Abstract
Myelination of axons by oligodendrocytes and Schwann cells in the central and peripheral nervous system, respectively, is essential for normal neuronal functions, and its failure results in devastating demyelinating diseases. During development, both oligodendrocyte and Schwann cell precursors undergo a temporally well-defined series of molecular and structural changes, ultimately culminating in the cessation of proliferation and the elaboration of a highly complex myelin sheath. Recent studies have demonstrated a critical role of microRNAs (miRNAs) in the progression of oligodendrocyte and Schwann cell precursors to the myelinating state—depletion of miRNAs from either cell type results in an arrest in differentiation and lack of myelination. Furthermore, these studies have begun to elucidate the dynamic regulation of miRNA expression and the complexity of miRNA-mediated gene regulation during differentiation of myelinating cells. In this review, the authors highlight the recent understanding of functional links of miRNAs to regulatory networks for central and peripheral myelination, as well as perspectives on the role of miRNAs in demyelinating diseases.
Keywords: microRNA, myelination, oligodendrocyte, Schwann cells, transcription factors
Oligodendrocytes in the CNS and Schwann cells (SCs) in the PNS produce multilamellar myelin sheaths wrapping around axons, which are essential for axonal insulation and saltatory conduction of action potentials in vertebrate nervous systems. These myelinating cells are specified from neural stem/progenitor cells, proliferate and differentiate, and uniquely interact with axons to establish myelin sheath (Nave and Trapp 2008; Richardson and others 2006). Abnormal development and/or maintenance of myelin sheaths may impair nerve conduction and lead to progressive axonal degeneration, leading to acquired or hereditary neurological disorders, including multiple sclerosis, leukodystrophies, periventricular leukomalacia, schizophrenia, and tumors in the CNS (Fields 2008; Trapp and Nave 2008), as well as motor and sensory disabilities in the PNS (Scherer and Wrabetz 2008).
A series of intrinsic and extrinsic regulators (Jessen and Mirsky 2005; Li and others 2009; Peru and others 2008; Wegner 2008) have been shown to positively and negatively control myelinating cell differentiation in a spatiotemporally specific manner. Oligodendrocyte differentiation is regulated by a cohort of promoting factors, including Sox10, Nkx2.2, Olig1, Zfp488, YY1, MRF, and Zfp191, and inhibiting factors, including Id2, Id4, Sox5, and Sox6, and the effectors of Notch signaling, Hes1 and Hes5 (Emery and others 2009; Li and others 2009). Similarly, in the PNS, a transcriptional cascade has been uncovered in SC maturation and myelination (Jessen and Mirsky 2005), including positive regulators that promote SC specification and differentiation (e.g., Sox10, NF-κB, Oct6/SCIP/Pou3f1, Brn2, Nab1/2, and Egr2/Krox20; Jessen and Mirsky 2005) as well as negative regulatory factors such as Notch1, ID2, ID4, c-Jun, Sox 2, and Sox4 (Le and others 2005; Mager and others 2008; Mirsky and others 2008; Parkinson and others 2008; Potzner and others 2010). The myelination processes in the CNS and PNS are regulated through conserved but distinct molecular mechanisms. In both oligodendrocytes and SCs, negative regulators are typically down-regulated, whereas positive regulators are induced during differentiation. This balance of positive and negative regulatory circuitry likely controls the timing of differentiation of myelinating cells.
The discovery of functional small noncoding RNAs (e.g., miRNAs) reveals novel posttranscriptional regulation that controls or fine-tunes the transcriptional output. Recent studies have revealed that complex miRNA regulatory machinery acts cooperatively and combinatorially to modulate the activities of known transcription regulators and signaling networks and thereby control neural cell fate specification and differentiation (Vo and others 2010). This review summarizes recent findings of miRNAs in regulating differentiation of myelinating cells in the CNS and PNS, with a focus on how miRNA pathways integrate into regulatory circuitries to control cell differentiation.
Expression and Function of miRNAs
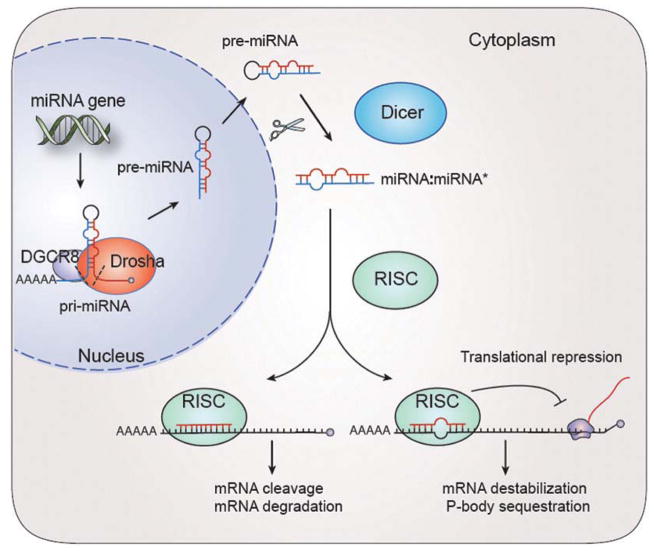
miRNAs are a class of ~21 to 23 nucleotide small non-coding RNAs that silence cognate targets by base-pairing with 3′ untranslated region (3′UTR) of mRNAs of protein-coding genes. The primary precursors of miRNAs (pri-miRNAs) are transcribed by RNA polymerase II. In animals, most pri-miRNAs are processed by the endo-nuclease Drosha and its cofactor DGCR8 into 60 to 100 hairpin-containing pre-miRNAs in the nucleus. After being exported to the cytoplasm by exportin 5, most pre-miRNAs are further cleaved by the RNaseIII-like endo-nuclease, Dicer1, to yield mature miRNAs (Jinek and Doudna 2009). Although this appears to be the norm, recent studies describe a few miRNAs that are processed in a Drosha-independent manner (Han and others 2009), whereas other miRNAs appear to be Dicer1 independent (Cao and others 2009; Chong and others 2010). The mature miRNAs are then incorporated into the RNA-induced silencing complex (RISC), which recognizes specific mRNAs and induces posttranscriptional gene silencing such as by inhibiting translational initiation, targeting mRNAs for destabilization or degradation through dead-enylation, or sequestering targets into cytoplasmic P bodies (Bartel 2009) (Fig. 1). A short “seed” sequence complementarity in 5′ miRNA (position 2–8) allows a single miRNA to pair with the 3′UTR of hundreds of mRNAs (Bartel 2009). To add a level of complexity, miRNAs are also found to recognize seedless sites, 5′UTRs, and coding regions of mRNAs (Asirvatham and others 2008; Lal and others 2009; Tay and others 2008).
