Abstract
Objective
Deleted in liver cancer 1 (DLC1) is a new candidate tumor suppressor gene, whose down-regulation or even silence will result from promoter hypermethylation in various human cancers including colorectal cancer (CRC). The aim of this study is toevaluate the diagnostic role of DLC1gene methylationin the serum DNA from CRC patients.
Methods
This study enrolled 85 CRC patients and 45 patients with benign colorectal diseases. Methylation-specific polymerase chain reaction (MSP) was used to determine the promoter methylation status of DLC1 gene in serum DNA. The combination of DLC1 methylation and conventional tumor markers was further analyzed.
Results
Hypermethylation of DLC1 was detected in 42.4% (36/85) of CRC serums, while seldom in the benign controls(8.9%, 4/45) (P<0.001). The aberrant DLC1 methylation in serum DNA was not associated with patients’ clinicopathological features and elevated CEA/CA19-9 levels. Furthermore, the combinational analysis of CEA, CA19-9 and DLC1 methylation showed a higher sensitivity and no reduced diagnostic specificity than CEA and CA19-9 combination for CRC diagnosis.
Conclusion
The serum DLC1 methylation may be a promising biomarker for the early detection of CRC, which will further increase the diagnostic efficiency in combination with CEA and CA19-9.
Key words: DLC1, Colorectal cancer, Methylation, Biomarker
INTRODUCTION
Colorectal cancer (CRC) is one of the most common cancers in the world, with approximately one million new cases occurring per year. Although endoscopic screening has been widely used, most patients can only be diagnosed at the middle or advanced stages, thus the options for curative resections are limited and survival rate remains low[1]. It is clearly imperative to develop more efficient diagnostic methods to realize the early detection of cancer. Molecular markers would provide an alternative approach and among them, DNA methylation alterations in the promoter region of tumor suppressor genes (TSGs) show great promise[2]. Many studies have shown that TSGs were frequently silenced by promoter hypermethylation in CRC as well as pre-malignant lesions, indicating that the aberrant methylation as a molecular marker system could be used for early detection of cancer[3,4]. Moreover, it has been demonstrated that methylated alteration also exists in “circulating DNA”, such as DNA from blood, stool or ascitic fluid, making it well- suited for non-invasive detection[5-7].
Deleted in liver cancer-1 (DLC1) gene, first identified as a rat p122RhoGAP gene homolog in hepatocellular carcinoma (HCC)[7], is considered to be a new candidate TSG. It is located on chromosome 8p21.3-22, a region frequently deleted in tumors[8]. Ectopic expression of DLC1 has been found to induce cell morphological changes or to suppress cell proliferation, migration, and colony formation in the cells of HCC, renal cancer, lung cancer and prostate cancer,etc[9-11]. In addition,promoter hypermethylation of DLC1 has been found in various human cancers including CRC, linked with the absence of the DLC1 gene expression[12, 13]. It has also been revealed that DLC1 methylation status in non-Hodgkin's lymphoma can be used as a diagnostic marker[14]. However, very few attempts have been made to clarify the value of DLC1 methylation in CRC diagnosis.
Therefore, this study tries to identify the methylation status of DLC1 in the serum DNA from CRC patients. The combination of DLC1 methylation and conventional tumor markers for CRC detection can efficiently increase the diagnostic rate.
Materials and Methods
Study Population
This study enrolled 85 CRC patients diagnosed at Departments of General Surgery of Jiangsu Cancer Hospital and AffiliatedZhongda Hospital, Southeast University from April, 2008 to April, 2011. None had received preoperative chemotherapy or radiation therapy. All patients were diagnosed adenocarcinoma based on pathological and/or cytological evidence. Tumor stage was determined according to the Duke’s staging system, and cellular differentiation was graded according to the Broders’ grading system. The control population consisted of 45 patients with benign colorectal diseases (benign polyp, nonmalignant adenoma, ulcerative colitis, etc.). Ethical approval was obtained from the hospital and fully informed consent from all patients prior to sample collection. Peripheral blood samples of about 5 ml were collected from each patient by using tubes containing clot activator. After standing for 2 h, the serum was isolated by centrifuging at 3,000 r/min for 10 min and stored at −80°C until use.
