, Preininger AM, Hubbell WL, Hamm HE. Mechanism of the receptor-catalyzed activation of heterotrimeric G proteins. Nat Struct Mol Biol. 2006;13:772–777. doi: 10.1038/nsmb1129. [DOI] [PubMed] [Google Scholar]
- 13.Natochin M, Moussaif M, Artemyev NO. Probing the mechanism of rhodopsin-catalyzed transducin activation. J Neurochem. 2001;77:202–210. doi: 10.1046/j.1471-4159.2001.t01-1-00221.x. [DOI] [PubMed] [Google Scholar]
- 14.Codina J, Birnbaumer L. Requirement for intramolecular domain interaction in activation of G protein α subunit by aluminum fluoride and GDP but not by GTPγS. J Biol Chem. 1994;269:29339–29342. [PubMed] [Google Scholar]
- 15.Markby DW, Onrust R, Bourne HR. Separate GTP binding and GTPase activating domains of a Gα subunit. Science. 1993;262:1895–1901. doi: 10.1126/science.8266082. [DOI] [PubMed] [Google Scholar]
- 16.Remmers AE, Engel C, Liu M, Neubig RR. Interdomain interactions regulate GDP release from heterotrimeric G proteins. Biochemistry. 1999;38:13795–13800. doi: 10.1021/bi990887f. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Q, Dickson A, Doupnik CA. Gβγ-activated inwardly rectifying K+ (GIRK) channel activation kinetics via Gαi and Gαo-coupled receptors are determined by Gα-specific interdomain interactions that affect GDP release rates. J Biol Chem. 2004;279:29787–29796. doi: 10.1074/jbc.M403359200. [DOI] [PubMed] [Google Scholar]
- 18.Iiri T, Farfel Z, Bourne HR. G-protein diseases furnish a model for the turn-on switch. Nature. 1998;394:35–38. doi: 10.1038/27831. [DOI] [PubMed] [Google Scholar]
- 19.Willard FS, Siderovski DP. Purification and in vitro functional analysis of the Arabidopsis thaliana regulator of G-protein signaling-1. Methods Enzymol. 2004;389:320–338. doi: 10.1016/S0076-6879(04)89019-0. [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 21.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 23.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, III, Snoeyink J, Richardson JS, Richardson DC. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holm L, Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sondek J, Lambright DG, Noel JP, Hamm HE, Sigler PB. GTPase mechanism of G proteins from the 1.7-Å crystal structure of transducin α-GDP-AIF4−. Nature. 1994;372:276–279. doi: 10.1038/372276a0. [DOI] [PubMed] [Google Scholar]
- 27.Teotico DG, Frazier ML, Ding F, Dokholyan NV, Temple BR, Redinbo MR. Active nuclear receptors exhibit highly correlated AF-2 domain motions. PLoS Comput Biol. 2008;4:e1000111. doi: 10.1371/journal.pcbi.1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryan MM, Temple BR, Phillips SE, Bankaitis VA. Conformational dynamics of the major yeast phosphatidylinositol transfer protein Sec14p: Insight into the mechanisms of phospholipid exchange and diseases of Sec14p-like protein deficiencies. Mol Biol Cell. 2007;18:1928–1942. doi: 10.1091/mbc.E06-11-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 30.Sharma S, Ding F, Dokholyan NV. Multiscale modeling of nucleosome dynamics. Biophys J. 2007;92:1457–1470. doi: 10.1529/biophysj.106.094805. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Solvent accessibility of guanine nucleotides in plant and animal G protein α subunits.
Fig. S2. B value analysis of AtGPA1.
Fig. S3. Nucleotide exchange by G protein α subunits.
Fig. S4. CD spectra of GDP-bound G protein α subunits.
Table S1. Data collection and refinement statistics for AtGPA1.
Table S2. GTP-binding rates for wild-type and mutant G protein α subunits.
Table S3. B value analysis of crystal structures of G protein α subunits.
Movie S1. Movie depicting domain motion in AtGPA1 generated from MD simulations.