Figure 1.
MicroRNA (miRNA) biogenesis and functions. miRNAs are transcribed from endogenous miRNA genes as double-strand primary precursors of mi-RNAs (pri-miRNAs). The pri-miRNAs are processed by Drosha and DGCR8 in the nucleus, producing precursor miRNA (pre-miRNA). The pre-miRNA is then exported to the cytoplasm, where Dicer excises pre-miRNA to yield a miRNA-miRNA* duplex. Typically, only the miRNA is incorporated into the RNA-induced silencing complex (RISC) and becomes functional. Seven- to 8-bp “seed” sequences in 5′ miRNAs are partially complementary to the 3′UTR of protein-coding mRNA targets, which induces posttranscriptional silencing through mechanisms such as mRNA destabilization and translational repression.
miRNAs mediate gene-regulatory events by targeting to the mRNAs of protein-coding genes to direct their repression mainly through either translational inhibition or mRNA destabilization. A recent study shows that in mammalian cells, most miRNAs execute their effects predominantly through mRNA destabilization rather than the conventional view of “translational repression” (Guo H and others 2010) (Fig. 1). Although most regulatory miRNAs appear to have a negative regulatory role, there are a few cases of miRNAs that execute activating functions to up-regulate translation (Schwartz and others 2008; Vasudevan and others 2007). Like transcriptional regulatory factors, the expression level of miRNAs is tightly regulated in a spatially and temporally controlled manner and responds to environmental cues and cellular states. Together, these properties allow miRNAs to act as “micromanagers” to regulate nearly every single process of development and disease (Bartel 2009; Liu and Olson 2010; Vo and others 2010).
miRNAs are shown to widely participate in neurogenesis, including cell fate determination, neural patterning, synaptic plasticity, activity-dependent regulation, and neurological diseases (Kosik 2006; Vo and others 2010). The function of individual miRNAs in neural cell–type differentiation appears to be context specific. A negative feedback control between miRNAs and transcriptional regulators has been shown as a common theme to regulate or refine cell lineage differentiation and neuronal activities (Kim and others 2007) (Fig. 2).
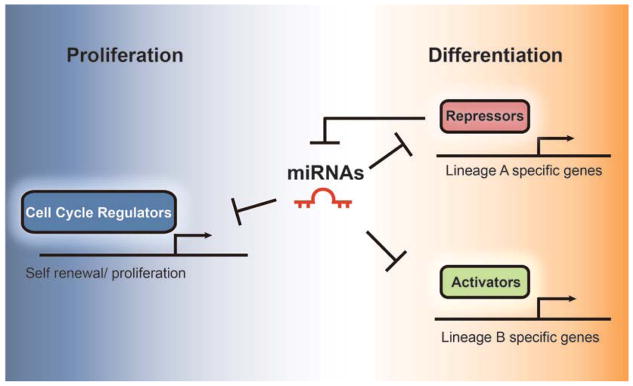
Figure 2.
Model for microRNA (miRNA)–mediated cell lineage progression and transition. Myelin-forming glial precursors (and any other stem cells) are characterized by their capacity to maintain the original cell pool through self-renewal and to generate progenies with a specific identity. miRNAs precisely control the transition from a proliferative state to terminal cellular differentiation. At proliferative stages, miRNAs are inhibited by repressors to antagonize precocious maturation. During differentiation, miRNAs modulate cell cycle regulators and inhibit precursor cell division. They silence repressors of a specific lineage and prevent the expression of activators in other lineages. All three scenarios restrict and reinforce cell lineage progression and block inappropriate cell fate choice.
Recently, the link between miRNAs and glial cell development has been emerging. Deletion of miRNA-processing enzyme Dicer in neural progenitor cells was found to control the switch of neurogenesis to gliogenesis (Kawase-Koga and others 2009; Zheng and others 2010). Overexpression of neurogenic miR-124a and miR-9 is found to inhibit glial fibrillary protein positive (GFAP+) astrocyte formation from embryonic stem (ES) cell–derived neural progenitors. These miRNAs appear to regulate the signaling pathway mediated by STAT3 by blocking STAT3 phosphorylation and activation but not STAT3 mRNA itself (Krichevsky and others 2006). Thus, miRNAs have critical roles in neuronal and astroglial cell fate decision. Besides neurons and astrocytes, the formation and maintenance of myelinating cells are controlled by the interplay of miRNAs with transcriptional and signaling networks (Emery 2010; Nave 2010; Yu and others 2010).