Determination of Carcinoembryonic Antigen (CEA) and Carbohydrate Antigen 19-9 (CA19-9)
The concentrations of CEA and CA19-9 of part of the patients came from their routine biochemical examination on the next day after admission, which was determined by an automated immunoassay system (Elecsys 2010, Roche Diagnostics, Mannheim, Germany) according to the manufacturer’s instructions. Serum levels of CEA greater than 5.0 ng/ml and CA19-9 greater than 37 U/ml were considered positive.
Methylation-specific Polymerase Chain Reaction (MSP)and Bisulphite Sequencing
Serum DNA was extracted by using QIAamp Blood Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s directions. All of the final 50μl eluted DNA was bisulphite-treated with EZ-DNA methylation Gold Kit immediately (Zymo Research, Orange, CA, USA). The bisulphite-modified DNA was resuspended in 10 μl of TE buffer. Then methylation status of DLC1 promoter was determined by MSP and the reaction system was performed in a 25 μl volume containing 5 μl of DNA template, 10×Buffer, 0.15 mmol/L dNTP, 0.1 mmol/L each primer and 0.5 U of Ex Taq Hot Start Version (Takara, Shiga, Japan). Primers used in the study have been listed in Table 1. Lymphocyte DNA, original or methylated in vitro by excessive CpG (SssI) methylase (New England Biolabs, Beverly, MA, USA), was used as unmethylation and methylation positive control. Water blank was used as a negative control. To verify the MSP results, a 292-bp fragment specific to the upstream region of the basic promoter of DLC1 was amplified, and the PCR product was sequenced.
Table 1. List of all primers used and conditions of PCR amplification.
| Primer | Sequence(5’-3’) | Product size | PCR condition |
|---|---|---|---|
| DLC1(M) | f: TTTAAAGATCGAAACGAGGGAGCG | 172 bp | 94°C 30s, 52°C 30s, |
| r: CCCAACGAAAAACCCGACTAACG | 72°C 30s, 45 cycles | ||
| DLC1 | f: TTTTTTAAAGATTGAAATGAGGGAGTG | 178 bp | 94°C 30s, 52°C 30s, |
| r: AAACCCAACAAAAAAACCCAACTAACA | 72°C 30s, 45 cycles | ||
| DNA (PCR) | f: GTTTTTAGTTAGGATATGGT | 292 bp | 94°C 30s, 52°C 30s, |
| r: CTTCTTTCTACACATCAAACA | 72°C 30s, 45 cycles |
M, methylation-specific primers; U, unmethylation-specific primers; f, forward; r, reverse
Statistical Analysis
Differences in frequency were assessed by Chi-square test or Fisher’s exact test. Statistical analyses were performed using SPSS version 12.0 for windows (SPSS Inc., Chicago, IL, USA). P<0.05 was considered statistically significant.
RESULTS
DLC1 Methylation Profile in Serum DNA of CRC Patients
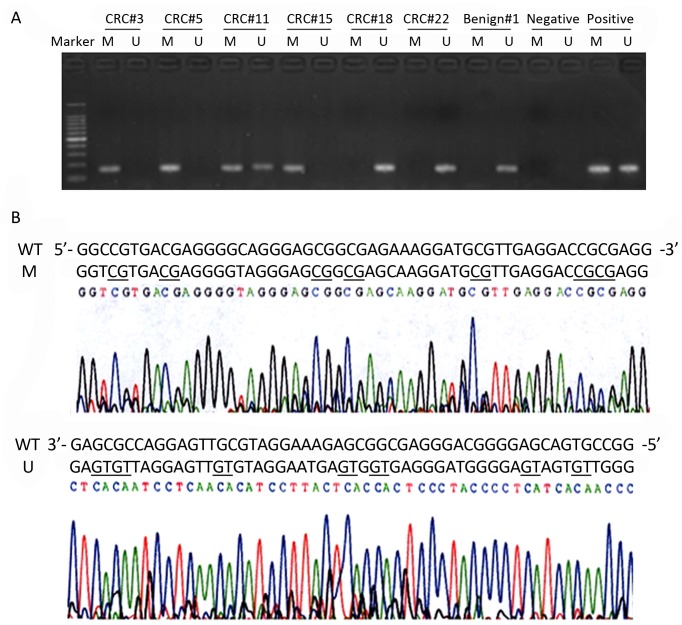
Firstly the methylation status of DLC1 in serum DNA was determined in cancer and benign control groups. The results showed that, hypermethylation of the DLC1 promoter was detected in 42.4% (36/85) of CRC, while in control group was 8.8% (4/45), and the difference was all statistically significant (P<0.001). The representative agarose gel electrophoresis results are shown in Figure 1A. Attentions should be paid to some especial results, if the methylated and unmethylated bands were both detected in a sample, it was incomplete methylation (calculated as methylation); if the methylated and unmethylated bands were both not detected in a sample (calculated as unmethylation), it might be caused by the extremely low DNA content in serum.
