Abstract
In this study, we characterized ceramide synthase (CerS) of the protozoan parasite Trypanosoma cruzi at the molecular and functional levels. TcCerS activity was detected initially in a cell-free system using the microsomal fraction of epimastigote forms of T. cruzi, [3H]dihydrosphingosine or [3H]sphingosine, and fatty acids or acyl-CoA derivatives as acceptor or donor substrates, respectively. TcCerS utilizes both sphingoid long-chain bases, and its activity is exclusively dependent on acyl-CoAs, with palmitoyl-CoA being preferred. In addition, Fumonisin B1, a broad and well-known acyl-CoA-dependent CerS inhibitor, blocked the parasite’s CerS activity. However, unlike observations in fungi, the CerS inhibitors Australifungin and Fumonisin B1 did not affect the proliferation of epimastigotes in culture, even after exposure to high concentrations or after extended periods of treatment. A search of the parasite genome with the conserved Lag1 motif from Lag1p, the yeast acyl-CoA-dependent CerS, identified a T. cruzi candidate gene (TcCERS1) that putatively encodes the parasite’s CerS activity. The TcCERS1 gene was able to functionally complement the lethality of a lag1Δlac1Δ double deletion yeast mutant in which the acyl-CoA-dependent CerS is not detectable. The complemented strain was capable of synthesizing normal inositol-containing sphingolipids and is 10 times more sensitive to Fumonisin B1 than the parental strain.
Index keywords: Ceramide synthase, Trypanosoma cruzi, Fumonisin B1, sphingolipid biosynthetic pathway, Chagas disease
1. Introduction
The protozoan parasite Trypanosoma cruzi is the causative agent of Chagas disease (American trypanosomiasis), which affects millions of individuals in endemic areas of Latin America [1]. The acute phase of Chagas disease often shows parasitemia prior to onset of a chronic phase that may have varying clinical features including myocarditis or pathological abnormalities of the digestive and peripheral nervous systems; alternatively, patients may remain asymptomatic [1].
In all life cycle stages of T. cruzi that involve the triatomine vector and a mammalian host [2], most of the parasite’s surface is covered by glycoconjugates attached to the plasma membrane. The attachment occurs via glycosylphosphatidylinositol (GPI) anchors, including glycoinositolphospholipids (GIPLs) and several GPI-anchored glycoproteins [3,4]. We have previously shown that T. cruzi GPI-protein anchor precursors are assembled in the endoplasmic reticulum (ER) by the sequential transfer of monosaccharides and ethanolamine-phosphate to phosphatidylinositol (PI), which is composed of alkyl-acyl-glycerol [5]. The alkyl-acyl-glycerol chain present in GPI-protein anchors [6–11] or GIPLs [12] from T. cruzi is homogeneous and always composed of sn-1-O-hexadecyl-2-O-acyl-glycerol (HDG). When replacing HDG, T. cruzi expresses GIPLs and GPI-anchors with ceramide composed of dihydrosphingosine (DHS) that is N-acylated with palmitic (C16:0) or lignoceric (C24:0) acids [8,10,13–15]. It is likely that these surface glycoconjugates participate in the complex interaction processes that compose the parasite repertoire of survival strategies [16]. The function of GIPLs in the biology of T. cruzi and their role as virulence factors have not been fully defined, although these molecules are antigenic [4]. Studies on cells of the host immune system have shown that T. cruzi GIPLs are bifunctional molecules, with the lipid and glycan elements eliciting different biological responses. While the ceramide-containing lipid moiety modulates T lymphocytes and phagocytes, the glycan chain stimulates NK cell activity and antibody production [17]. Among the surface GPI-anchored protein components of T. cruzi, two groups of mucin-like glycoproteins have been identified as the most predominant [18,19], and they are also involved in the induction/regulation of immune responses and inflammation during T. cruzi infection [20].
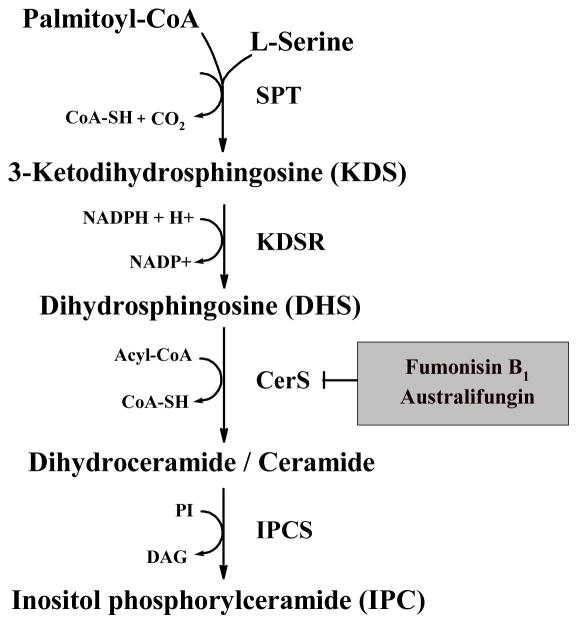
In eukaryotes, sphingolipids are synthesized de novo in the ER (Figure 1) through the initial condensation of palmitoyl-CoA and serine to form 3-ketodihydrosphingosine (KDS) and CO2, a reaction catalyzed by serine palmitoyltransferase (SPT). In yeast, SPT is a heterodimer made from Lcb1p and Lcb2p [21]. After forming KDS, Tsc10p reduces KDS to DHS, which is then amide-linked to a C26:0 fatty acid by the ceramide synthases (CerS) Lag1p and Lac1p [22,23], thereby yielding dihydroceramide (DHCer). In yeast, the CerS complex also includes the regulatory protein Lip1p [24]. The major differences in the synthesis of sphingolipids in mammals and fungi are the main types of ceramide produced de novo and the polar head group added to ceramide. In mammals, the ceramide is N-acyl sphingosine, and the head group is phosphocholine or carbohydrates [25]. In contrast, fungi transfer inositol-phosphate to the C1-hydroxyl group of DHCer or phytoceramide (C-4 hydroxylated DHCer) to form IPC, a reaction catalyzed by the unique enzyme IPC synthase [26]. Mutants of S. cerevisiae that do not synthesize sphingolipids are not viable, and pathogenic fungi are killed when treated with inhibitors of the sphingolipid biosynthetic pathway (SBP), particularly Australifungin that targets CerS [27] and the IPC synthase inhibitors Rustmicin, Khafrefungin and Aureobasidin A [27,28].
Fig. 1.
The initial steps in the biosynthesis of sphingolipids in yeast. The enzymes are: SPT, serine palmitoyl transferase; KDSR, 3-ketodihydrosphingosine (KDS) reductase; CerS, ceramide synthase; IPCS, inositolphosphrylceramide (IPC) synthase. Intermediates and co-factors include: PI, phosphatidylinositol; DAG, diacylglycerol. The inhibitors of CerS Fumonisin B1 and Australifungin are in a grey box.
Recent studies in Leishmania and T. brucei parasites have identified differences in sphingolipid metabolism among eukaryotes. Targeted deletion of the subunit 2 of SPT (SPT2) demonstrated that de novo sphingolipid synthesis is essential for differentiation but not growth in Leishmania, as the spt2− null-mutants lacked sphingolipids but grew well as promastigotes, and the parasites retained their lipid rafts [29]. Similar phenotypes were observed in mutant parasites lacking the degradation enzyme sphingosine-1-phosphate lyase [29]. Although the spt2− null-mutants did not differentiate into infective metacyclic forms, they retained the ability to enter macrophages silently and inhibit their activation. Further, in a mouse infection model, amastigotes recovered from delayed lesions were fully infective and virulent to macrophages and mice [29]. In addition, the spt2− amastigotes contained high levels of IPC, and inhibition studies using Myriocin (an SPT inhibitor) and Fumonisin B1 (a CerS inhibitor) indicated that Leishmania is able to salvage host complex sphingolipids through ‘head group’ remodeling [29]. Recent studies identified an inositolphosphosphingolipid phospholipase C-like (ISCL) that is most likely involved in the degradation of host-derived sphingomyelin, which is essential for Leishmania virulence [29]. Unlike Leishmania, pharmacological and genetic approaches demonstrated that de novo sphingolipid synthesis is essential for viability, kinetoplastid segregation, and cytokinesis in T. brucei [30,31]. While procyclic forms of T. brucei contain IPC and sphingomyelin, bloodstream forms contain sphingomyelin and ethanolamine phosphorylceramide (EPC), but not IPC [32]. These lipids are synthesized by a trypanosome sphingolipid synthase (SLS) gene family [32,33] that is orthologous to Leishmania IPC synthase [34]. In T. cruzi, metabolic incorporation studies using [3H]palmitic acid suggest that SPT, KDS reductase, CerS and IPC synthase activities are present in T. cruzi [35,36], but no further studies on SPT, KDS reductase and CerS have been performed. The single T. cruzi SBP enzyme that has been characterized is IPC synthase, which has been detected in microsomal membranes of all forms of the parasite and is not inhibited by the fungicidal and anti-fungal IPC synthase inhibitors Rustmicin or Aureobasidin A [37]. In a recent study using a cell-free synthesis approach, Sevova and colleagues [38] completed a functional characterization of all SLSs from parasitic trypanosomatid protozoa and confirmed each of their substrate specificities and previous data showing that Aureobasidin A does not significantly inhibit any of the trypanosome’s SLS activities.
The increasing number of unraveled genome sequences from pathogenic microorganisms provides new approaches for the identification of new drug targets against infectious diseases. Here, we characterized T. cruzi ceramide synthase (TcCerS) at the biochemical and molecular levels to evaluate and identify novel drug targets that specifically participate and interfere with the SBP of this parasite. Our results indicate that although insensitive to the action of the antifungal CerS inhibitors Australifungin and Fumonisin B1, the parasite contains an acyl-CoA dependent CerS, and the expression of the putative TcCERS1 gene rescued the lethality of a lag1Δ lac1Δ double deletion yeast mutant.
2. Materials and methods
2.1. Materials
Australifungin was kindly provided by Dr. Suzanne Mandala, Merck Research Laboratories (New Jersey, U.S.A), and Miltefosine (1-O-hexadecylphosphocholine) was provided by Zentaris (Frankfurt, Germany). The Protease Inhibitor Cocktail Set III was purchased from Calbiochem (San Diego, U.S.A.). The following were purchased from Sigma (St. Louis, U.S.A.): Dihydrosphingosine (DSH), Dithiothreitol (DTT), phenylmethanesulfonyl fluoride (PMSF), EDTA, EGTA, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), Sucrose, Sorbitol, Leupeptin, Soybean trypsin inhibitor, Dextrose, Galactose, Yeast nitrogen base without amino acids, L-amino acids, Casein hydrolysate, NaF, Ficoll 400, C3:0-C20:0-acyl-CoA derivatives, ATP, Platinum oxide, Palmitic acid, NADPH, CoA, Triton X-100, CHAPS, fatty acid-free BSA, 5-fluorootic acid (FOA), Zymolyase and Fumonisin B1. The C22:0-C26:0-acyl-CoA derivatives were from Avanti Polar Lipids Inc. (Alabaster, U.S.A.), and fetal calf serum (FCS) was from Gibco-Invitrogen (Carlsbad, U.S.A.). Peptone, yeast extract and brain-heart infusion (BHI) were from Acumedia (Lansing, U.S.A.). Culture plastic labware was from Techno Plastic Products AG (Trasadingen, Switzerland). All solvents and glass-backed (10 × 20 cm) Silica Gel 60 TLC plates were from Merck (Frankfurt, Germany). D-erythro-[4,5-3H]dihydrosphingosine (3H-DSH) (30 Ci·mmol−1) was purchased from American Radiolabeled Chemicals, Inc. (St. Louis, U.S.A.), and [1,2-3H]myo-inositol (74.5 Ci·mmol−1) and D-erythro-[3-3H]sphingosine (3H-SPH) (23 Ci.mmol−1) were purchased from PerkinElmer (Boston, U.S.A.). Alternatively, a total of 50 μCi (0.651 mg, w/v) of [3-3H]SPH was submitted to catalytic hydrogenation in 500 μL ethanol (0.1 mCi·mL−1) containing 1 mg of Platinum oxide (Sigma-Aldrich, U.S.A.) by sparging with H2 for 2 min followed by constant stirring under a saturated atmosphere of H2 for an additional 5 h at room temperature [39]. The conversion of [3-3H]SPH into [3-3H]DHS was monitored by autoradiography after fractionation on 10 × 20 cm glass-backed Silica Gel 60 HPTLC plates (Merck, Frankfurt, Germany) using chloroform/methanol/2 M NH4OH (40:10:1, v/v) as the solvent system. [3-3H]DHS was purified as described by Hirschberg et al. [40].
2.2. Trypanosoma cruzi, Cryptococcus neoformans, Crithidia fasciculata and Saccharomyces cerevisiae
T. cruzi (Y and Dm28c strain) parasites were obtained from the Instituto Oswaldo Cruz culture collection (IOC, Rio de Janeiro-RJ, Brazil). Epimastigotes were axenically cultured in brain-heart infusion (BHI) supplemented with 10 mg·L−1 hemin and 5% heat-inactivated FCS (BHI-FCS medium) at 28°C with shaking (~80 rev·min−1) as described previously [37]. The cultures were used for evaluating the effects of Fumonisin B1 and Australifungin on the proliferation of the epimastigotes or as a source of parasites for the preparation of microsomal membranes and total DNA (see below).