miRNAs Maintain the Oligodendrocyte Identity
MiRNA expression profiling analysis of different stages of oligodendrocyte lineage cells sorted from neonatal rat brains reveals a cohort of 43 miRNAs whose expression dynamically changes during the transition from oligodendrocyte precursor cells (OPCs) to premyelinating oligodendrocytes (Lau and others 2008). Similarly, profiling of distinct stages in oligodendroglial cells derived from human ES cells shows a number of temporally regulated miRNAs during progression of the oligodendrocyte lineage (Letzen and others 2010). Although a few miRNAs in these two microarray screens are common, the majority of them are not overlapping. This may due to the difference between human and mouse samples and cell sources. miR-9 is found highly abundant in mouse OPCs and targets the 3′UTR of an mRNA encoding peripheral myelin protein Pmp22. Although its transcript is present in oligodendrocytes, Pmp22 protein is not detectable in oligodendrocytes. Together with inhibition of astrocytic GFAP by miR-9 (Krichevsky and others 2006), these results suggest that miRNAs such as miR-9 may repress expression of non-lineage-related proteins in oligodendrocytes and function as a guardian to maintain oligodendroglial identity. Mir 9 loss-of-function experiments are required to confirm this hypothesis.
Other miRNAs may function to maintain myelin structure. MiR-23 is a negative regulator of lamin B1 (Lin and Fu 2009). Excessive lamin B1 expression has been shown to cause severe CNS myelin loss in adult-onset autosomal dominant leukodystrophy patients by repressing production of myelin proteins such as myelin basic protein (MBP), proteolipid protein (PLP), and myelin oligodendrocyte glycoprotein (MOG). This suggests that miRNAs such as miR-23 could play an important role in myelin maintenance.
Dicer1-Mediated miRNA Processing Is Required for Oligodendrocyte Differentiation
The role of miRNAs in oligodendrocyte differentiation has been assessed using mouse models in which the Dicer1 gene, responsible for miRNA processing, has been specifically deleted in oligodendrocyte lineage cells. Inactivation of Dicer1 by Cre recombinase directed by Olig1, Olig2, or CNP promoters in the oligodendrocyte lineage beginning at early embryonic stages causes severe dysmyelination and motor behavior deficits, including tremors and seizures (Dugas and others 2010; Zhao and others 2010). Dicer1 deletion leads to a substantial increase of OPC proliferation and a drastic reduction in myelination, suggesting that miRNAs are required for normal OPC cell cycle exit and differentiation.
Although mice with Dicer1 deletion mediated by Olig1-Cre or Olig2-Cre exhibit similar dysmyelination, the former mutants lose the majority of myelin in the CNS and die around postnatal week three, whereas the latter show a developmental delay but ultimate recovery of myelination in adulthood. A possible explanation is that floxed Dicer1 allele is not completely removed by Olig2-Cre in a population of immature oligodendrocytes. Those OPCs that escape Cre excision expand and eventually restore near-normal levels of myelination in adulthood.
Deletion of Dicer1 in adult mice by inducible oligodendrocyte-expressing PLPCreERT resulted in demyelination and progressive axonal degeneration, leading to shorter animal life span (Shin and others 2009). This study suggests that miRNAs are required for not only oligodendrocyte differentiation but also myelin maintenance and homeostasis. Axonal degeneration observed in these mice suggests a neurotrophic function of myelin. However, given that the PLP promoter may also direct CreERT expression in neuronal cells (Guo F and others 2009), it would be interesting to investigate whether axonal degeneration is caused by secondary effects due to the loss of myelin or direct loss of Dicer1 in neurons.
Stage-Specific Regulation of Oligodendrocyte Development by miRNAs
To identify the specific miRNAs responsible for oligodendrocyte differentiation, several groups have performed miRNA microarray profiling of miRNAs on isolated OPCs and oligodendrocytes (Dugas and others 2010; Lau and others 2008) or on the CNS tissues of wild-type and myelin-deficient Dicer1 knockout mice (Shin and others 2009; Zhao and others 2010). These studies identify a cohort of miRNAs that are preferentially enriched in mature oligodendrocytes or down-regulated in myelin-deficient Dicer1 mutants. Among them, several miRNAs appear in common across the different profiling data. Most notably, miR-219 and miR-338 are substantially increased at the onset of oligodendrocyte myelination and in mature oligodendrocytes.
Mature oligodendrocyte-expressing miRNAs play a positive role in promoting OPC differentiation. Expression of stable miRNA mimics for either miR-219 or miR-338-5p and miR-338-3p together can promote OPC differentiation and partially rescue the differentiation deficit of Dicer1-deleted OPCs in vitro (Dugas and others 2010; Zhao and others 2010). Overexpression of miR-219 or miR-338 miRNA is sufficient to promote precocious expression of oligodendrocyte lineage markers in the developing chick and mouse CNS (Zhao and others 2010). Consistently, knockdown of these two miRNAs in cultured OPCs (Dugas and others 2010; Zhao and others 2010) or miR-219 in zebrafish embryos (note that miR-338 is not detectable in the CNS of zebrafish embryos; Zhao and others 2010) inhibits OPC differentiation, suggesting that miR-219 is both sufficient and essential for oligodendrocyte differentiation.