Figure 1.
Methylation status of DLC1 gene in serum DNA of CRC patients.A.Typical agarose gel electrophoresis of MSP results. CRC#3, #5 and #15 are examples of methylation (M), CRC#18, #22 and Benign#1 are examples of unmethylation (U), and CRC#11 is an example of incomplete methylation (analyzed as methylation). Lymphocyte DNA, original or methylated in vitro by excessive CpG (SssI) methylase was used as unmethylation and methylation positive control. Water blank was used as a negative control.B. Bisulfite sequencing of DLC1 methylation, a 292-bp fragment of the DLC-1 promoter region was sequenced. Examples of a highly methylated CpG island from the CRC#5 (M) and an unmethylatedDLC-15’CpG island from the CRC#18 (U). Underlining indicates the positions of CpGdinucleotides.
To verify the MSP results, we performed high-resolution bisulphate genomic sequencing in the stochastic samples. In agreement with the results of the MSP, six CpGdinucleotides of DLC1 gene in the sample #5 showed extensive hypermethylation, whereas sample #18 was unmethylated at these CpGdinucleotides (Figure 1B).
Clinicopathological Correlation of DLC1 Methylation in Serum DNA of CRC
The relationship between DLC1 promoter methylation status in CRC serum and patients’ clinicopathological features was further analyzed. As showed in Table 2, DLC1 methylation was not correlated with patients’ sex, age, tumor site, differentiation grade, stage, lymph metastasis and distant metastasis. There was also no correlation of DLC1 gene methylation status and serum levels of CEA and CA19-9.
Table 2. Association between the DLC1 methylation in CRC serum and clinicopathologicalfeatures (n).
| Characteristics | No. |
DLC1 methylation |
P | ||
|---|---|---|---|---|---|
| M | U | ||||
| Age (year) | <60 | 45 | 17 | 28 | 0.365 |
| ≥60 | 40 | 19 | 21 | ||
| Gender | Male | 48 | 19 | 29 | 0.556 |
| Female | 37 | 17 | 20 | ||
| Proximal | 24 | 8 | 16 | ||
| Tumor site | Distal | 25 | 13 | 12 | 0.415 |
| Rectum | 36 | 15 | 21 | ||
| Well/moderate | 51 | 19 | 32 | ||
| Differentiation | Poor | 34 | 17 | 0.244 | |
| Undifferentiation | 17 | ||||
| Stage | I/II | 39 | 14 | 25 | 0.267 |
| III/IV | 46 | 22 | 24 | ||
| Lymph Metastasis | No | 40 | 14 | 26 | 0.196 |
| Yes | 45 | 22 | 23 | ||
| Distant Metastasis | No | 66 | 26 | 40 | 0.303 |
| Yes | 19 | 10 | 9 | ||
| CEA | ≥5.0 ng/ml | 33 | 15 | 18 | 0.645 |
| <5.0 ng/ml | 52 | 21 | 31 | ||
| CA19-9 | ≥37 U/ml | 29 | 11 | 18 | 0.553 |
| <37 U/ml | 56 | 25 | 31 | ||
Combination of DLC1 Methylation and Conventional Tumor Markers
The overall detection rate of the DLC1 methylation in CRC had no difference with the positive rate of CEA (38.8%, 33/85) and CA19-9 (34.1%, 29/85). However, the combinational analysis of CEA, CA19-9 and DLC1 methylation showed a higher sensitivity than conventional combination of CEA and CA19-9 for CRC detection (70.6%, 60/85 vs. 52.9%, 45/85; P=0.018). Furthermore, the diagnostic specificity of tripartite combination was not reduced, compared with CEA and CA19-9 combination (60%, 27/45 vs. 66.7%, 30/45; P=0.512) (Table 3).