An encapsulated form of C. neoformans (strain 444) originally isolated from a patient with AIDS who developed cryptococcal meningitis was provided by Prof. L.R. Travassos, Universidade Federal de São Paulo, São Paulo-SP, Brazil. C. neoformans was grown on Sabouraud dextrose agar medium at 37°C for 24 h and maintained at 4°C. For dose-dependent growth inhibition studies, yeast cells were grown in a defined liquid medium at 37°C in a shaker (~100 rev·min−1). After 5 days, the cells were centrifuged (7,000 g, 15 min, 4°C), washed three times with cold 150 mM NaCl pH 7.4 and transferred to BHI-FCS medium containing serial dilutions of Fumonisin B1, Australifungin or methanol.
Strain TCC030 of Crithidia fasciculata was kindly provided by Dr. Lucia Mendonça-Previato (IBCCF-UFRJ, Rio de Janeiro-RJ, Brazil). Choanomastigote forms of the parasite were grown in BHI-FCS medium as described above for T. cruzi epimastigotes.
S. cerevisiae strains (Table 1) were kindly provided by Dr. Andreas Conzelmann (Department of Medicine, University of Fribourg, Switzerland). The cells were grown at 30°C in solid (2% agar, w/v) rich media [YPD, 1% yeast extract, 2% peptone, 2% dextrose (w/v) or YPG; same as YPD but containing 2% galactose as the carbon source] or synthetic minimal media (0.67 % yeast nitrogen base without amino acids, 0.002% L-adenine and 0.003% L-lysine [w/v]) containing 2% (w/v) dextrose (SD) or galactose (SGal), and L-uracil, L-histidine, L-tryptophan or L-leucine as indicated [41]. For metabolic labeling experiments, the cells were grown in a synthetic minimal medium (SC) containing salts, vitamins (but no myo-inositol), trace elements and 2% glucose as the carbon source [41]. The cells grown in liquid medium in a shaker (~150 rev·min−1) at 30°C for different time-periods were collected by centrifugation (7,000 g, 15 min, 4°C), washed with cold PBS (150 mM NaCl, 10 mM NaH2PO4·H2O and 10 mM Na2HPO4·7H2O, pH 7.4) and used for transformations/selections, DNA extraction or metabolic incorporations with [3H]myo-inositol (see below). For the functional complementation assays, the cells were plated on SD or SG solid medium containing 1 mg·mL−1 5-fluorootic acid (FOA).
Table 1.
Yeast Saccharomyces cerevisiae strains
| Strains | Genotype | Reference |
|---|---|---|
| YPK9 | MATa ade2-101ochre his3-Δ200 leu2-Δ1 lys2-801amber trp1-Δ63 ura3-52 | [55] |
| YPK9 p423TEF | Same as YPK9, but containing p423TEF(a) | This study |
| YPK9.2Δ.LAG1 | Same as YPK9, but lac1::LEU2 lag1::TRP1 and containing pBM150:LAG1(b) | [56] |
| YPK9.2Δ.LAG1 p423TEF | Same as YPK9.2Δ.LAG1, but containin p423TEF | This study |
| YPK9.2Δ.LAG1 p423TEF:TcCERS1 | Same as YPK9.2Δ.LAG1, but containing pBM150:LAG1 and p423TEF:TcCERS1(c) | This study |
| YPK9.2Δ.TcCERS1 | Same as YPK9, but lac1::LEU2 lag1::TRP1 containing p423TEF:TcCERS1 | This study |
2.3. Treatment of C. neoformans, S. cerevisiae, C. fasciculata and T. cruzi epimastigotes with Fumonisin B1, Australifungin or Miltefosine in vitro
Fumonisin B1, Australifungin and Miltefosine were kept as stock solutions (10 mg·mL−1) in methanol and were diluted in BHI-FCS medium just before use. Drug-free control medium always contained comparable final concentrations of methanol. Yeast forms of C. neoformans (1 × 10−5 cells·mL−1), choanomastigotes of C. fasciculata (5 × 10−5 cells·mL−1) or epimastigotes of T. cruzi (5 × 10−5 cells·mL−1) were incubated in BHI-FCS medium (final volume of 200–1,000μL) containing increasing concentrations of drugs or equivalent amounts of methanol only in 96 (200 μL) or 48 (1000 μL) multi-well flat bottom plates as described previously [37,42]. After 48 h at 37°C, growth of C. neoformans was estimated by absorbance at 560 nm (ELISA Plate Reader, BioRad, U.K.) after homogenization of each culture. The numbers of parasites per mL (C. fasciculata and T. cruzi) were determined by direct counting using a Neubauer chamber or a Z2™ Coulter Counter (Beckman Coulter, Inc.) after incubations (48 h or longer) at 28°C. For S. cerevisiae, yeast cells (5 × 10−4 cells·mL−1) were incubated with variable amounts of Fumonisin B1 in liquid SD medium in a shaker (~80 rev·min−1) at 30°C for 4 days, and then growth was estimated by measuring absorbance at 560 nm as detailed above. Miltefosine (1-O-hexadecylphosphocholine) was used as a positive control for growth inhibition of T. cruzi [42] and yeast [43]. The minimum inhibitory concentrations (MICs) were estimated as described previously [44], and the 50% inhibitory concentration (IC50) values were determined by linear regression analysis [42].
2.4. Metabolic incorporation of [3H]myo-inositol in S. cerevisiae in vivo and lipid extraction and analysis by TLC
The strains of S. cerevisiae were grown overnight in SC medium lacking inositol. Exponentially growing cells (10 OD600 units) were labeled with 50 μCi of [1,2-3H]myo-inositol in 1 mL of SC for 18 h at 30°C [45]. Whole cell lipid extractions, desalting and analysis were performed as described by Schorling et al. [23].
2.5. Membrane preparations
Microsomal fractions from T. cruzi epimastigotes were obtained as described by Figueiredo et al. [37]. Briefly, epimastigotes grown in BHI-FCS (2 × 1011) were collected by centrifugation (1,500 g, 10 min) and washed 2x with cold PBS. The pellet obtained from each culture preparation (approximately 8–9 g of wet weight) was washed in 10–20 mL of 25 mM Tris-HCl pH 7.4, 250 mM sucrose and 1 mM EDTA (STE buffer), centrifuged and suspended in 4–5 mL STE containing 10 mM 2-mercaptoethanol, 0.1 mM PMSF and Protease Inhibitor Cocktail Set III. Homogenates were obtained by grinding the washed parasite pellets in liquid nitrogen. After dilution of 10–20 mL of STE containing 10 mM 2-mercaptoethanol and 0.1 mM PMSF, a post-nuclear fraction devoid of unbroken cells, nuclei, and debris was obtained by centrifugation (1,500 g, 10 min). The post-nuclear fraction was then centrifuged at 5,000 g for 10 min to remove large granules (mostly composed of mitochondria), and the supernatant was further centrifuged at 27,000 g for 10 min. The supernatant was then centrifuged 100,000 g for 90 min at 4°C, and the resulting pellet was suspended in 50 mM Tris-HCl pH 7.4 containing 5 mM mercaptoethan-2-ol, 1 mM PMSF and Protease Inhibitor Cocktail Set III (20–30 mg of protein.mL−1). Aliquots of 50 μL were then prepared and kept frozen at −80°C until use.
Alternatively, we prepared whole cell extracts of Dm28c epimastigotes and metacyclic trypomastigotes obtained after differentiation under chemically defined conditions [45] to compare the acyl-CoA preferences of their CerS activities. A total of 4 × 108 cell equivalents were suspended in 2 mL of cold water containing Protease Inhibitor Cocktail Set III, incubated on ice for 15 min and submitted to 3 cycles of freeze-thawing in liquid N2. After centrifugation at 48,000 g for 30 min at 4°C, pellets were suspended in 200 μL 200 mM Tris-HCl, pH 7.5, containing 2 mM MgCl2, 2 mM DTT, 2 mM NADPH, and 0.4% (w/v) CHAPS (2x concentrated) and used for CerS activity measurements as below.
2.6. Assay for ceramide synthase
Ceramide synthase activity was detected using an in vitro cell-free system using reaction conditions described previously [40,47–49]. For the assay, 0.05–1 μCi (final) of [3H]DHS (23–30 Ci mmol−1) or [3H]SPH (23 Ci mmol−1) were initially dried into each test tube under N2 prior to the addition of one of the following buffers: 100 mM Tris-HCl, pH 7.5, containing 1 mM MgCl2, 1 mM DTT, 1 mM NADPH, 4 mM NaF and 0.2% (w/v) CHAPS or 50 mM potassium phosphate, pH 7.5, containing 1 mM MnCl2, 0.2% (w/v) CHAPS, and 2–200 μM of the acyl-CoA donor. Microsomal membrane protein (25–250 μg or 1 × 108 cell equivalents) was added in a final volume of 100 μL. In the absence of exogenously added acyl-CoA donor, assay mixtures were prepared containing 1–5 mM fatty acids alone or with 1–10 mM ATP and 1 mM CoA [39]. pH optimal conditions were tested using Tris-maleate buffer in the range of 6.0 to 9.5 [37]. In the presence of exogenously added acyl-CoA donor, 1 μM of Fumonisin B1 was dried under N2 after the [3H]DHS/[3H]SPH (see above) and preincubated with microsomal membranes for 15 min at 28°C prior to the addition of acyl-CoA derivatives. Incubations were performed at 4°C, 14°C, 28°C, 37°C or 42°C for various times, and each reaction was terminated by addition of 375 μl of chloroform/methanol (1:2, v/v) followed by lipid extraction as described by Bligh and Dyer [50]. The reaction products were extracted in saturated 1-butanol, washed with water saturated with 1-butanol as described by Heise et al. [5] and separated on glass-backed Silica Gel 60 TLC plates using chloroform/methanol/2 N NH4OH (65:25:4, v/v) as the solvent system. The standards with relative migrations (RF) of ~0.7–0.8, corresponding to C16-ceramide (C16-Cer) and C24-ceramide (C24-Cer) (Sigma, St. Louis, U.S.A.), were visualized under a saturated atmosphere of iodine. The unreacted [3H]DHS/[3H]SPH (RF ~0.3–0.4) and the [3H]DHCer/[3H]Cer (RF ~0.7–0.8) products were detected by fluorography after spraying the TLC plate with EN3HANCE (NEN™ Life Science Products, Boston, U.S.A.) using Kodak MXG-Plus (Eastman Kodak Company, Rochester, U.S.A.) or Hyperfilm™ ECL (GE Healthcare, Buckinghamshire, U.K.) film exposed at −80°C as described previously [15]. The films were photographed with a Kodak Digital Science™ DC 40 Camera. The [3H]DHCer/[3H]Cer products were quantified by densitometry of underexposed films using the image software 1D Kodak Digital Science™ or ImageJ and plotted as pixel units using the software GraphPrism 5.0.
2.7. Mild alkaline hydrolysis
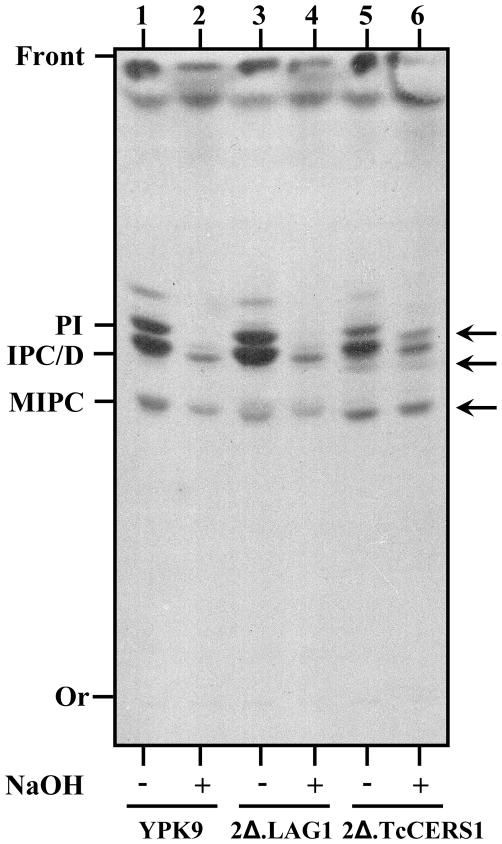
Aliquots containing equal amounts of dried total lipids obtained after [3H]myo-inositol metabolic incorporation by strains of S. cerevisiae were incubated in 20 μL of 50 mM NaOH/90% methanol or 50 mM NaCl/90% methanol for 40 min at 37°C [8]. The treatments were terminated with 80 μL of 20% acetic acid followed by extraction in butan-1-ol saturated with water. Separation was performed by TLC using chloroform/methanol/H2O (10:10:3, v/v) as the solvent system and detected by autoradiography as described above.
2.8. Cloning of TcCERS1
Dm28c total DNA was prepared from 2.5 × 108 epimastigotes by the phenol/chloroform method as described by Sambrook and Russel [51]. TcCERS1 DNA was amplified by PCR in 50 μL of 1x PCR Platinum buffer (Invitrogen, Carlsbad, U.S.A.) supplemented with 1.5 mM MgCl2, 100 ng Dm28c total DNA, 200 μM dNTPs, 1 μM of primers LAGTCS (5′-TCGGATCCATGGGGACGTTGCGTTTCAC; BamHI site underlined) and LAGTCAS (5′-TCGAATTCTTAGAAATCTTCCTCATCATCC; EcoRI site underlined), and 1.5 U of Platinum Taq DNA polymerase (Invitrogen, Carlsbad, U.S.A.). The full-length TcCERS1 coding region was amplified with an initial step of 94°C for 5 min, followed by 25 cycles of 94°C for 60 s, 55°C for 60 s, 72°C for 60 s, and a final incubation at 72°C for 10 min. The amplified product was cloned into pCR2.1-TOPO (Invitrogen, Carlsbad, U.S.A.). The product (1212 bp) was sequenced and found to match sequences in the Sanger genome database. After digestion with BamHI and EcoRI (New England BioLabs, Ipswich, MA, U.S.A.), the DNA was ligated into similarly digested p423TEF [52], thereby yielding p423TEF:TcCERS1 (Table 1).