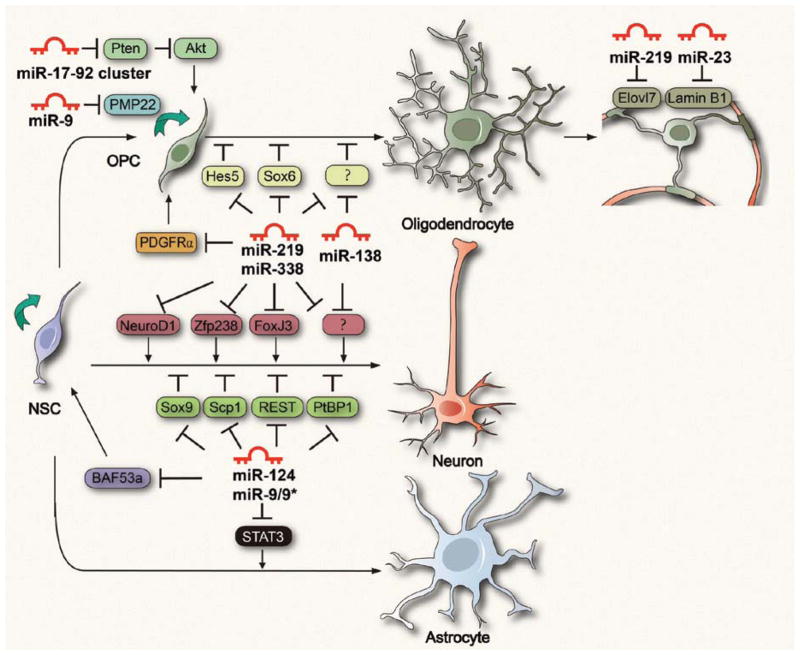
To identify the targets of these miRNAs that are relevant to oligodendrocyte development, a series of computational prediction algorithms reveal miR-219 binding sites in the 3′UTR of a number of genes that have previously been shown to be involved in inhibiting OPC differentiation and maintaining OPCs in the proliferative state, including PDGFRα, Hes5, and Sox6 (Dugas and others 2010; Zhao and others 2010). Despite the lack of sequence homology between miR-219 and miR-338, both miRNAs are predicted to target a disproportionately large number of the same genes involved in negative regulation of oligodendrocyte differentiation such as Hes5 and Sox6 (Fig. 3). This prediction has been demonstrated at least by in vitro luciferase assays in heterologous cells, wherein miR-219 and miR-338 are sufficient to repress constructs harboring the 3′UTR of Hes5 and Sox6. Thus, miR-219 and miR-338 may act synergistically to promote OPC differentiation by alleviating the brakes of cell differentiation (Nave 2010). Interestingly, miR-219 may also target other transcription factors such as Zfp238, FoxJ3, NeuroD1, Isl1, and Otx2 that are potentially involved in neurogenesis (Dugas and others 2010; Zhao and others 2010) (Fig. 3). Overexpression of these proneural factors is able to inhibit OPC differentiation. miRNA-mediated inhibition of the factors may restrict neural progenitor differentiation to the oligodendrocyte lineage. Other differentially regulated miRNAs such as miR-138 exhibit transient expression in early postmitotic oligodendrocytes. Expression of miR-138 stimulates early differentiation but inhibits oligodendrocyte terminal differentiation (Dugas and others 2010), although the targets of miR-138 have not been identified yet. Together, a network of miRNAs individually or cooperatively regulates distinct stages of oligodendrocyte lineage progression, as well as safeguards against the expression of neuronal and SC lineage genes (Figs. 2 and 3).
Figure 3.
MicroRNAs (miRNAs) in regulating neural fate choice and oligodendrocyte differentiation. Neural stem cells (NSCs) proliferate and give rise to neurons, astrocytes, and oligodendrocyte precursor cells (OPCs). Neuronal specific miRNAs inhibit genes required for stem cell self-renew, neuronal repressors, and astrocyte-specific transcriptional factors, providing permission for the onset of neuronal differentiation and preventing alternative fate choice. Similarly, oligodendrocyte-expressing miRNAs such as miR-219 and miR-338 repress negative regulators of differentiation (e.g., Hes5 and Sox6), block PDGFRα signaling, and inhibit proneuronal gene expression, thereby promoting oligodendrocyte lineage progression and myelination.
Besides regulating the differentiation program, miR-219 also regulates a lipid metabolism such as by targeting fatty acid elongase ELOVL7, which synthesizes a very long chain of fatty acid. Accumulation of fatty acids was observed in PLP-CreERT Dicer-floxed mice (Shin and others 2009), suggesting miRNAs such as miR-219 also play a role in myelin membrane homeostasis.
Control of Oligodendroglial Cell Number by miRNAs
The miR-17-92 miRNA cluster encoding miR-19b and miR-17 is identified as highly enriched in oligodendrocyte lineage cells, including A2B5+ OPCs and GalC+ OL (Budde and others 2010; Lau and others 2008). Targeted inactivation of the miR-17-92 cluster in oligodendrocytes directed by an oligodendrocyte-expressing CNP-Cre line resulted in ~25% reduction of Olig2-positive cells in the brain (Budde and others 2010), although the function of this miRNA cluster in OL differentiation and myelination is not described for the mutant mice. miR-19b appears to target the tumor suppressor gene Pten. Expression of miR-19b in OPCs leads to activation of Akt signaling, a downstream target of Pten, and promotes OPC proliferation. Consistent with the role of this cluster (also known as Oncomir-1) in promoting cell survival (Ventura and others 2008), the miR-17-92 cluster plays an important role in oligodendrocyte precursor survival and proliferation. Collectively, miRNAs control oligodendrocyte differentiation by inactivating transcriptional repressors while promoting OPC cell cycle exit and survival in a temporally specific fashion.