Table 3. Comparison of DLC1 gene methylation rates and positive rates of tumor markers in serum.
| Patients | No. | DLC1 methylation | CEA | CA19-9 | CEA+CA19-9 | DLC1 methylation+CEA+CA19-9 |
|---|---|---|---|---|---|---|
| CRC | 85 | 36(42.4%) | 33(38.8%) | 29(34.1%) | 45(52.9%) | 60(70.6%)* |
| Controls1 | 45 | 4(8.8%) | 10(22.2%) | 8(17.8%) | 15(33.3%) | 18(40.0%)△ |
1Controls were 40 patients with benign colorectal diseases. *P<0.05, △P>0.05 (vs. CEA+CA19-9), Chi-square test
DISCUSSION
DNA methylation is an important regulatory mechanism of epigenetic modification. Hypermethylation will lead to the down-regulation or silence of TSGs and then contribute to carcinogenesis. Given the DNA methylation usually happens before genic changes, the abnormal methylation changes at CpGisland of TSGs may be used as one of the available means for the early detection of tumor patients[15].
The new candidate tumor suppressor gene DLC1 was isolated from human HCC by a PCR-based subtractive hybridization approach[7]. Determination of the DLC1cDNA sequence shows that it is the human homologue of rat p122, which has been found to act as a Rho GTPaseactivating protein (RhoGAP)[16]. Thus, the DLC1 gene is considered to be a negative regulator of the Rho protein family of small GTPasesand to control cytoskeletal rearrangement, membrane trafficking, gene expression, cell proliferation, malignant transformation, and metastatis[17,18]. Considerable evidence has accumulated to prove that DLC1 acts as a bona fide TSG, for DLC1 is widely expressed in many adult human tissues, but it is frequently downregulatedby genomic deletions or DNA methylation in various human cancers, including HCC, breast cancer, ovarian cancer, uterine cancer, gastric cancer, lung cancer, pancreatic cancer, prostate cancer, renal cancer, nasopharyngeal cancer and colon cancer [12, 13, 19-21]. In addition, several studies show that reactivation of DLC1 function results in suppression of tumor cell proliferation and induces caspase-3-mediated apoptosis in vitro as well as abolishes or reduces tumorigenicityin vivo[7, 8]. In CRC, our previous data suggest that DLC1 gene is silenced by promoter methylation in HT29 cell line, and restoration of DLC1 expression in HT29 cells inhibits cell proliferation and migration significantly, flow cytometry also shows DLC1 transfected HT29 cells are induced apoptosis and cell cycle arrest [22]. Undoubtedly, DLC1 methylation plays an important role in CRC development, this study currently investigates the diagnostic role of DLC1 methylation in CRC.
It is well known that double-strand DNA fragments often appear in considerable quantities in the serum or plasma of cancer patients, which maybe arise from lysis of tumor cells[23]. Enriched DNA makes it possible to detect tumor-specific DNA alteration in the peripheral blood of patients. In the present study, we found a higher methylation frequency of DLC1 gene promoter in the serum DNA from CRC patients, whereas it was rare in the serum of benign disease. The sensitivity, as well as the specificity wassatisfactory compared with several other frequently methylated loci identified in plasma/serum of CRC, for example p15, p16, APC, hMLH1, MGMT, RASSF1A, RUNX3, SFRP1 and 2[24-26]. Additionally, consistent with Zhang, et al.[21], we did not find any correlation between DLC1 methylation and clinicopathological features in CRC, which suggested DLC1 methylation might be a relatively early event during tumorigenesis. Thus, DLC1 methylation may be a new ideal serum biomarker for CRC early detection.