2.9. Phylogenetic analysis
Protein sequences were aligned with CLUSTAW [53] and positions with gaps were removed. Phylogenetic analyses were carried out by the neighbour-joining methods using the program MEGA 5.0 [54] withh 10,000 bootstrap samplings.
2.10. Functional complementation of an S. cerevisiae mutant with TcCERS1
The conditional expression of TcCERS1 in yeast was achieved after transformation of different yeast strains by the lithium-acetate method as described by Ausubel et al. [41] using p423TEF:TcCERS1 (see above). The strains used are listed in Table 1 and include the wild-type YPK9 [55] and the double mutant YPK9.2Δ.LAG1 [56]. Strain YPK9.2Δ.LAG1 contains the S. cerevisiae LAG1 gene cloned into the centromeric vector pBM150 (with the URA3 marker) under the control of the GAL1-10 promoter [57]. Vector p423TEF contains the HIS3 gene marker and the strong TEF promoter [52]. During the functional complementation test, yeast cells were transformed with p423TEF or p423TEF:TcCERS1 followed by plating onto SD or SG agar containing 1 mg·mL−1 FOA and 50 μg·mL−1 of uracil as described by Guillas et al. [22]. The presence of the desired genes in each of the selected transformants was confirmed by PCR. The amplification reaction was performed in 50 μL of 1x PCR Platinum buffer with 1.5 mM MgCl2, 200 μM dNTPs, 1 μM of primers and 100 ng of DNA extracted from YPK9, 2Δ.LAG1, 2Δ.LAG1 p423TEF, 2Δ.LAG1 p423TEF:TcCERS1 or 2ΔTcCERS1 before and after selection in FOA [22]. The following pairs of primers were used: ScLAG1F (5′-TGTTGTAATTCGACCATTCA) and ScLAG1R (5′-TCTTGAACAACCACAAATCA) for amplification of a 155 bp fragment of the S. cerevisiae LAG1 gene [45] or Internal_TcCerSF (5′-GACTCAGCTGTGGCTTGCCGTG) and Internal_TcCerSR (5′-GCATGATGTCACTGGCGTCG) for amplification of a 357 bp fragment of the coding region of TcCERS1. PCR reactions began with a hot-start and addition of 1.5 U of Platinum Taq DNA polymerase. Then, reaction conditions were as follows: an initial step of 94°C for 5 min; 30 cycles of 94°C for 60 s; 55°C (for primers ScLAG1F and ScLAG1R) or 61°C (for primers Internal_TcCerSF and Internal_TcCerSR) for 60 s; 72°C for 60 s; and a final incubation at 72°C for 10 min. Amplified products were visualized after separation on 1.5% agarose gels as described above.
3. Results
3.1. Effects of Australifungin and Fumonisin B1 on the proliferation of T. cruzi epimastigote forms
The SBP is essential in eukaryotic organisms and is a target for chemotherapy against several human and animal diseases caused by fungi [26] and protozoan parasites [34,58,59]. Natural product inhibitors directed against the initial steps of SBP have been described, and among those are two mycotoxins that are well-known acyl-CoA-dependent CerS activity inhibitors (Figure 1), Fumonisin B1 [48] and Australifungin [27]. In a previous study, Osborne et al. [60] showed that Fumonisin B1 (50 μM or 36 μg.mL−1) was unable to reduce the motility of trypomastigote forms of T. cruzi in vitro, and a 48 h pre-treatment did not affected their capacity to infect mammalian host cells or the replication of intracellular amastigotes. However, there was 50% inhibition of epimastigotes growth after 24 days of treatment with 5–10 μM (3.6–7.2 μg·mL−1) of Fumonisin B1. The growth rate decreased beginning on day 9 after the start of treatment when compared to untreated controls [60].
To confirm the previous data obtained by Osborne [60] and to validate T. cruzi CerS as a potential target for the action of these SBP inhibitors, we compared the effects of increasing amounts of Fumonisin B1 and of Australifungin on the in vitro proliferation of epimastigotes of T. cruzi. As controls, we used yeast forms of Saccharomyces cerevisiae and Cryptococcus neoformans (non-pathogenic and pathogenic fungi, respectively) and choanomastigote forms of Crithidia fasciculata, a monogenetic non-human infective trypanosomatid that parasitizes several species of mosquito. Although toxic to fungi as expected [27,61], both compounds were unable to inhibit the proliferation of epimastigotes of T. cruzi at concentrations far above the IC50 described for fungal cells (Supplementary Figure 1, A–B). Epimastigotes had MICs for Australifungin and Fumonisin B1 above 50 and 500 μg·mL−1, respectively (Supplementary Figure 1, A–B). This lack of growth inhibition of epimastigotes by Fumonisin B1 is in disagreement with data reported previously by Osborne et al. [60]. Miltefosine was used as a positive control for growth inhibition under our conditions. As expected, it proved effective against T. cruzi and fungi (Supplementary Figure 1C), with the same IC50 described previously for each of these microorganisms [42,43]. Australifungin was also unable to inhibit growth of choanomastigotes of Crithidia fasciculata (Supplementary Figure 1A), a trypanosomatid known to synthesize phytosphingosine as a major branched long-chain sphingolipid base linked to their GIPLs [62], the same base that fungal cells attach to their major structural phosphorylated sphingolipids [63,64].
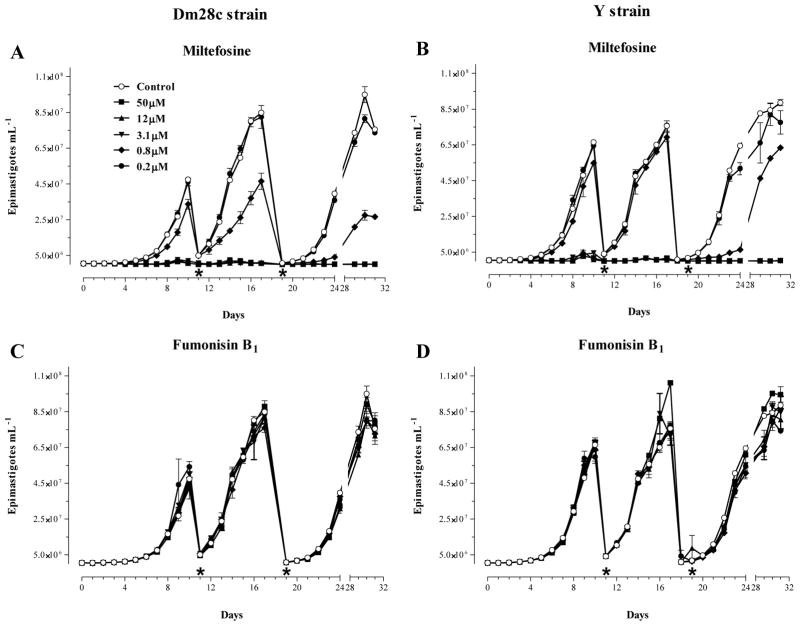
Because the 50% growth-inhibition described by Osborne et al. [60] was only achieved after a long period of treatment (24 days with 5–10 μM of Fumonisin B1), another set of experiments was performed comparing the effects of a longer treatment with increasing amounts (0.2–50 μM) of Fumonisin B1 on the proliferation of epimastigotes of Dm28c and Y strain. As shown in Figure 2, after 9 days of culture, parasites were collected by centrifugation and suspended at the same original cell density using fresh medium alone (Control) or medium containing the same original concentration of either Miltefosine (Fig. 2, A–B) or Fumonisin B1 (Fig. 2, C–D). This procedure was repeated once, and the proliferation of the parasites in each culture was monitored until day 32. As expected, Miltefosine reduced the proliferation rate of parasites in a dose-dependent manner and with a continuous drop in the concentration needed to block growth after sub-culturing (Fig. 2, A–B). However, this effect was not observed with Fumonisin B1, despite the fact that a high concentration (50 μM) was continuously used after each sub-culture (Fig. 2, C–D).
Fig. 2.
Effects of Miltefosine and Fumonisin B1 on the proliferation of epimastigote forms of T. cruzi in vitro. Cultures of epimastigotes (5 × 10−5.mL−1) of Dm28c (A and C) and Y (B and D) strains were prepared at day 0 in the absence (Control) or presence of increasing amounts of Miltefosine (A, B) and Fumonisin B1 (C, D), and the number of cells was determined daily by direct counting. At days 10 (*) and 19 (*), parasites were collected by centrifugation and adjusted to their starting densities (5 × 10−5·mL−1) using fresh media containing the same original concentrations of Miltefosine or Fumonisin B1. Each subculture was followed daily by counting for a total of 32 days as indicated at the bottom of each graph. The results shown are the means ± standard error of two sets of independent experiments.
3.2. Identification and preliminary characterization of the CerS activity in microsomes of T. cruzi epimastigotes
In eukaryotes, ceramide is synthesized mainly from fatty acyl-CoA and a long-chain sphingoid base by a CoA-dependent CerS [49]. However, the synthesis of ceramide via the condensation of sphingosine and a free fatty acid, which is likely associated with a reverse ceramidase action, has been observed [65,66]. Because the latter CerS activity is independent of CoA and is not inhibited by Fumonisin B1, this pathway could be operational in T. cruzi and thus support parasite growth resistance to the classical CerS inhibitors Australifungin (Supplementary Figure 1A) and Fumonisin B1 ((Supplementary Figure 1B and Fig. 2, C–D). Therefore, we decided to analyze which of the above CerS pathways were active in T. cruzi.
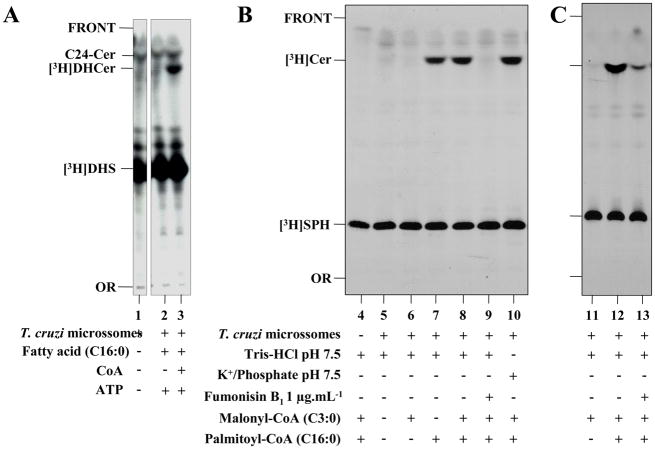
The identification and partial biochemical characterization of the T. cruzi CerS was performed using a cell-free system composed of microsomal membranes (prepared from epimastigote forms), a radioactive long-chain base, a fatty acid or acyl-CoA donor, and a variety of previously optimized conditions used to measure CerS activities in other systems [40,47–49]. As described in section 2.6, reaction conditions for the measurement of T. cruzi CerS were initially compared by incubating microsomal membranes in the presence of 3H-DHS, a Tris-HCl based buffer containing Mg+2, DTT, NaF and the Zwittergent CHAPS, and either palmitic acid (C16:0) and ATP or C16:0, ATP and CoA. The latter condition is an in vitro acyl-CoA generation system that is known to be effective in parasite microsomal membranes [39]. As observed in Figure 3A, epimastigote microsomes cannot promote generation of any radiolabeled ceramide species when incubated in the presence of [3H]DHS alone or [3H]DHS, C16:0 and ATP (Fig. 3A, lanes 1 and 2). However, if C16:0, CoA and ATP were included in the reaction mix, a radiolabeled ceramide product ([3H]DHCer) was formed (Fig. 3A, lane 3). This [3H]DHCer product migrated close to a palmitoyl-sphingosine (C16-Cer) standard but was far slower than the lignoceroyl-sphingosine (C24-Cer) standard (Fig. 3A, lane 3). Thus, in our reaction conditions, no reverse ceramidase activity was detected in T. cruzi microsomal fractions (Fig. 3A, compare lanes 2 and 3).
Fig. 3.
The effects of different substrates and Fumonisin B1 on T. cruzi CerS activity in vitro. Microsomal membranes from epimastigote forms (50 μg.mL−1) of Y (A and B) and Dm28c (C) strain were assayed in the presence of 0.5 μCi [3H]DHS (A) or [3H]SPH (B, C) at 28 °C. (A) Assays were performed in 100 μL of 100 mM Tris-HCl pH 7.5 (lanes 1–3) in the absence (−) or presence (+) of 150 μM palmitic acid (C16:0), 1 mM CoA and 1 mM ATP, as indicated at the bottom of the figure. (B, C) Assays were performed in 100 μL of 100 mM Tris-HCl pH 7.5 containing 1 mM MgCl2, 1 mM DTT, 1 mM NADPH, 4 mM NaF and 0.2% (w/v) CHAPS (lanes 4–9, and 11–13) or 50 mM Potassium Phosphate pH 7.5 containing 1 mM MnCl2 and 0.2% (w/v) CHAPS (lane 10), in the absence (−) or presence (+) of microsomes (50 μg·mL−1), 1 μg·mL−1 Fumonisin B1, and 75 μM of palmitoyl-CoA and/or malonyl-CoA, as indicated. After 4 h incubation, the lipids were extracted, separated by TLC using chloroform/methanol/2 N NH4OH (65:25:4, v/v) and visualized after autoradiography as described in item 2.6. The relative positions of synthesized ceramides ([3H]DHCer or [3H]Cer) labeled with [3H]DHS or [3H]SPH, respectively, are indicated on the left of each panel, together with the non-reactive excess of each substrate, a standard of lignoceroyl-sphingosine (C24-Cer), the origin (OR) and front (FRONT) of the chromatograms.