Requirement of miRNAs for Schwann Cell Myelination Revealed by Dicer1 Deletion
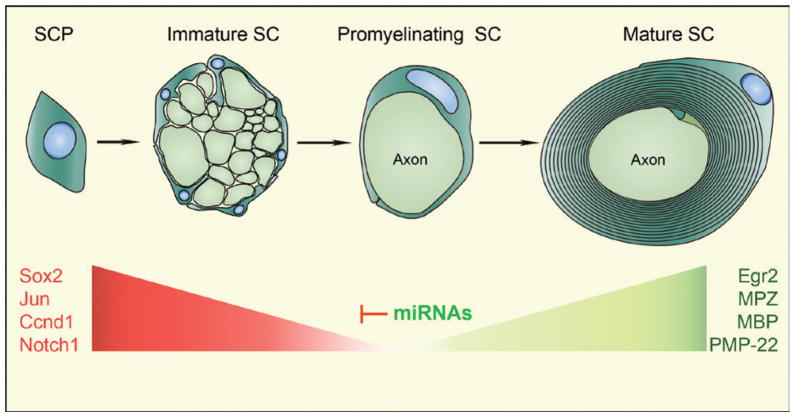
SC development is a particularly good model to study the role of miRNAs because SCs undergo a molecularly and ultrastructurally well-defined, multistage process of differentiation over a protracted period of time. During embryonic development, the neural crest gives rise to SC precursors, which then transition to an immature SC state (Jessen and Mirsky 2005). Most immature SCs originally surround large numbers of axons but then proceed to establish a one-to-one relation with a single axon to become promyelinating SC. Finally, promyelinating cells transition to the myelinating state and ensheath the axon with the multilamellar myelin sheath. Critical for the transition from one developmental stage to the next is the repression of genes involved in the antecedent stage (e.g., Sox2, Notch1, and c-Jun), as well as the activation of new genes that will define the next stage (e.g., Egr2/Krox20, Mbp, and Mpz). Recent studies suggest that miRNAs, by abetting the repression of antecedent negative regulators, play a critical role in the transition of an immature undifferentiated SC to a myelinating SC.
The critical role of miRNAs in SC differentiation and myelination has been revealed by a flurry of studies using Dicer1 depletion both in vitro and in conditional knockout mice (Bremer and others 2010; Pereira and others 2010; Verrier and others 2010; Yun and others 2010). Conditional deletion of Dicer1, using a transgenic mouse expressing Cre recombinase under the control of SC lineage-specific promoters such as Dhh– (Bremer and others 2010; Pereira and others 2010) or P0– gene promoters (Yun and others 2010), results in congenital hypo-myelination and consequent behavioral phenotypes, including severe hindlimb paralysis. Similarly, Dicer1 knockdown by lentivirus-mediated short hairpin RNA (shRNA) leads to the failure of SC differentiation in vitro (Verrier and others 2010).
In Dicer1 conditional knockout (cKO) sciatic nerves, the most prominent phenotype is an arrest of SC differentiation at the promyelinating state, similar to other congenital hypomyelination mutants (Le and others 2005; Topilko and others 1994). Most SCs have completed the process of radial sorting, although some unsorted bundles are observed (Bremer and others 2010; Pereira and others 2010; Yun and others 2010). Most SCs reach the promyelinating state, establishing a one-to-one relationship with axons but failing to myelinate. Very few compact myelin sheaths were observed, and many of these appeared thinner than usual; it is unclear whether these cells represent true outliers or underwent recombination at a slightly later time than the rest of the cohort, thereby bypassing arrest at the promyelinating state. The number of myelinated axons, albeit very few, displays some regional specificity, being more common in the quadriceps nerve than in the saphenous nerve (Pereira and others 2010). The regional specificity may be due to intrinsic differences among SC subpopulations or the distinct influences or activities exerted by motor versus sensory axons. Finally, nonmyelinating (Remak) SCs also display aberrant morphology in these mutants (Bremer and others 2010). In contrast to normal Remak bundle formation, in which SC squeezes its cytoplasm between the axons, mutant SCs engulf groups of small-caliber axons as a whole without axon sorting by their processes (mesaxons; Bremer and others 2010).
Consistent with an arrest in differentiation, deletion of Dicer1 results in a drastic reduction of mature SC-specific genes (e.g., Mbp, Plp1, Mag, Mpz, Pmp22) and the positive master regulatory transcription factor, Egr2/Krox20 (Pereira and others 2010; Yun and others 2010). At early stages, neuregulin/Akt signaling is also reduced in mutants and could underlie the reduced thickness of the occasionally occurring myelin sheaths (Bremer and others 2010; Pereira and others 2010). In contrast, in Dicer1-depleted SCs, mRNAs of antecedent stages (e.g., Ccnd1, Ngfr, Notch1, Hes1, Sox2, and, to a lesser extent, c-Jun) fail to be repressed and are thus elevated compared to age-matched controls (Pereira and others 2010; Verrier and others 2010; Yun and others 2010). De-repression of Sox2 was not observed in one study (Bremer and others 2010), likely because of the younger age of nerve used, when Sox2 levels were relatively high even in controls. In accordance with the molecular and ultrastructural analysis, proliferation is increased in Dicer1-depleted SCs (Bremer and others 2010; Verrier and others 2010; Yun and others 2010), suggesting a failure to appropriately exit the cell cycle. These observations suggest that during development, miRNAs normally suppress negative regulators of myelination and promote cell cycle exit, similar to their role in CNS myelination (Fig. 3) (Dugas and others 2010; Zhao and others 2010).
miRNA-Mediated Regulation of PNS Myelination
Two studies have carried out miRNA profiling analysis and uncovered a cohort of miRNAs that are significantly increased during SC development and down-regulated in Dicer1 mutants in vivo (Bremer and others 2010; Yun and others 2010). A third study profiled miRNAs in cultured SC (Verrier and others 2009; Verrier and others 2010). These studies show that miRNAs are dynamically expressed during development, as well as in different culture conditions. In Dicer1 mutants, the expression of several miRNAs is drastically reduced. Interestingly, in Dicer1 mutants, even more than 2 weeks after recombination of the Dicer1 allele, several miRNAs persist at high levels (Yun and others 2010). This could be due to the extraordinary stability of miRNAs (Jung and others 2010) or to an enticing new possibility that some miRNAs are synthesized by a Dicer1-independent pathway (Chong and others 2010).