Conventional tumor markers have been widely used in clinical diagnosis. At present, considerable evidence reveals the advantages of DNA methylation over protein-based markers in many aspects, such as excellent specificity, relative stability and high sensitivity based on PCR approaches[27]. According to our data, the overall detection rate of the DLC1 methylation in CRC patients was paralel to that of CEA and CA19-9. However, we found no correlation between DLC1 methylation and elevated levels of CEA or CA19-9[28-30], which demonstrated that DNA methylation and conventional tumor markers could serve as complementary diagnosis markers. As was expected, the tripartite combination of CEA, CA19-9 and DLC1 methylation showed a significantly higher sensitivity and no reduced diagnostic specificity than the bipartite combination of CEA and CA19-9. Thus, a combinational analysis of CEA, CA19-9 and DLC1 methylation may be an intensive screening for CRC diagnosis. Meanwhile, other biomarkes are still under investigation for gastrointestinal cancers. This combination may be further improved with the occurance of other biomarkers.
In conclusion, our study showed DLC1 gene promoter methylation has a higher detection rate in CRC serum. Its diagnostic value for CRC will be further confirmed with the increase of diagnostic efficiency when combined with CEA and CA19-9.
REFERENCES
- 1.Wu PP, Wu P, Huang PL, et al. Stanniocalcin-1 detection of peripheral blood in patients with colorectal cancer. Chin J Cancer Res 2010;22:274-79 [Google Scholar]
- 2.Watanabe Y, Maekawa M.Methylation of DNA in cancer. Adv Clin Chem 2010;52:145-67 [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Wang R, Song H, et al. Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Lett 2011;303:21-8 [DOI] [PubMed] [Google Scholar]
- 4.Kim HC, Roh SA, Ga IH, et al. CpG island methylation as an early event during adenoma progression in carcinogenesis of sporadic colorectal cancer. J Gastroenterol Hepatol 2005;20:1920-6 [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Ye X, Geng J, et al. Epigenetic inactivation of deleted in lung and esophageal cancer 1 gene by promoter methylation in gastric and colorectal adenocarcinoma. Hepatogastroenterology 2010;57:1614-9 [PubMed] [Google Scholar]
- 6.Huang ZH, Li LH, Yang F, et al. Detection of aberrant methylation in fecal DNA as a molecular screening tool for colorectal cancer and precancerous lesions. World J Gastroenterol 2007;13:950-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan BZ, Miller MJ, Keck CL, et al. Cloning, characterization, and chromosomal localization of a gene frequently deleted in human liver cancer (DLC-1) homologous to rat RhoGAP. Cancer Res 1998;58:2196-9 [PubMed] [Google Scholar]
- 8.Xue W, Krasnitz A, Lucito R, et al. DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma. Genes Dev 2008;22:1439-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim TY, Lee JW, Kim HP, et al. DLC-1, a GTPase-activating protein for Rho, is associated with cell proliferation, morphology, and migration in human hepatocellular carcinoma. Biochem Biophys Res Commun 2007;355:72-7 [DOI] [PubMed] [Google Scholar]
- 10.Zhang T, Zheng J, Jiang N, et al. Overexpression of DLC-1 induces cell apoptosis and proliferation inhibition in the renal cell carcinoma. Cancer Lett 2009;283:59-67 [DOI] [PubMed] [Google Scholar]
- 11.Guan M, Tripathi V, Zhou X, et al. Adenovirus-mediated restoration of expression of the tumor suppressor gene DLC1 inhibits the proliferation and tumorigenicity of aggressive, androgen-independent human prostate cancer cell lines: prospects for gene therapy. Cancer Gene Ther 2008;15:371-81 [DOI] [PubMed] [Google Scholar]