Next, [3H]SPH was used as the radioactive long-chain sphingoid base acceptor instead of [3H]DHS. [3H]SPH was incubated in the absence or presence of microsomal membranes, fatty acyl-CoA donors, and Fumonisin B1. As observed in Figure 3B and 3C, [3H]Cer products could be observed if membranes were incubated in the presence of [3H]SPH and palmitoyl-CoA but not in samples without [3H]SPH and palmitoyl-CoA or with the shorter malonyl-CoA as the acyl-CoA donor (Fig. 3B, lanes 4–8). Most importantly, [3H]Cer synthesis was blocked if membranes were pre-incubated with Fumonisin B1 (Fig. 3B, compare lanes 8 and 9 and 3C, compare lanes 12 and 13). As observed previously with [3H]DHS (Fig. 3A), the newly synthesized [3H]Cer products were observed as a single band on TLC, which migrated very closely to a C16-Cer standard. Finally, the change of a Tris-Mg+2-DTT-NaF-NADPH-CHAPS buffer to a Phosphate/K+-Mn+2-CHAPS buffer slightly improved the efficiency of [3H]Cer formation (Fig. 3B, compare lanes 8 and 10). However, no activity was detected if [3H]DHS was previously complexed with fatty acid free BSA [45] or the assay was performed without detergent (Supplementary Figure 2, A and B). TcCerS activity was also effective in the absence of Mg+2, Mn+2, DTT, NADPH, ATP and NaF; furthermore, the activity required CHAPS (Supplementary Figure 2, A and B) and had an optimal pH between 7.0 and 7.5 (Supplementary Figure 3B). Thus, TcCerS is similar to the mouse brain and yeast CerS in that it does not require a metal cofactor, ATP, NADPH or a thiol-protecting agent [45,47].
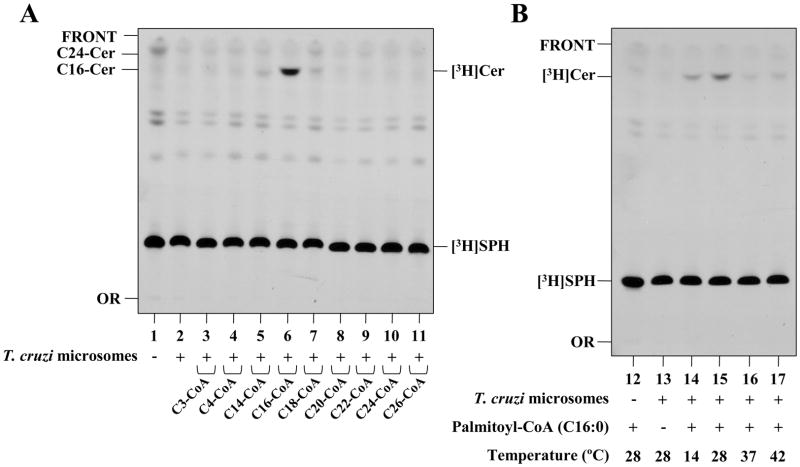
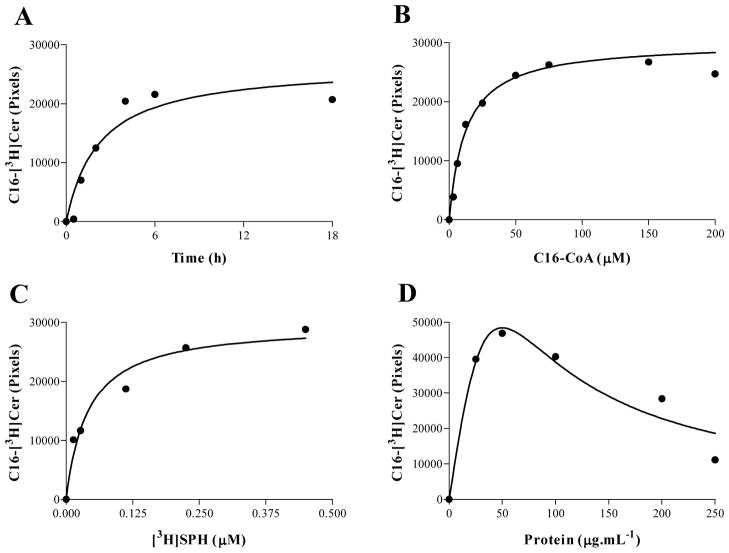
To better characterize TcCerS, experiments were performed to test its preference for different chain-length fatty acyl-CoA donors and temperatures. As shown in Figure 4 and Supplementary Figure 3A, TcCerS from Y and Dm28c strain epimastigotes has a preference for palmitoyl-CoA, with low activity towards myristoyl-CoA and stearoyl-CoA, and almost no CerS activity associated with all other acyl-CoAs tested, which ranged from C3, C4 and to C20 up to C26 (Fig. 4A and Supplementary Figure 3A). In case of strain Dm28c, we tested the acyl-CoA preference of the CerS activity in whole cell membranes obtained from metacylcic and epimastigote forms, but activity was only detected with epimastigotes when using palmitoyl-CoA as donor (Supplementay Figure 4). This preference for palmitoyl-CoA distinguishes TcCerS from the yeast enzyme, which uses very-long-chain acyl-CoAs [67]. As observed in Fig. 4B, TcCerS from Y strain has an optimal activity that increases between 14°C and 28°C, but decreases at 37°C and 42°C. Based on these observations, several kinetic parameters were used to set up optimal assay conditions (Figure 5). The TcCerS reaction was linear for times over 4 h (Fig. 5A), and [3H]Cer formation was dependent on the presence of palmitoyl-CoA (Fig. 5B) and [3H]SPH (Fig. 5C). As observed in Fig. 5D, when the amount of microsomal membranes was varied in our assay and the assay contained 50 μg·mL−1 of protein, a maximal amount of [3H]Cer was generated. At higher protein concentrations, the amount of [3H]Cer product formed decreased. Similar findings were also observed for the microsomal CerS of the mouse brain [47] and yeast [45]. Under these optimal conditions for TcCerS, we observed an apparent Km of 12.4 ± 2.9 μM (mean ± standard error, n = 2, r2 = 0.9588) with palmitoyl-CoA.
Fig. 4.
Acyl-CoA donor specificity and temperature preference of Y-strain epimastigotes of T. cruzi CerS in vitro. (A) [3H]SPH was incubated in 100 μL of 100 mM Tris-HCl pH 7.5 containing 1 mM MgCl2, 1 mM DTT, 4 mM NaF and 0.2% (w/v) CHAPS at 28 °C (lanes 1–11) in the absence (−) or presence of epimastigote microsomes (50 μg·mL−1) and in the absence (−) or presence (+) of various acyl-CoAs of different lengths (100 μM). (B) Lanes 12–17 are the same as lanes 1–11, but only palmitoyl-CoA was used as the acyl-CoA substrate donor; 200 μg·mL−1 of microsomes were present, and the temperature was varied between 14 °C, 28 °C and 37 °C. After 4 h incubation, lipids were extracted and analyzed by TLC as in Figure 2. The relative positions of palmitoyl-sphingosine (C16-Cer) and lignoceroyl-sphingosine (C24-Cer) standards are indicated on the left of Panel A.
Fig. 5.
Characterization of the T. cruzi microsomal CerS assay. (A) Assays were performed in 100 μL of 100 mM Tris-HCl pH 7.5 containing 1 mM MgCl2, 1 mM DTT, 4 mM NaF, 0.2% (w/v) CHAPS, 0.5 μCi [3H]SPH and 75 μM pamitoyl-CoA at 28 °C for varying periods of time. (B) Measurements were performed as in (A), but all incubations were performed for 4 h; the pamitoyl-CoA concentrations were varied as indicated. (C) Assays were performed as in (B) but with 75 μM pamitoyl-CoA and varied amounts of [3H]SPH. (D) The experiments were performed as in (A) but with incubation for 4 h and varied amounts of microsomes. Lipid extractions and quantification of [3H]Cer were performed as described in item 2.6.
Together, the results demonstrated that T. cruzi microsomes harbor a Fumonisin B1-sensitive acyl-CoA-dependent CerS activity that uses both DHS and SPH as long-chain sphingoid base acceptors and palmitoyl-CoA as the principal fatty-acyl-CoA substrate donor.
3.3. Functional characterization of the T. cruzi TcCERS1 gene
Based on the biochemical evidence of an exclusive acyl-CoA dependent CerS activity in T. cruzi, we next searched for putative CERS gene(s) in the genome of the parasite [68]. Using BLAST analysis [69] with the amino acid sequences of Lag1p and Lac1p of S. cerevisiae [56] and the 52 amino acids of the Lag1 motif [56,70], we identified an open reading frame in the CL-Brener genome (named TcCERS1) of the Wellcome Trust Sanger Institute Pathogen Sequencing Unit (www.genedb.org/). The predicted TcCERS1 is represented in the GeneDB database by two genomic fragments (Tc00.1047053510087.30 and Tc00.1047053507395.10) whose nucleotide and amino acid sequences are 98% identical. These probably represent the two haplotypes present in the hybrid CL-Brener strain [68]. So far, no homologs of the yeast YPC1 [65] or YDC1 [66] that encode ceramidases with CoA-independent and Fumonisin B1-insensitive CerS reverse activities have been found in GeneDB (N. Heise, personal communication).
Once we identified a putative TcCERS1 gene, we next used specific oligonucleotides to amplify the entire open reading frame from the Dm28c strain genomic DNA by PCR. The single amplified band was cloned, and the nucleotide sequence of 1,212 base pairs (bp) (GenBank Accession Number HQ845264) was 96% and 98% identical to Tc00.1047053510087.30 and Tc00.1047053507395.10, respectively (not shown). The predicted amino acid sequence (403 aa) of TcCerS1p from Dm28c has a theoretical molecular mass of 45,185 Da (pI = 8.91) and is only 22% identical to the Lag1p of yeast. Identity was highest at positions R-225, D-227, L-241, S-245, G-254, K-272 and Y-276, which are fully conserved in the Lag1 motif consensus, and positions H-234, H-235, D-262, and D-265, which are thought to be critical for catalytic activity [71,72]. TopPred analysis [73] of the TcCerS1p protein from the Dm28c strain indicated a predicted hydrophobicity plot that is compatible to the plots of Lag1p and Lac1p from yeast [56], with 6 predicted transmembrane helices (see Graphical Abstract). Similar to the other CerS gene candidates from lower eukaryotes, TcCERS1 does not contain a DNA-interacting Hox-domain [74]. Except for Trypanosoma brucei, which contains two paralogs of the yeast Lag1p (Q57V92_9TRYP and Q583F9_9TRYP), only a single ortholog has been found in other lower eukaryotes. This group includes the social amoeba Dictyostelium discoideum (Q54S87_DICDI), the enteropathogenic amoeba Entamoeba histolytica (C4LXF4_ENTHI), the Apicomplexa Plasmodium falciparum (C0H4D2_PLAF7), P. vivax (A5KB10_PLAVI), P. knowlesi (B3L6I7_PLAKH), P. chabaudi (Q4Y1U1_PLACH), Toxoplasma gondii (B9PSW9_TOXGO), Cryptosporidium muris (B6AA96_9CRYT), other trypanosomatids with sequenced genomes [Leishmania major (Q4Q684_LEIMA), L. brasiliensis (A4HJG0_LEIBR), and L. infantum (A4I6V5_LEIIN)]. Among the trypanoisomatids, the Tb927.4.4740 (Q583F9_9TRYP) is syntenic with all of the single CerS sequences. A phylogenetic analysis showed that the trypanosomatid proteins group together, and are more related to fungal CerS than other sequences that do not contain the Hox-domain (Supplementary Figure 5).