Although most miRNAs up-regulated in differentiated SCs are not overlapping between in vitro and vivo studies, several key miRNAs are common between two in vivo studies. Despite the fact that slightly different ages of animals and different selection criteria are used, miR-138 and miR-338 are identified in two studies as potentially important for SC development (Bremer and others 2010; Yun and others 2010). In heterologous cell assays, miR-138 and its reverse strand, miR138*, modestly repress three genes characterizing the undifferentiated SC: Ccnd1 (cyclinD1), a cell cycle gene in G1/S transition, and Jun and Sox2, two negative regulators of SC differentiation (Yun and others 2010). Ccnd1 could exert its effects on SC proliferation, at least in part, by transcriptionally enhancing expression of Notch1, a negative regulator of myelination (Bienvenu and others 2010; Woodhoo and others 2009). Thus, miR-138 may regulate cell cycle exit and myelination by inhibiting the Ccnd1 and Notch signaling pathways, as well as negative regulators of myelination such as Sox2 and c-Jun (Fig. 4). miR-338 is predicted to target Jun, Ccnd1, and Ngfr. Further in vivo experiments are required to confirm the activity of miR-138 on its targets, as well as explore additional potential targets of miR-138 and miR-338.
Figure 4.
Functions of microRNAs (miRNAs) in Schwann cell lineage progression and terminal differentiation. Schematic illustration depicts the main stages of PNS myelination by Schwann cells (SCs). Loss of Dicer1 in SC precursors (SCPs) results in the failure of PNS myelination, with many SCs being arrested at the promyelinating stage. During development, miRNAs are likely to repress negative regulators of Schwann cell myelination, which, in turn, allows myelination to proceed.
Besides miR-138 and miR-338, other miRNAs are likely to be important in SC differentiation. miR-145 is modestly down-regulated in Dicer1 mutants in vivo (Yun and others 2010) and in vitro (Verrier and others 2010). This miRNA was shown to target Sox2 to repress pluripotency in human embryonic stem cells (Xu and others 2009) and may potentially do the same in SCs. miR-204 increases during development, is reduced in Dicer 1 mutant nerves, and is also predicted to target Sox2 (Targetscan algorithm). Because Sox2 is a potent negative regulator of Egr2 and myelination (Kao and others 2009; Le and others 2005), targeting Sox2 by multiple miRNAs such as miR-138, miR-145, and miR204 may be crucial to allow SC differentiation. In addition, other miRNAs that target the Notch pathway could also be important for myelination. miR-34a (Bremer and others 2010) could be relevant to SC proliferation and differentiation because miR-34a inhibits cell proliferation by repressing Notch and mitogen-activated protein kinase (MAPK) signaling pathways (Ichimura and others 2010; Pang and others 2010). miR-146b is also predicted to target Notch1 (Targetscan) and hence could be involved in suppression of Notch signaling (Fig. 4). Finally, unrelated to repressing negative regulators, miR-29a was proposed to repress Pmp22 expression at early stages. In this regard, two studies show contradictory expression profiles of miR-29a expression (Verrier and others 2010; Yun and others 2010). Further work is required to elucidate the expression and functional role of each of these miRNAs in SC differentiation.
Given the wealth of miRNA profiling studies in myelinating cells in the CNS and PNS, it is now possible to compare these cell types. Interestingly, several miRNAs identified in SCs are also expressed in differentiating oligodendrocytes (Bremer and others 2010; Lau and others 2008; Yun and others 2010). For instance, in CNS and PNS studies, miR-138 and miR-338 increase during development and are drastically decreased in Dicer1 conditional mutants (Bremer and others 2010; Dugas and others 2010; Yun and others 2010; Zhao and others 2010). In contrast, miR-219, a critical miRNA implicated in oligodendrocyte differentiation (Dugas and others 2010; Zhao and others 2010), is not present in SCs (Yun and others 2010), suggesting that myelinating cells in the CNS and PNS are regulated by partly overlapping but distinct miRNA networks. It would be significant in the future to identify the miRNA(s) that promotes SC differentiation and myelination.