- 12.Ullmannova V, Popescu NC. Expression profile of the tumor suppressor genes DLC-1 and DLC-2 in solid tumors. Int J Oncol 2006;29:1127-32 [PubMed] [Google Scholar]
- 13.Liao YC, Lo SH. Deleted in liver cancer-1 (DLC-1): a tumor suppressor not just for liver. Int J Biochem Cell Biol 2008;40:843-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi H, Guo J, Duff DJ, et al. Discovery of novel epigenetic markers in non-Hodgkin's lymphoma. Carcinogenesis 2007;28:60-70 [DOI] [PubMed] [Google Scholar]
- 15.Sugimura T, Ushijima T.Genetic and epigenetic alterations in carcinogenesis. Mutat Res 2000;462:235-46 [DOI] [PubMed] [Google Scholar]
- 16.Zheng SL, Mychaleckyj JC, Hawkins GA, et al. Evaluation of DLC1 as a prostate cancer susceptibility gene: mutation screen and association study. Mutat Res 2003;528:45-53 [DOI] [PubMed] [Google Scholar]
- 17.Lahoz A, Hall A.DLC1: a significant GAP in the cancer genome. Genes Dev 2008;22:1724-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim TY, Vigil D, Der CJ, et al. Role of DLC-1, a tumor suppressor protein with RhoGAP activity, in regulation of the cytoskeleton and cell motility. Cancer Metastasis Rev 2009;28:77-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guan M, Zhou X, Soulitzis N, et al. Aberrant methylation and deacetylation of deleted in liver cancer-1 gene in prostate cancer: potential clinical applications. Clin Cancer Res 2006;12:1412-9 [DOI] [PubMed] [Google Scholar]
- 20.Peng D, Ren CP, Yi HM, et al. Genetic and epigenetic alterations of DLC-1, a candidate tumor suppressor gene, in nasopharyngeal carcinoma. Acta Biochim Biophys Sin 2006;38:349-55 [DOI] [PubMed] [Google Scholar]
- 21.Zhang Q, Ying J, Zhang K, et al. Aberrant methylation of the 8p22 tumor suppressor gene DLC1 in renal cell carcinoma. Cancer Lett 2007;249:220-6 [DOI] [PubMed] [Google Scholar]
- 22.Wu PP, Jin YL, Shang YF, et al. Restoration of DLC1 gene inhibits proliferation and migration of human colon cancer HT29 cells. Ann Clin Lab Sci 2009;39:263-9 [PubMed] [Google Scholar]
- 23.Lecomte T, Ceze N, Dorval E, et al. Circulating free tumor DNA and colorectal cancer. Gastroenterol Clin Biol 2010;34:662-81 [DOI] [PubMed] [Google Scholar]
- 24.Wang YC, Yu ZH, Liu C, et al. Detection of RASSF1A promoter hypermethylation in serum from gastric and colorectal adenocarcinoma patients. World J Gastroenterol 2008;14:3074-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishio M, Sakakura C, Nagata T, et al. RUNX3 promoter methylation in colorectal cancer: its relationship with microsatellite instability and its suitability as a novel serum tumor marker. Anticancer Res 2010;30:2673-82 [PubMed] [Google Scholar]
- 26.Nakayama G, Hibi K, Kodera Y, et al. P16 methylation in serum as a potential marker for the malignancy of colorectal carcinoma. Anticancer Res 2007;27:3367-70 [PubMed] [Google Scholar]
- 27.Koike H, Ichikawa D, Ikoma H, et al. Comparison of serum aberrant methylation and conventional tumor markers in gastric cancer patients. Hepatogastroenterology 2005;52:1293-6 [PubMed] [Google Scholar]
- 28.Nielsen HJ, Brünner N, Jorgensen LN, et al. Plasma TIMP-1 and CEA in detection of primary colorectal cancer: a prospective, population based study of 4509 high-risk individuals. Scand J Gastroenterol 2011;46:60-9 [DOI] [PubMed] [Google Scholar]
- 29.Meller B, Rave-Fränck M, Breunig C, et al. Novel Carcinoembryonic- Antigen-(CEA)-Specific Pretargeting System to Assess Tumor Cell Viability after Irradiation of Colorectal Cancer Cells. Strahlenther Onkol 2011;187:120-6 [DOI] [PubMed] [Google Scholar]
- 30.Yakabe T, Nakafusa Y, Sumi K, et al. Clinical significance of CEA and CA19-9 in postoperative follow-up of colorectal cancer. Ann Surg Oncol 2010;17:2349-56 [DOI] [PubMed] [Google Scholar]