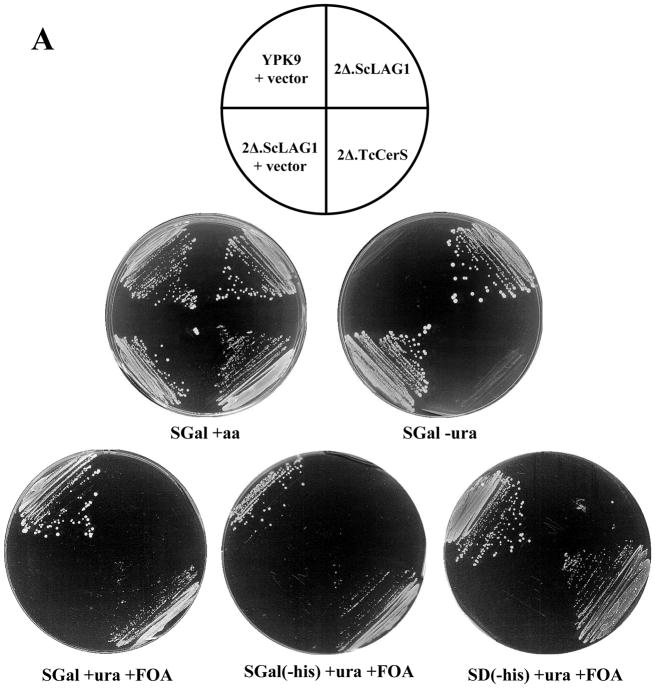
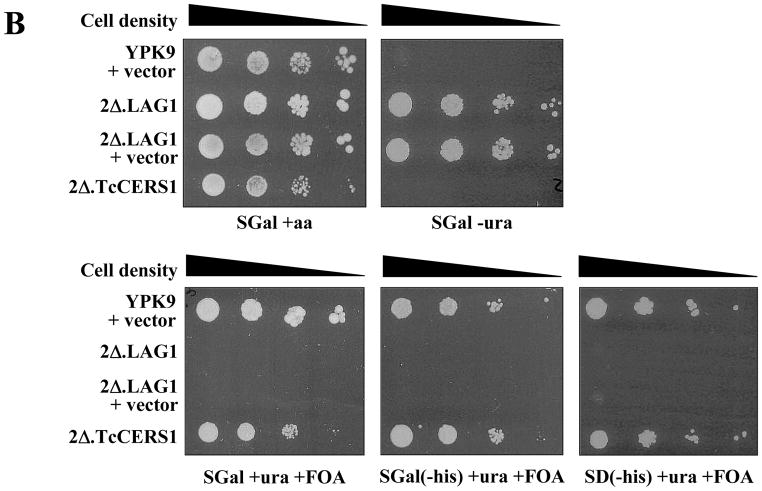
To determine if the putative Dm28c TcCERS1 gene encodes a ‘bonafide’ CerS, we used the parasite sequence to functionally complement the lethality of a lag1Δ lac1Δ double deletion mutant [22,23], despite the low overall sequence identity to the yeast Lag1p and Lac1p. This approach has been used to functionally characterize the Caenorhabditis elegans and human CerS gene sequences as encoding for genuine CerSs [45]. The TcCERS1 gene from the Dm28c strain was cloned into the 2μ p423TEF expression vector under the control of the TEF promoter and with a HIS3 marker for auxothrophy [52] as described in section 2.8, followed by transfection into the YPK9-based 2Δ.LAG1 strain (Table 1). This null mutant strain for genomic copies of LAG1 and LAC1 contains a centromere plasmid pBM150 with an extrachromosomal copy of LAG1 under the expression control of the GAL1-10 promoter [22,56]. This vector contains a CEN4-ARS1 replication origin for stable maintenance at low copy numbers and URA3 as the auxotrophic marker [57]. Controls included the transfection of the empty p423TEF vector into the YPK9 and 2Δ.LAG1 strains. After transfection, cells were selected on minimal synthetic medium containing galactose (SGal) in the absence of uracil or histidine and uracil. Selected colonies were then plated in the presence of uracil and FOA and in the absence or presence of histidine. As observed in Figure 6A and 6B, all strains grew normally without any nutritional restriction (SGal + aa). In the absence of uracil (SGal - ura), only strains that harbor the pBM150 plasmid are able to grow as expected (2Δ.LAG1 and 2Δ.LAG1 + vector). However, these strains are unable to grow on FOA because FOA kills cells harboring an intact URA3 gene and eliminates all cells that cannot survive without the LAG1-containing pBM150 plasmid that carries URA3 [45]. However, YPK9 cells transfected with p423TEF alone and 2Δ.LAG1 cells transfected with p423TEF-TcCerS were able to grow on SGal plates containing uracil and FOA but no histidine (selection marker for p423TEF). The results suggested that the rescue of the lethal phenotype of the lag1Δlac1Δ double deletion mutant was only possible after substitution of the centromeric LAG1 gene copy by p423TEF, which contains the TcCERS1 gene (2Δ.TcCERS1). The growth of the rescued cells was similar to the wild-type YPK9 parental cell line (Fig. 6B).
Fig. 6.
Rescue of YPK9 lag1Δlac1Δ by overexpression of TcCERS1. (A) YPK9 (wild-type) cells containing the empty histidine-based plasmid p423TEF vector (YPK + vector), YPK9 lag1Δlac1Δ transformed with pBM150:LAG1 (LAG1 in uracil-based plasmid) (2Δ.LAG1), 2Δ.LAG1 containing the empty p423TEF vector (2Δ.LAG1 + vector), and 2Δ.LAG1 containing the p423TEF:TcCERS1 vector (2Δ.TcCERS1) were plated on minimal media containing galactose (SGal) or glucose (SD), amino acids (aa) and FOA (1 mg·mL−1) as indicated beneath each plate. The cells were streaked heavily, and the plates were photographed after 5 days incubation at 30°C. (B) The experiment was performed as in (A), but the transformants were diluted to a final concentration of 1 × 107 cells·mL−1; 3 μL of 10-fold dilutions (from left to right) were spotted onto the plates and incubated for 5 days at 30 °C.
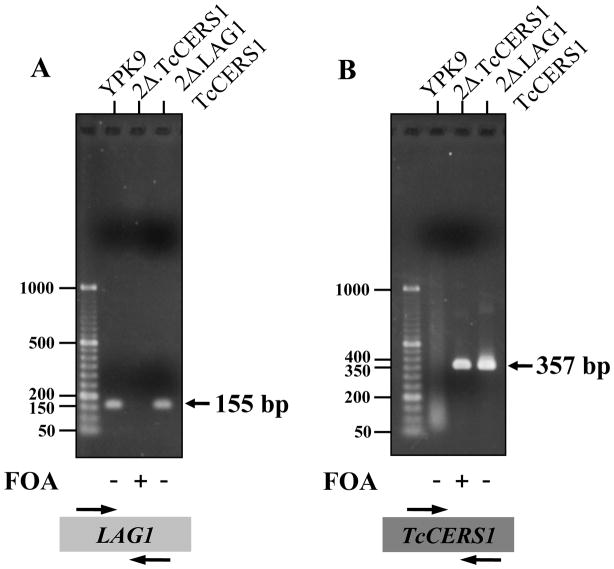
The absence of LAG1 sequences in 2Δ.TcCerS cells was confirmed by whole cell PCR as shown in Figure 7. The expected 155 bp fragment amplified from the LAG1 template [45] was only found in YPK9 and 2Δ.LAG1.TcCERS1 cells before selection on FOA, but not in 2Δ.TcCERS1 cells (Fig. 7A). On the other hand, the expected 357 bp fragment amplified from TcCERS1 template was observed in 2Δ.LAG1.TcCERS1 cells before selection on FOA and in 2Δ.TcCERS1 cells but not in wild-type YPK9 cells (Fig. 7B). The results indicated that the 2Δ.TcCERS1 strain lost the plasmid that contains yeast LAG1 and that there is no other LAG1 sequence present in these cells.
Fig. 7.
Confirmation of rescue of YPK9 lag1Δlac1Δ by overexpression of TcCERS1 using PCR. Colonies of YPK9 containing the empty p423TEF vector (YPK9), YPK9.2Δ.TcCERS1 (2Δ.TcCERS1) and YPK9.2Δ.LAG1 containing the p423TEF:TcCERS1 (2Δ.LAG1.TcCERS1) vector either before (−) or after (+) growth on FOA (1 mg·mL−1) were tested for the presence of LAG1 (A) or TcCERS1 (B) by PCR as described in item 2.9. Markers (left) are 50–1000 bp GeneRuler™ DNA Ladders from Fermentas.
Because expression of TcCERS1 rescues the lethality of lag1Δ lac1Δ double deletion mutant cells after they have lost the extra-copy of LAG1, we examined the inositol-containing lipid profile of the rescued 2Δ.TcCERS1 strain. As shown in Figure 8, TcCERS1 was able to restore inositol-containing phospholipids that include the major phosphatidylinositol (PI) and inositolphosphorylceramide (IPC) species, thus showing a labeling profile quantitatively and qualitatively similar to the wild-type YPK9 and 2Δ.ScLAG1 cells (Fig. 8, compare lanes 5–6 versus 1–2 and 3–4, respectively). The TcCERS1-complemented strain produced extra and stronger base-resistant sphingolipid species (Fig. 8, arrows) that were not observed in the other yeast strains. These bands migrated at positions expected for IPC species containing fatty acids that are shorter than C26. These differences could reflect some fatty acid preferences of the TcCERS1 when overexpressed in yeast, but the exact chemical nature of those species was not determined. In conclusion, TcCERS1 can replace the activity of LAG1 and induce significant ceramide-containing sphingolipid synthesis in yeast.
Fig. 8.
Metabolic labeling of parental and complemented yeast cells using [3H]myo-inositol. YPK9, YPK9.2Δ.LAG1 (2Δ.LAG1) and YPK9.2Δ.TcCERS1 (2Δ.TcCERS1) growing exponentially on SC were labeled with [3H]myo-inositol as described in item 2.4. Lipids were extracted, deacylated with a mild-base (NaOH) as indicated and analyzed by TLC using chloroform/methanol/H2O (10:10:3, v/v) followed by autoradiography. The major inositol-containing phospholipids (PI, IPC/D and MIPC) typically made by yeast and abnormal lipids (arrows) made by YPK9.2Δ.TcCERS1 are indicated.
In this study, T. cruzi epimastigotes were completely resistant to Fumonisin B1 treatment, without perturbation of cell proliferation after long-term treatment with up to 500 μg·mL−1 (Fig. 2 and Supplementary Figure 1). However, we observed that TcCerS activity was blocked by this compound when assayed in vitro (Fig. 3B and 3C). Fumonisins have very poor activity against whole cell fungal sphingolipid synthesis, probably because of limited uptake [27]. Although our yeast cells were quite resistant to Fumonisin B1, we detected a measurable IC50 of 250 μg·mL−1 for growth inhibition (not shown), which is similar to the described dosage used by others [61]. Thus, we compared the effects of this CerS inhibitor on the proliferation of the wild-type YPK9, 2Δ.ScLAG and 2Δ.TcCERS1 strains. As shown in Figure 9, both YPK9 and 2Δ.ScLAG1 strains showed an IC50 of 250 μg·mL−1 after 4 days of treatment, but the 2Δ.TcCERS1 strain was 10 times more sensitive to Fumonisin B1 than the other two strains. Together with the previous results of [3H]myo-inositol metabolic labeling (Fig. 8), this result strongly supports the hypothesis that TcCERS1 rescued the lethality of lag1Δ lac1Δ double deletion mutant cells by replacing the LAG1 activity and generating significant ceramide synthesis.
Fig. 9.
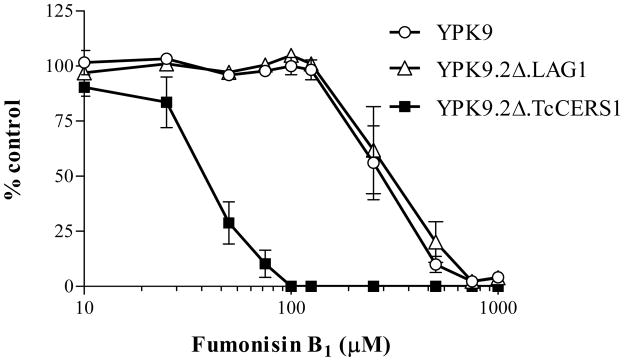
Sensitivity of parental and functionally complemented lag1Δlac1Δ cells to Fumonisin B1. YPK9, YPK9.2Δ.LAG1 and YPK9.2Δ.TcCERS1 growing exponentially on SD were incubated in liquid SD containing increasing amounts of Fumonisin B1 as indicated. After 4 days at 30°C, the number of cells was determined as described in section 2.3, and the growth rates were compared to untreated controls. Each experimental point corresponds to the mean ± standard error for duplicates of two sets of independent experiments.
4. Discussion
In this work, we demonstrate for the first time the CerS activity of a trypanosomatid member, the human pathogenic parasite Trypanosoma cruzi. The TcCerS activity was detected in microsomal membranes of epimastigote forms and was able to use both [3H]DHS and [3H]SPH as long-chain sphingoid base acceptors and palmitoyl-CoA as the preferred acyl-CoA donor substrate. In agreement with this finding, the TcCerS activity was blocked by the potent mycotoxin Fumonisin B1, a well-known acyl-CoA dependent CerS inhibitor [48]. The optimal assay conditions for TcCerS indicated that it was not dependent on Mg+2, Mn+2, ATP or a thiol-protecting agent. The activity was similar to the CerS of rat brain [47] and yeast [45] but completely different from CerS of rat liver, whose activity is activated by Mg+2 and strongly inhibited by Mn+2 [40]. Most of the TcCerS kinetic parameters were comparable to those of mammalian and yeast CerS, including the reverse effect observed at higher protein concentrations, when the amount of product formed decreased. The hydrophobic nature of the substrates used in the CerS reaction assays prompted the establishment of a detergent-free system in which the [4,5-3H]DHS was introduced as a complex with BSA [40]. Following these conditions, dihydroceramide synthesis was shown to be limited neither by substrate availability nor by amounts of microsomal protein or reaction time. However, in a study using rat liver microsomes, there was strong CerS inhibition when membranes were incubated in higher concentrations (> 75 μM) of palmitoyl-CoA, probably because of excess of free substrate [40]. We did not observe this inhibitory effect on palmitoyl-CoA in our system. More recently, optimization of the detergent-free assay allowed the determination of Km values of mammalian CerS on DHS in the range of 2–5 μM [75]. In our system however, we did not detect any CerS activity when using the DHS/BSA complex, and the inclusion of CHAPS in our conditions was pivotal for the successful establishment of the TcCerS assay (Supplementary Figure 2).