miRNAs in Demyelinating Diseases
miRNAs are key players not only during development but also during disease formation. They have been implicated in many neurological diseases, including brain tumors (gliomas; Lawler and Chiocca 2009; Novakova and others 2009) and sporadic Alzheimer (Ho and others 2010; Maes and others 2009) and Parkinson disease (Hebert and De Strooper 2007; Santosh and others 2009). Recently, miRNA profiles from active and inactive demyelinating lesions (Junker and others 2009) and peripheral blood cells (Du and others 2009; Keller and others 2009; Otaegui and others 2009) from patients with multiple sclerosis (MS) have been established. Because of substantial gliosis and immune cell infiltration in demyelinating lesions, strongly up-regulated miRNAs in tissue lesions are mainly assigned to astrocytes, T cells, and monocytes (Junker and others 2009). Several miRNAs up-regulated in active MS lesions, including miR-34a, miR-155, and miR-326, target to mRNA of CD47, which functions as an inhibiting “don’t eat me” signal for macrophage activity (Oldenborg and others 2000). Thus, miRNAs may reduce CD47 expression and permit macrophages to execute phagocytosis of myelin debris in MS lesions. Strikingly, mature oligodendrocyte-enriched miRNAs such as miR-219 and miR-338 are the least detectable miRNAs in chronic MS lesions (Junker and others 2009), suggesting that these miRNAs may also function in human oligodendrocyte maturation and myelin repair. Other miRNAs such as miRNA-23 may contribute to demyelinating diseases such as adult-onset autosomal dominant leukodystrophy by targeting lamin B1 (Lin and Fu 2009). An elevated level of lamin B1 by gene duplications preferentially leads to myelin loss in the CNS. Further miRNA profiling in different demyelinating diseases to identify miRNAs that reflect the fundamental feature of demyelinating pathology may point to new biomarkers showing a beneficial effect in therapy for demyelinating diseases such as MS.
Conclusion and Perspectives
The discovery of miRNAs enables a deeper insight into the complexity of gene regulatory networks during the myelination process. Distinct patterns of miRNA expression during normal myelinating cell development and different phases of demyelinating diseases such as MS have been revealed recently. The functional significance of some of these miRNAs is beginning to emerge—miRNAs play multiple roles in oligodendrocytes and SCs, including cell proliferation, differentiation, and myelin homeostasis. A consensus based on the data from the CNS and PNS is that miRNAs control the myelination program by repressing negative regulators of myelination, inhibiting neuronal cell lineages, and promoting cell cycle exit of the precursors (Fig. 2). A challenge remains to understand the individual components of the entire miRNA network and their functional interplay with regulatory circuitry that controls the myelination program during development and myelin regeneration after injury.
Finally, understanding miRNA signatures during development and in disease states will be valuable both for diagnostic and therapeutic purposes. miRNAs have an advantage over mRNAs as they are more stable and do not undergo a significant decay during the tissue sample processing (Jung and others 2010). The robustness of profiling data suggests that miRNAs are potential biomarkers for diagnosis or monitoring demyelinating diseases. Further uncovering key functions for miRNAs will illuminate important aspects in regulating myelination processes. New knowledge about these small regulatory molecules will offer novel therapeutic interventions by which disease-related miRNAs could be antagonized or functionally restored for myelin repair. The future challenge will be to translate this knowledge into improved outcome for patients with demyelinating diseases.
Footnotes
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interests with respect to the authorship and/or publication of this article.
Reprints and permission: http://www.sagepub.com/journalsPermissions.nav
Financial Disclosure/Funding
The author(s) disclosed receipt of the following financial support for the research and/or authorship of this article: The authors acknowledge the support by the US National Multiple Sclerosis Society RG3978 to QRL and the US National Institutes of Health NS050389 to QRL and R21NS063138 to RA.
References
- Asirvatham AJ, Gregorie CJ, Hu Z, Magner WJ, Tomasi TB. MicroRNA targets in immune genes and the Dicer/Argonaute and ARE machinery components. Mol Immunol. 2008;45(7):1995–2006. doi: 10.1016/j.molimm.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer J, O’Connor T, Tiberi C, Rehrauer H, Weis J, Aguzzi A. Ablation of Dicer from murine Schwann cells increases their proliferation while blocking myelination. PLoS One. 2010;5(8):e12450. doi: 10.1371/journal.pone.0012450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde H, Schmitt S, Fitzner D, Opitz L, Salinas-Riester G, Simons M. Control of oligodendroglial cell number by the miR-17-92 cluster. Development. 2010;137(13):2127–32. doi: 10.1242/dev.050633. [DOI] [PubMed] [Google Scholar]
- Cao F, Li X, Hiew S, Brady H, Liu Y, Dou Y. Dicer independent small RNAs associate with telomeric heterochromatin. RNA. 2009;15(7):1274–81. doi: 10.1261/rna.1423309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong MM, Zhang G, Cheloufi S, Neubert TA, Hannon GJ, Littman DR. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010;24(17):1951–60. doi: 10.1101/gad.1953310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10(12):1252–9. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- Dugas JC, Cuellar TL, Scholze A, Ason B, Ibrahim A, Emery B, et al. Dicer1 and miR-219 are required for normal oligodendrocyte differentiation and myelination. Neuron. 2010;65(5):597–611. doi: 10.1016/j.neuron.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery B. Transcriptional and post-transcriptional control of CNS myelination. Curr Opin Neurobiol. 2010;20(5):601–7. doi: 10.1016/j.conb.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Emery B, Agalliu D, Cahoy JD, Watkins TA, Dugas JC, Mulinyawe SB, et al. Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell. 2009;138(1):172–85. doi: 10.1016/j.cell.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields RD. White matter matters. Sci Am. 2008;298(3):42–9. [PubMed] [Google Scholar]