The putative T. cruzi CerS gene (TcCERS1) was previously identified as a TLC member, a family of membrane-associated proteins with lipid-sensing domains related to human TRAM, yeast Lag1p and mammalian CLN8 [76]. Here, we showed that the TcCERS1 gene sequence of the Dm28c strain of T. cruzi was able to rescue the lethality of lag1Δlac1Δ double deletion mutant cells by replacing the LAG1 activity and inducing significant ceramide synthesis in the rescued cells (Fig. 6–9). However, as described before for other human ‘Lag1 motif-containing’ sequences [45], we cannot ensure that this phenotype would be displayed if TcCERS1 was not overexpressed.
In mammals, there are six different isoforms of CerS (CerS1-6; [77]), and each one presents a distinct specificity towards the acyl-CoA donor substrate. For instance, while CerS1p was shown to preferably use stearoyl-CoA [70], CerS5p prefers palmitoyl-CoA [78], and CerS2p predominately utilizes lignoceroyl-CoA [79]. The two isoforms of CerS found in yeast (Lag1p and Lac1p) are redundant and prefer hexacosanoyl-CoA as an acyl donor [45]. Here, we demonstrated that TcCerS1 uses only palmitoyl-CoA (Fig. 3–5 and Supplementary Figure 3) with almost no detectable CerS activity in vitro when using lignoceroyl-CoA (Fig. 4A and Supplementary Figure 3). Lignoceric acid is the major fatty acid that is linked to DHS of the T. cruzi GIPLs [4] and GPI-anchored proteins [8,10], but we currently do not know how the parasite incorporates this fatty acid into ceramides. One possibility is that the elongation of palmitoyl-CoA up to lignoceroyl-CoA is mediated by the newly described trypanosomal microsomal elongases that use malonyl-CoA as acyl donors to synthesize fatty acids de novo [80]. However, inclusion of malonyl-CoA and NADPH in our cell-free system was not sufficient to generate [3H]Cer species that were longer than palmitoyl-SPH (Fig. 3B and 3C). The same negative result was obtained when the system was primed with butyryl-CoA or acetyl-CoA (not shown). Therefore, either an additional co-factor is needed to allow the elongation process from palmitoyl-CoA that was missing in our system [81] or the substrate is another fatty acid that could be elongated and desaturated further to generate very-long-chain fatty acids, such as arachidonate (C20:4 from extracellular sources) [80]. It has been suggested that T. cruzi ceramides can be remodeled by de-N-acylation/re-N-acylation [5,36] and this process could be intensified in metacyclic trypomastigotes [10]. However, when using membranes obtained from whole cell extracts of metacyclics, we were unable to detect any CerS activity with palmitoyl-CoA or lignoceroyl-CoA (Supplementary Figure 4).
One method used to determine the acyl-CoA specificity of TcCerSp was the direct CerS assay on microsomes made from three different yeast cell lines. However, we were not successful in detecting any CerS activity using yeast microsomal membranes (not shown). As an alternative, we studied [1,2-3H]myo-inositol metabolic incorporation to determine the phosphosphingolipids that are being synthesized by these yeast cells. The results (Fig. 8) indicate that the yeast cell mutants rescued with TcCERS1 can produce typical IPCs and MIPC with a few more intense base-resistant bands, but without any apparent species with shorter ceramides. Although these results did not reveal the acyl-CoA specificity of TcCerSp, they clearly show that TcCERS1 is the gene responsible for this activity and that it is able to functionally complement a lag1Δlac1Δ double deletion yeast mutant.
The mycotoxins Australifungin and Fumonisin B1, which are the two best-known acyl-CoA dependent CerS inhibitors, are able to inhibit the growth of fungal cells [27,61,82]. Osborne et al. [60] demonstrated that 5–10 μM Fumonisin B1 inhibited the proliferation of epimastigotes from the Brazil strain of T. cruzi by 50%. However, in our study, these two compounds were not capable of inhibiting the proliferation of the Dm28c (Fig. 2C) and Y (Fig. 2D) strains. Strain variations could explain differences in drug sensitivity [83]. For instance, the IC50 for Fumonisin B1 in the YPK9 S. cerevisiae strain (Fig. 9) was similar (250 μg·mL−1) to that for strains used before [61,82]. However, in another study, the IC50 of Australifungin in S. cerevisiae (strain BY4741) was 1000x higher (>200 μg·mL−1) than our values for YPK9 and C. neoformans strain 444 (Supplementary Figure 1A) and those of Mandala et al. [27]. No information is available on drug-sensitivity differences between the Brazil, Y and Dm28c strains of T. cruzi. One possibility is the presence of multidrug resistance (MDR) transporters that actively exclude the drugs from the cellular environment. This type of transporter has been described in several human protozoan parasites, including T. cruzi [84]. However, no experimental evidence correlates Fumonisin B1 resistance with MDR transporters. Additional experiments are needed to confirm that Fumonisin B1 or the sphingosine analog FTY720 [85] can enter the parasites and inhibit TcCerS in vivo. Alternatively, T. cruzi may compensate the blockage of the de novo ceramide synthesis by a salvage pathway using exogenous sources of sphingolipids as suggested for Leishmania [29].
Supplementary Material
Suppl. Fig. 1. Dose-dependent inhibition of epimastigotes viability by Australifungin, Fumonisin B1, and Miltefosine in axenic culture. C. neoformans (open squares), T. cruzi epimastigotes from strain Y (closed triangles) or Dm28c (closed circles), and C. fasciculata (closed diamonds) were cultivated in absence or presence of increasing amounts of Australifungin (A), Fumonisin B1 (B) or Miltefosine (C) as described in M&M. Viability was determined by direct counting and expressed as the percentage of the control cells cultivated in absence of drugs. Each experimental point corresponds to the mean ± standard error for duplicates of two sets of independent experiments.
Suppl. Fig. 2. Effects of different conditons on T. cruzi CerS activity in vitro. Microsomal membranes from epimastigote forms (50 μg.mL−1) of Y (A) and Dm28c (B) strain were assayed in the presence of 0.5 μCi [3H]DHS at 28 °C. Assays were performed in 100 μL of 100 mM Tris-HCl pH 7.5 in the absence (−) or presence (+) of 150 μM palmitic acid (C16:0), 1 mM MgCl2, 1 mM MnCl2, 1 mM DTT, 4 mM NaF, 1 mM NADPH, 1 mM ATP, 0.2% (w/v) CHAPS and/or a mixture of BSA:DHS (1:1) as indicated. After 4 h incubation, the lipids were extracted, separated by TLC using chloroform/methanol/2 N NH4OH (65:25:4, v/v) and visualized after autoradiography as described in item 2.6. The relative positions of synthesized [3H]Cer labeled with [3H]SPH is indicated on the left of each panel, together with the non-reactive excess of each substrate, the origin (OR) and front (FRONT) of the chromatograms.
Suppl. Fig. 3. Acyl-CoA donor specificity and pH preference of Dm28c-strain epimastigotes of T. cruzi CerS in vitro. (A) [3H]SPH was incubated in 100 μL of 100 mM Tris-HCl pH 7.5 containing 1 mM MgCl2, 1 mM DTT, 4 mM NaF and 0.2% (w/v) CHAPS at 28 °C (lanes 1–14) in the absence (−) or presence of epimastigote microsomes (50 μg·mL−1) and in the absence (−) or presence (+) of various acyl-CoAs of different lengths (100 μM). (B) Lanes 1–7 are the same as lanes 1–14, but only palmitoyl-CoA was used as the acyl-CoA substrate donor; 200 μg·mL−1 of microsomes were present, and the pH was varied between 6.0–9.5 as indicated. After 4 h incubation, lipids were extracted and analyzed by TLC as in Figure 3. The relative positions of palmitoyl-sphingosine (C16-Cer) and lignoceroyl-sphingosine (C24-Cer) standards are indicated on the left of Panel A.
Suppl. Fig. 4. Acyl-CoA donor specificity of epimastigotes and metacyclics Dm28c T. cruzi CerS in vitro. Total membrane pellets obtained from metacyclic trypomastigotes (left) and epimastigotes (right) of Dm28c-strain were incubated in 100 μL of 100 mM Tris-HCl pH 7.5 containing 1 mM MgCl2, 1 mM DTT, 4 mM NaF and 0.2% (w/v) CHAPS at 28 °C in the absence (−) or presence of palmitoyl-CoA or lignoceroyl-CoA as indicated. After 4 h incubation, lipids were extracted and analyzed by TLC as in Figure 3. The relative positions of palmitoyl-sphingosine (C16-Cer) and lignoceroyl-sphingosine (C24-Cer) standards are indicated on the right.
Suppl. Fig. 5. Phylogenetic analysis of ceramide synthases. The phylogenetic tree was created using the neighbour-joining method, with 10,000 replicates, with MEGA-5 [54]. The amino acid sequences include S. cerevisiae LAG1 (LAG1_YEAST) and LAC 1 (LAC1_YEAST), Schizosaccharomyces pombe LAG1 (LAG1_SCHPO) and LAC 1 (LAC1_SCHPO), Homo sapiens LASS1 (LASS1_HUMAN), the social amoeba Dictyostelium discoideum (Q54S87_DICDI), the enteropathogenic amoeba Entamoeba histolytica (C4LXF4_ENTHI), the Apicomplexa Plasmodium falciparum (C0H4D2_PLAF7), P. vivax (A5KB10_PLAVI), P. knowlesi (B3L6I7_PLAKH), P. chabaudi (Q4Y1U1_PLACH), Toxoplasma gondii (B9PSW9_TOXGO), Cryptosporidium muris (B6AA96_9CRYT), and the trypanosomatids Leishmania major (Q4Q684_LEIMA), L. brasiliensis (A4HJG0_LEIBR), L. mexicana (E9B1X8_LEIMU), L. infantum (A4I6V5_LEIIN), Trypanosoma vivax (G0TUW$_TRYVI), T. brucei (Q57V92_9TRYP and Q583F9_9TRYP), T. cruzi CL-Brener (Q4CUX8_TRYCR and Q4DB03_TRYCR) and T. cruzi Dm28c (G0YYC4_TRYCR). Numbers represent bootstrap values. The bar represents the percentage of substitutions.
Research Highlights.
We characterized the T. CRUZI ceramide synthase (TcCerS) at the biochemical level.
TcCerS is fully dependent on palmitoyl-CoA donor and is inhibited by Fumonisin B1.
TCCERS1 gene was able to rescue the lethality of a CERS deletion yeast mutant.
At the molecular level, TCCERS1 encodes a ‘bona-fide’ acyl-CoA dependent CerS.
Acknowledgments
We thank Dr. Ulysses Lins (Instituto de Microbiologia Prof. Paulo de Góes - UFRJ) for reading the manuscript. We are grateful to Dr. Alcides José Monteiro da Silva (Núcleo de Pesquisas em Produtos Naturais - UFRJ) for assistance with the catalytic hydrogenation of [3-3H]SPH.
Funding
This work was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (research grant 477124/2009-7 to N.H.), Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) (research grant E-26/110.917/2009 to N.H) and National Institutes of Health (NIH) (research grant AG006168 to S.M.J.). J.M.F and D.C.R. were PhD fellows from the Coordenadoria de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). R.C.M.C.S is the recipient of a master fellowship from CAPES, and C.M.K is a PhD fellow from CNPq.
Abbreviations
- BHI
brain-heart infusion
- Cer
ceramide
- CerS
ceramide synthase
- DHCer
dihydroceramide
- DHS
dihydrosphingosine; endoplasmic reticulum
- DTT
dithiothreitol
- ER
endoplasmic reticulum
- EPC
ethanolamine phosphorylceramide
- FCS
fetal calf serum
- FOA
5-fluorootic acid
- HDG
1-O-hexadecyl glycerol
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- GIPL
glycoinositolphospholipid
- GPI
glycosylphosphatidylinositol
- IPC
inositol phosphorylceramide
- IPCS
IPC synthase
- KDS
3-ketodihydrosphingosine
- MIC
minimum inhibitory concentration
- PI
phosphatidylnositol
- PMSF
phenylmethanesulfonyl fluoride
- SBP
sphingolipid biosynthetic pathway
- SLS
sphingolipid synthase
- SPH
sphingosine
- SPT
serine palmitoyl transferase
- YPD
culture medium composed of 1% yeast extract, 2% peptone and 2% dextrose
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Coura JR, Viñas PA. Chagas disease: a new worldwide challenge. Nature. 2010;465:S6–S7. doi: 10.1038/nature09221. [DOI] [PubMed] [Google Scholar]
- 2.Brener Z. Biology of Trypanosoma cruzi. Ann Rev Microbiol. 1973;27:347–482. doi: 10.1146/annurev.mi.27.100173.002023. [DOI] [PubMed] [Google Scholar]
- 3.Cardoso de Almeida ML, Heise N. Proteins anchored via GPI and solubilizing phospholipases in Trypanosoma cruzi. Biol Res. 1993;26:285–312. [PubMed] [Google Scholar]
- 4.Previato JO, Wait R, Jones C, Dos Reis GA, Todeschini AR, Heise N, Mendonça-Previato L. Glycoinositolphospholipids (GIPL) from Trypanosoma cruzi: structure, biosynthesis and immunology. Adv Parasitol. 2004;56:1–41. doi: 10.1016/s0065-308x(03)56001-8. [DOI] [PubMed] [Google Scholar]
- 5.Heise N, Raper J, Buxbaum LU, Peranivich TMS, Cardoso de Almeida ML. Identification of complete precursors for the glycosylphosphatidylinositol protein anchors of Trypanosoma cruzi. J Biol Chem. 1996;271:16877–87. doi: 10.1074/jbc.271.28.16877. [DOI] [PubMed] [Google Scholar]
- 6.Almeida IC, Ferguson MAJ, Schenkman S, Travassos LR. Lytic anti-α-galactosyl antibodies from patients with chronic Chagas disease recognize novel O-linked oligosaccharides on mucin-like glycosylphosphatidylinositol anchored glycoproteins of Trypanosoma cruzi. Biochem J. 1994;304:793–802. doi: 10.1042/bj3040793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shenkman S, Ferguson MAJ, Heise N, Cardoso de Almeida ML, Mortara RA, Yoshida N. Mucin-like glycoproteins linked to the membrane by glycosylphosphatidylinositol anchor are the major acceptors of sialic acid in a reaction catalyzed by trans-sialidase in metacyclic forms of Trypanosoma cruzi. Mol Biochem Parasitol. 1993;59:293–304. doi: 10.1016/0166-6851(93)90227-o. [DOI] [PubMed] [Google Scholar]
- 8.Heise N, Cardoso de Almeida ML, Ferguson MAJ. Characterization of the lipid moiety of the GPI anchor of Trypanosoma cruzi 1G7-antigen. Mol Biochem Parasitol. 1995;70:71–84. doi: 10.1016/0166-6851(95)00009-p. [DOI] [PubMed] [Google Scholar]
- 9.Previato JO, Jones C, Xavier MT, Wait R, Travassos LR, Parodi AJ, Mendonça-Previato L. Structural characterization of the major glycosylphosphatidylinositol membrane-anchored glycoprotein from epimastigote forms of Trypanosoma cruzi Y-strain. J Biol Chem. 1995;270:7241–50. doi: 10.1074/jbc.270.13.7241. [DOI] [PubMed] [Google Scholar]
- 10.Serrano AA, Schenkman S, Yoshida N, Mehlert A, Richardson JM, Ferguson MAJ. The lipid structure of the GPI-anchored mucin-like sialic acid acceptors of Trypanosoma cruzi changes during parasite differentiation from epimastigotes to infective metacyclic trypomastigote forms. J Biol Chem. 1995;270:27244–53. doi: 10.1074/jbc.270.45.27244. [DOI] [PubMed] [Google Scholar]
- 11.Almeida IC, Camargo MM, Procópio DO, Silva LS, Mehlert A, Travassos LR, Gazzinelli RT, Ferguson MAJ. Highly purified glycosylphosphatidylinositols from Trypanosoma cruzi are potent proinflammatory agents. EMBO J. 2000;19:1476–85. doi: 10.1093/emboj/19.7.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lederkremer RM, Lima CE, Ramirez MI, Gonçalves MF, Colli W. Hexadecylpalmitoylglycerol or ceramide is linked to similar glycophosphoinositol anchor-like structures in Trypanosoma cruzi. Eur J Biochem. 1993;218:929–36. doi: 10.1111/j.1432-1033.1993.tb18449.x. [DOI] [PubMed] [Google Scholar]
- 13.Previato JO, Gorin PA, Mazurek M, Xavier MT, Fournet B, Wierusxesk JM, Mendonça-Previato L. Primary structure of the oligosaccharide chain of lipopeptidophosphoglycan of epimastigote forms of Trypanosoma cruzi. J Biol Chem. 1990;265:2518–26. [PubMed] [Google Scholar]
- 14.Uhrig ML, Couto AS, Colli W, Lederkremer RM. Characterization of inositolphospholipids in Trypanosoma cruzi trypomastigote forms. Biochim Biophys Acta. 1996;1300:233–9. doi: 10.1016/0005-2760(96)00021-5. [DOI] [PubMed] [Google Scholar]
- 15.Agusti R, Couto AS, Colli W, Lederkremer RM. Structure of the glycosylphosphatidylinositol-anchor of the trans-sialidase from Trypanosoma cruzi metacyclic trypomastigote forms. Mol Biochem Parasitol. 1998;97:123–31. doi: 10.1016/s0166-6851(98)00137-6. [DOI] [PubMed] [Google Scholar]
- 16.Burleigh BA, Andrews NW. The mechanisms of Trypanosoma cruzi invasion of mammalian cells. Annu Rev Microbiol. 1995;49:175–200. doi: 10.1146/annurev.mi.49.100195.001135. [DOI] [PubMed] [Google Scholar]
- 17.DosReis GA, Peçanha LM, Bellio M, Previato JO, Mendonça-Previato L. Glycoinositolphospholipids from Trypanosoma cruzi transmit signals to the cells of the host immune system through both ceramide and glycan chains. Microbes Infect. 2002;4:1007–13. doi: 10.1016/s1286-4579(02)01616-7. [DOI] [PubMed] [Google Scholar]
- 18.Acosta-Serrano A, Almeida IC, Freitas-Junior L, Yoshida N, Schenkman S. The mucin-like glycoprotein super-family of Trypanosoma cruzi: structure and biological roles. Mol Biochem Parasitol. 2001;114:143–50. doi: 10.1016/s0166-6851(01)00245-6. [DOI] [PubMed] [Google Scholar]
- 19.Buscaglia CA, Campo VA, Frasch ACC, Di Noia JM. Trypanosoma cruzi surface mucins: host-dependent coat diversity. Nat Rev Microbiol. 2006;4:229–36. doi: 10.1038/nrmicro1351. [DOI] [PubMed] [Google Scholar]
- 20.Gazzinelli RT, Rodrigues MM, Almeida IC, Travassos LR. Role of parasite surface glycoconjugates on induction/regulation of immune responses and inflammation, elicited during Trypanosoma cruzi infection: potential implications on pathphysiology of Chagas’ disease. Ci Cult J Braz Assoc Adv Sci. 1999;51:411–24. [Google Scholar]
- 21.Hanada K. Serine palmitoyltransferase, a key enzyme of sphingolipid metabolism. Biochim Biophys Acta. 2003;1632:16–30. doi: 10.1016/s1388-1981(03)00059-3. [DOI] [PubMed] [Google Scholar]
- 22.Guillas I, Kirchman PA, Chuard R, Pfefferli M, Jiang JC, Jazwinski SM, Conzelmann A. C26-CoA-dependent ceramide synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1p. EMBO J. 2001;20:2655–65. doi: 10.1093/emboj/20.11.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schorling S, Vallée B, Barz WP, Rizman H, Oesterhelt D. Lag1p and Lac1p are essential for the acyl-CoA-dependent ceramide synthase reaction in Saccharomyces cerevisiae. Mol Biol Cell. 2001;12:3417–27. doi: 10.1091/mbc.12.11.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vallée B, Riezman H. Lip1p: a novel subunit of acyl-CoA ceramide synthase. EMBO J. 2005;24:730–41. doi: 10.1038/sj.emboj.7600562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hannun YA, Obeid LM. Principles of bioactive lipid signaling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–50. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 26.Dickson RC. Sphingolipid functions in Saccharomyces cerevisiae: comparison to mammals. Annu Rev Biochem. 1998;67:27–48. doi: 10.1146/annurev.biochem.67.1.27. [DOI] [PubMed] [Google Scholar]
- 27.Mandala SM, Harris GH. Isolation and characterization of novel inhibitors of sphingolipid synthesis: australifungin, viridiofungins, rustmicin, and khafrefungin. Methods Enzymol. 1999;311:335–48. doi: 10.1016/s0076-6879(00)11094-8. [DOI] [PubMed] [Google Scholar]
- 28.Takesako K, Kuroda H, Inoue T, Haruna F, Yoshikawa Y, Kato I, et al. Biological properties of aureobasidin A, a cyclic depsipeptide antifungal antibiotic. J Antibiotics. 1993;46:1414–20. doi: 10.7164/antibiotics.46.1414. [DOI] [PubMed] [Google Scholar]
- 29.Zhang K, Beverley SM. Phospholipid and sphingolipid metabolism in Leishmania. Mol Biochem Parasitol. 2010;170:55–64. doi: 10.1016/j.molbiopara.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sutterwala SS, Creswell CH, Sanyal S, Menon AK, Bangs JD. De novo sphingolipid synthesis is essential for viability, but not for transport of glycosylphosphatidylinositol-anchored proteins in African trypanosomes. Eukaryot Cell. 2007;6:454–64. doi: 10.1128/EC.00283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fridberg A, Olson CL, Nakayasu ES, Tyler KM, Almeida IC, Engman DM. Sphingolipid synthesis is necessary for kinetoplast segregation and cytokinesis in Trypanosoma brucei. J Cell Sci. 2007;121:522–35. doi: 10.1242/jcs.016741. [DOI] [PubMed] [Google Scholar]
- 32.Sutterwala SS, Hsu F, Senova ES, Schwartz KJ, Zhang K, Key P, et al. Developmentally regulated sphingolipid synthesis in African trypanosomes. Mol Microbiol. 2008;70:281–96. doi: 10.1111/j.1365-2958.2008.06393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mina JG, Pan S, Wansadhipathi NK, Bruce CR, Shams-Eldin H, Schwarz RT, et al. The Trypanosoma brucei sphingolipid synthase, an essential enzyme and drug target. Mol Biochem Parasitol. 2009;168:16–23. doi: 10.1016/j.molbiopara.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 34.Denny PW, Shams-Eldin H, Price HP, Smith DF, Schwarz RT. The protozoan inositol phosphorylceramide synthase. A novel drug target that defines a new class of sphingolipid synthase. J Biol Chem. 2006;281:28200–9. doi: 10.1074/jbc.M600796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bertello LE, Andrews NW, Lederkremer RM. Developmentally regulated expression of ceramide in Trypanosoma cruzi. Mol Biochem Parasitol. 1996;79:143–51. doi: 10.1016/0166-6851(96)02645-x. [DOI] [PubMed] [Google Scholar]
- 36.Salto ML, Bertello LE, Vieira M, Docampo R, Moreno SNJ, Lederkremer RM. Formation and remodeling of inositolphosphoceramide during differentiation of Trypanosoma cruzi from trypomastigote to amastigote. Eukaryot Cell. 2003;2:756–68. doi: 10.1128/EC.2.4.756-768.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Figueiredo JM, Dias WB, Mendonça-Previato L, Previato JO, Heise N. Characterization of the inositol phosphorylceramide synthase activity from Trypanosoma cruzi. Biochem J. 2005;387:519–29. doi: 10.1042/BJ20041842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sevova ES, Goren MA, Schwartz KJ, Hsu F, Turk J, Fox BG, Bangs JD. Cell-free synthesis and functional characterization of sphingolipid synthases from parasitic trypanosomatid protozoa. J Biol Chem. 2010;285:20580–7. doi: 10.1074/jbc.M110.127662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heise N, Opperdoes FR. Localisation of a 3-hydroxy-3-methylglutaryl-coenzyme A reductase in the mitochondrial matrix of Trypanosoma brucei procyclics. Z Naturforsch. 2000;55c:473–7. doi: 10.1515/znc-2000-5-626. [DOI] [PubMed] [Google Scholar]
- 40.Hirschberg K, Rodger J, Futerman AH. The long-chain sphingoid base of sphingolipids is acylated at the cytosolic surface of the endoplasmic reticulum in rat liver. Biochem J. 1993;290:751–7. doi: 10.1042/bj2900751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ausubel FM, Brent R, Kingston RE, Morre DD, Seidman JG, Smith JA, Struhl K. Saccharomyces cerevisiae. Basic techniques of yeast genetics. In: Ausubel FM, editor. Short Protocols in Molecular Biology. New York: John Wiley and Sons; 1992. [Google Scholar]
- 42.Saraiva VB, Gibaldi D, Previato JO, Mendonça-Previato L, Bozza MT, Freire-de-Lima CG, Heise N. Proinflammatory and cytotoxic effects of hexadecylphosphocholine (Miltefosine) against drug-resistant strains of Trypanosoma cruzi. Antimicrob Agents Chemother. 2002;46:3472–7. doi: 10.1128/AAC.46.11.3472-3477.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Widmer F, Wright LC, Obando D, Handke R, Ganendren R, Ellis DH, Sorrell TC. Hexadecylphosphocholine (Miltefosine) has broad-spectrum fungicidal activity and is efficacious in a mouse model of crytococcosis. Antimicrob Agents Chemother. 2006;50:414–21. doi: 10.1128/AAC.50.2.414-421.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother. 2001;48:5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 45.Guillas I, Jiang JC, Vionnet C, Roubaty C, Uldry D, Chuard R, Wang J, Jazwinski SM, Conzelmann A. Human homologues of LAG1 reconstitute Acyl-CoA-dependent ceramide synthesis in yeast. J Biol Chem. 2003;278:37083–91. doi: 10.1074/jbc.M307554200. [DOI] [PubMed] [Google Scholar]
- 46.Contreras V, Salles J, Thomas N, Morel C, Goldenberg S. In vitro differentiation of Trypanosoma cruzi under chemically defined conditions. Mol Biochem Parasitol. 1985;16:315–27. doi: 10.1016/0166-6851(85)90073-8. [DOI] [PubMed] [Google Scholar]
- 47.Morell P, Radin NS. Specificity in ceramide biosynthesis from long chain bases and various fatty acyl coenzyme A’s by brain microsomes. J Biol Chem. 1970;245:342–50. [PubMed] [Google Scholar]
- 48.Wang E, Norred WP, Bacon CW, Riley RT, Merril AH., Jr Inhibition of sphingolipid biosynthesis by fumonisins. J Biol Chem. 1991;266:14486–90. [PubMed] [Google Scholar]
- 49.Wang E, Merril AH., Jr Ceramide synthase. Methods Enzymol. 1999;311:15–21. doi: 10.1016/s0076-6879(00)11062-6. [DOI] [PubMed] [Google Scholar]
- 50.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–7. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook J, Russell DW. Molecular Cloning: a laboratory manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 52.Mumberg D, Müller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–22. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 53.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA 5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kirchman PA, Kim S, Lai CY, Jazwinski SM. Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae. Genetics. 1999;152:179–90. doi: 10.1093/genetics/152.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang JC, Kirchman PA, Zagulski M, Hunt J, Jazwinski SM. Homologs of the yeast longevity gene LAG1 in Caenorhabditis elegans and human. Genome Res. 1998;8:1259–72. doi: 10.1101/gr.8.12.1259. [DOI] [PubMed] [Google Scholar]
- 57.Johnston M, Davis RW. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–8. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pankova-Kholmyansky I, Flescher E. Potential new antimalarial chemotherapeutics based on sphingolipid metabolism. Chemotherapy. 2006;52:205–9. doi: 10.1159/000093037. [DOI] [PubMed] [Google Scholar]
- 59.Suzuki E, Tanaka AK, Toledo MS, Levery SB, Straus AH, Takahashi HK. Trypanosomatid and fungal glycolipids and sphingolipids as infective factors and potential targets for development of new therapeutic strategies. Biochim Biophys Acta. 2008;1780:362–9. doi: 10.1016/j.bbagen.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 60.Osborne CD, Piiman Noblet G, Enongene EN, Bacon CW, Riley RT, Voc KA. Host resistance to Trypanosoma cruzi infection is enhanced in mice fed Fusarium verticillioides (= F. moniliforme) culture material containing fumonisins. Food Chem Toxicol. 2002;40:1789–98. doi: 10.1016/s0278-6915(02)00182-5. [DOI] [PubMed] [Google Scholar]
- 61.Wu WI, McDonough VM, Nickels JT, Jr, Ko J, Fischl AS, Vales TR, et al. Regulation of lipid biosynthesis in Saccharomyces cerevisiae by Fumonisin B1. J Biol Chem. 1995;270:13171–8. doi: 10.1074/jbc.270.22.13171. [DOI] [PubMed] [Google Scholar]
- 62.Routier F, Previato JO, Jones C, Wait R, Mendonça-Previato L. Glycoinositolphosplipids from members of the Trypanosomatidae family: investigation on the lipid moiety. J Braz Assoc Adv Sci. 1993;45:66–8. [Google Scholar]
- 63.Heise N, Gutierrez ALS, Mattos KA, Jones C, Wait R, Previato JO, Mendonça-Previato L. Molecular analysis of a novel family of complex glycoinositolphosphoryl ceramide from Cryptococcus neoformans: structural differences between encapsulated and acapsular yeast forms. Glycobiology. 2002;12:409–20. doi: 10.1093/glycob/cwf053. [DOI] [PubMed] [Google Scholar]
- 64.Cowart LA, Obeid LM. Yeast sphingolipids: recent developments in understanding biosynthesis, regulation, and function. Biochim Biophys Acta. 2007;1771:421–31. doi: 10.1016/j.bbalip.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mao C, Xu R, Bielawska A, Obeid LM. Cloning of an alkaline ceramidase from Saccharomyces cerevisiae An enzyme with reverse (CoA-independent) ceramide synthase activity. J Biol Chem. 2000a;275:6876–84. doi: 10.1074/jbc.275.10.6876. [DOI] [PubMed] [Google Scholar]
- 66.Mao C, Xu R, Bielawska A, Szulc ZM, Obeid LM. Cloning and characterization of a Saccharomyces cerevisiae alkaline ceramidase with specificity for dihydroceramide. J Biol Chem. 2000b;275:31369–78. doi: 10.1074/jbc.M003683200. [DOI] [PubMed] [Google Scholar]
- 67.Cerantola V, Vionnet C, Aebischer OF, Jenny T, Knudsen J, Conzelmann A. Yeast sphingolipids do not need to contain very long chain fatty acids. Biochem J. 2007;401:205–16. doi: 10.1042/BJ20061128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran AN, et al. The genome sequence of Trypanosoma cruzi, ethiologic agent of Chagas disease. Science. 2005;309:409–15. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 69.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1999;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 70.Venkataraman K, Futerman AH. Do longevity assurance genes containing Hox domains regulate cell development via ceramide synthesis? FEBS Lett. 2002;528:3–4. doi: 10.1016/s0014-5793(02)03248-9. [DOI] [PubMed] [Google Scholar]
- 71.Kageyama-Yahara N, Riezman H. Transmembrane topology of ceramide synthase in yeast. Biochem J. 2006;398:585–93. doi: 10.1042/BJ20060697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Spassieva S, Seo JG, Jiang JC, Bielawski J, Alvarez-Vasquez F, Jazwinski SM, et al. Necessary role of for the Lag1p motif in (dihydro)ceramide synthase activity. J Biol Chem. 2006;281:33931–8. doi: 10.1074/jbc.M608092200. [DOI] [PubMed] [Google Scholar]
- 73.von Heijne G. Membrane protein structure prediction: hydrphobicity analysis and the ‘positive inside’ rule. J Mol Biol. 1992;225:487–94. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 74.Mesika A, Ben-Dor S, Laviad EL, Futerman AH. A new functional motif in Hox domain-containing ceramide synthases. Identification of a novel region flanking the Hox and TLC domains essential for activity. J Biol Chem. 2007;282:27366–73. doi: 10.1074/jbc.M703487200. [DOI] [PubMed] [Google Scholar]
- 75.Lahiri S, Lee H, Mesicek J, Fuks Z, Haimovitz-Friedman A, Kolesnick RN, Futerman AH. Kinetic characterization of mammalian ceramide synthases: determination of Km values towards sphinganine. FEBS Lett. 2007;581:5289–94. doi: 10.1016/j.febslet.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 76.Winter E, Ponting CP. TRAM, LAG1 and CLN8: members of a novel family of lipid-sensing domains? Trends Biochem Sci. 2002;27:381–3. doi: 10.1016/s0968-0004(02)02154-0. [DOI] [PubMed] [Google Scholar]
- 77.Pewzner-Jung Y, Ben-Dor S, Futerman AH. When do Lasses (Longevity Assurance genes) become CerS (ceramide synthases)? Insights into the regulation of ceramide synthesis. J Biol Chem. 2006;281:25001–5. doi: 10.1074/jbc.R600010200. [DOI] [PubMed] [Google Scholar]
- 78.Lahiri S, Futerman AH. LASS5 is a bona fide dihydroceramide synthase that selectively utilizes palmitoyl-CoA as acyl donor. J Biol Chem. 2005;280:33735–8. doi: 10.1074/jbc.M506485200. [DOI] [PubMed] [Google Scholar]
- 79.Mizutani Y, Kihara A, Igarashi Y. Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem J. 2005;390:263–71. doi: 10.1042/BJ20050291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee SH, Stephens JL, Englund PT. A fatty-acid synthesis mechanism specialized for parasitism. Nat Rev Microbiol. 2006;5:287–97. doi: 10.1038/nrmicro1617. [DOI] [PubMed] [Google Scholar]
- 81.Schulz A, Mousallem T, Venkataramani M, Persaud-Sawin DA, Zucker A, Luberto C, et al. The CLN9 protein, a regulator of dihydroceramide synthase. J Biol Chem. 2006;281:2784–94. doi: 10.1074/jbc.M509483200. [DOI] [PubMed] [Google Scholar]
- 82.Kobayashi SD, Nagiec MM. Ceramide/long-chain base phosphate rheostat in Saccharomyces cerevisiae: regulation of ceramide synthesis by Elo3p and Cka2p. Eukaryotic Cell. 2003;2:284–94. doi: 10.1128/EC.2.2.284-294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Murta SM, Gazzinelli RT, Brener Z, Romanha AJ. Molecular characterization of susceptible and naturally resistant strains of Trypanosoma cruzi to benznidazole and nifurtimox. Mol Biochem Parasitol. 1998;93:203–14. doi: 10.1016/s0166-6851(98)00037-1. [DOI] [PubMed] [Google Scholar]
- 84.Dallagiovanna B, Camarro F, Castanys S. Molecular characterization of a P-glycoprotein- related tcpgp2 gene in Trypanosoma cruzi. Mol Biochem Parasitol. 1996;75:145–157. doi: 10.1016/0166-6851(95)02519-7. [DOI] [PubMed] [Google Scholar]
- 85.Lahiri S, Park H, Laviad EL, Lu X, Bittman R, Futerman AH. Ceramide synthesis is modulated by the sphingosine analog FTY720 via a mixture of uncompetitive and noncompetitive inhibition in an acyl-CoA chain-length-dependent manner. J Biol Chem. 2009;284:16090–8. doi: 10.1074/jbc.M807438200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Fig. 1. Dose-dependent inhibition of epimastigotes viability by Australifungin, Fumonisin B1, and Miltefosine in axenic culture. C. neoformans (open squares), T. cruzi epimastigotes from strain Y (closed triangles) or Dm28c (closed circles), and C. fasciculata (closed diamonds) were cultivated in absence or presence of increasing amounts of Australifungin (A), Fumonisin B1 (B) or Miltefosine (C) as described in M&M. Viability was determined by direct counting and expressed as the percentage of the control cells cultivated in absence of drugs. Each experimental point corresponds to the mean ± standard error for duplicates of two sets of independent experiments.
Suppl. Fig. 2. Effects of different conditons on T. cruzi CerS activity in vitro. Microsomal membranes from epimastigote forms (50 μg.mL−1) of Y (A) and Dm28c (B) strain were assayed in the presence of 0.5 μCi [3H]DHS at 28 °C. Assays were performed in 100 μL of 100 mM Tris-HCl pH 7.5 in the absence (−) or presence (+) of 150 μM palmitic acid (C16:0), 1 mM MgCl2, 1 mM MnCl2, 1 mM DTT, 4 mM NaF, 1 mM NADPH, 1 mM ATP, 0.2% (w/v) CHAPS and/or a mixture of BSA:DHS (1:1) as indicated. After 4 h incubation, the lipids were extracted, separated by TLC using chloroform/methanol/2 N NH4OH (65:25:4, v/v) and visualized after autoradiography as described in item 2.6. The relative positions of synthesized [3H]Cer labeled with [3H]SPH is indicated on the left of each panel, together with the non-reactive excess of each substrate, the origin (OR) and front (FRONT) of the chromatograms.
Suppl. Fig. 3. Acyl-CoA donor specificity and pH preference of Dm28c-strain epimastigotes of T. cruzi CerS in vitro. (A) [3H]SPH was incubated in 100 μL of 100 mM Tris-HCl pH 7.5 containing 1 mM MgCl2, 1 mM DTT, 4 mM NaF and 0.2% (w/v) CHAPS at 28 °C (lanes 1–14) in the absence (−) or presence of epimastigote microsomes (50 μg·mL−1) and in the absence (−) or presence (+) of various acyl-CoAs of different lengths (100 μM). (B) Lanes 1–7 are the same as lanes 1–14, but only palmitoyl-CoA was used as the acyl-CoA substrate donor; 200 μg·mL−1 of microsomes were present, and the pH was varied between 6.0–9.5 as indicated. After 4 h incubation, lipids were extracted and analyzed by TLC as in Figure 3. The relative positions of palmitoyl-sphingosine (C16-Cer) and lignoceroyl-sphingosine (C24-Cer) standards are indicated on the left of Panel A.
Suppl. Fig. 4. Acyl-CoA donor specificity of epimastigotes and metacyclics Dm28c T. cruzi CerS in vitro. Total membrane pellets obtained from metacyclic trypomastigotes (left) and epimastigotes (right) of Dm28c-strain were incubated in 100 μL of 100 mM Tris-HCl pH 7.5 containing 1 mM MgCl2, 1 mM DTT, 4 mM NaF and 0.2% (w/v) CHAPS at 28 °C in the absence (−) or presence of palmitoyl-CoA or lignoceroyl-CoA as indicated. After 4 h incubation, lipids were extracted and analyzed by TLC as in Figure 3. The relative positions of palmitoyl-sphingosine (C16-Cer) and lignoceroyl-sphingosine (C24-Cer) standards are indicated on the right.
Suppl. Fig. 5. Phylogenetic analysis of ceramide synthases. The phylogenetic tree was created using the neighbour-joining method, with 10,000 replicates, with MEGA-5 [54]. The amino acid sequences include S. cerevisiae LAG1 (LAG1_YEAST) and LAC 1 (LAC1_YEAST), Schizosaccharomyces pombe LAG1 (LAG1_SCHPO) and LAC 1 (LAC1_SCHPO), Homo sapiens LASS1 (LASS1_HUMAN), the social amoeba Dictyostelium discoideum (Q54S87_DICDI), the enteropathogenic amoeba Entamoeba histolytica (C4LXF4_ENTHI), the Apicomplexa Plasmodium falciparum (C0H4D2_PLAF7), P. vivax (A5KB10_PLAVI), P. knowlesi (B3L6I7_PLAKH), P. chabaudi (Q4Y1U1_PLACH), Toxoplasma gondii (B9PSW9_TOXGO), Cryptosporidium muris (B6AA96_9CRYT), and the trypanosomatids Leishmania major (Q4Q684_LEIMA), L. brasiliensis (A4HJG0_LEIBR), L. mexicana (E9B1X8_LEIMU), L. infantum (A4I6V5_LEIIN), Trypanosoma vivax (G0TUW$_TRYVI), T. brucei (Q57V92_9TRYP and Q583F9_9TRYP), T. cruzi CL-Brener (Q4CUX8_TRYCR and Q4DB03_TRYCR) and T. cruzi Dm28c (G0YYC4_TRYCR). Numbers represent bootstrap values. The bar represents the percentage of substitutions.