- Guo F, Ma J, McCauley E, Bannerman P, Pleasure D. Early postnatal proteolipid promoter-expressing progenitors produce multilineage cells in vivo. J Neurosci. 2009;29(22):7256–70. doi: 10.1523/JNEUROSCI.5653-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, et al. Posttranscriptional cross regulation between Drosha and DGCR8. Cell. 2009;136(1):75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert SS, De Strooper B. Molecular biology. miRNAs in neurodegeneration. Science. 2007;317(5842):1179–80. doi: 10.1126/science.1148530. [DOI] [PubMed] [Google Scholar]
- Ho L, Fivecoat H, Wang J, Pasinetti GM. Alzheimer’s disease biomarker discovery in symptomatic and asymptomatic patients: experimental approaches and future clinical applications. Exp Gerontol. 2010;45(1):15–22. doi: 10.1016/j.exger.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura A, Ruike Y, Terasawa K, Shimizu K, Tsujimoto G. MicroRNA-34a inhibits cell proliferation by repressing mitogen-activated protein kinase kinase 1 during mega-karyocytic differentiation of K562 cells. Mol Pharmacol. 2010;77(6):1016–24. doi: 10.1124/mol.109.063321. [DOI] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R. The origin and development of glial cells in peripheral nerves. Nat Rev Neurosci. 2005;6(9):671–82. doi: 10.1038/nrn1746. [DOI] [PubMed] [Google Scholar]
- Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457(7228):405–12. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- Jung M, Schaefer A, Steiner I, Kempkensteffen C, Stephan C, Erbersdobler A, et al. Robust microRNA stability in degraded RNA preparations from human tissue and cell samples. Clin Chem. 2010;56(6):998–1006. doi: 10.1373/clinchem.2009.141580. [DOI] [PubMed] [Google Scholar]
- Junker A, Krumbholz M, Eisele S, Mohan H, Augstein F, Bittner R, et al. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain. 2009;132(Pt 12):3342–52. doi: 10.1093/brain/awp300. [DOI] [PubMed] [Google Scholar]
- Kao SC, Wu H, Xie J, Chang CP, Ranish JA, Graef IA, et al. Calcineurin/NFAT signaling is required for neuregulin-regulated Schwann cell differentiation. Science. 2009;323(5914):651–4. doi: 10.1126/science.1166562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase-Koga Y, Otaegi G, Sun T. Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev Dyn. 2009;238(11):2800–12. doi: 10.1002/dvdy.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Leidinger P, Lange J, Borries A, Schroers H, Scheffler M, et al. Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS One. 2009;4(10):e7440. doi: 10.1371/journal.pone.0007440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, et al. A microRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317(5842):1220–4. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7(12):911–20. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24(4):857–64. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O’Day E, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′UTR microRNA recognition elements. Mol Cell. 2009;35(5):610–25. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P, Verrier JD, Nielsen JA, Johnson KR, Notterpek L, Hudson LD. Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. J Neurosci. 2008;28(45):11720–30. doi: 10.1523/JNEUROSCI.1932-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler S, Chiocca EA. Emerging functions of microRNAs in glioblastoma. J Neurooncol. 2009;92(3):297–306. doi: 10.1007/s11060-009-9843-2. [DOI] [PubMed] [Google Scholar]
- Le N, Nagarajan R, Wang JY, Araki T, Schmidt RE, Milbrandt J. Analysis of congenital hypomyelinating Egr2Lo/Lo nerves identifies Sox2 as an inhibitor of Schwann cell differentiation and myelination. Proc Natl Acad Sci U S A. 2005;102(7):2596–601. doi: 10.1073/pnas.0407836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzen BS, Liu C, Thakor NV, Gearhart JD, All AH, Kerr CL. MicroRNA expression profiling of oligodendrocyte differentiation from human embryonic stem cells. PLoS One. 2010;5(5):e10480. doi: 10.1371/journal.pone.0010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, He Y, Richardson WD, Casaccia P. Two-tier transcriptional control of oligodendrocyte differentiation. Curr Opin Neurobiol. 2009;19(5):479–85. doi: 10.1016/j.conb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin ST, Fu YH. miR-23 regulation of lamin B1 is crucial for oligodendrocyte development and myelination. Dis Model Mech. 2009;2(3–4):178–88. doi: 10.1242/dmm.001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev Cell. 2010;18(4):510–25. doi: 10.1016/j.devcel.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes OC, Chertkow HM, Wang E, Schipper HM. MicroRNA: implications for Alzheimer disease and other human CNS disorders. Curr Genomics. 2009;10(3):154–68. doi: 10.2174/138920209788185252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager GM, Ward RM, Srinivasan R, Jang SW, Wrabetz L, Svaren J. Active gene repression by the Egr2.NAB complex during peripheral nerve myelination. J Biol Chem. 2008;283(26):18187–97. doi: 10.1074/jbc.M803330200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirsky R, Woodhoo A, Parkinson DB, Arthur-Farraj P, Bhaskaran A, Jessen KR. Novel signals controlling embryonic Schwann cell development, myelination and dedifferentiation. J Peripher Nerv Syst. 2008;13(2):122–35. doi: 10.1111/j.1529-8027.2008.00168.x. [DOI] [PubMed] [Google Scholar]
- Nave KA. Oligodendrocytes and the “micro brake” of progenitor cell proliferation. Neuron. 2010;65(5):577–9. doi: 10.1016/j.neuron.2010.02.026. [DOI] [PubMed] [Google Scholar]
- Nave KA, Trapp BD. Axon-glial signaling and the glial support of axon function. Annu Rev Neurosci. 2008;31:535–61. doi: 10.1146/annurev.neuro.30.051606.094309. [DOI] [PubMed] [Google Scholar]
- Novakova J, Slaby O, Vyzula R, Michalek J. MicroRNA involvement in glioblastoma pathogenesis. Biochem Biophys Res Commun. 2009;386(1):1–5. doi: 10.1016/j.bbrc.2009.06.034. [DOI] [PubMed] [Google Scholar]
- Oldenborg PA, Zheleznyak A, Fang YF, Lagenaur CF, Gresham HD, Lindberg FP. Role of CD47 as a marker of self on red blood cells. Science. 2000;288(5473):2051–4. doi: 10.1126/science.288.5473.2051. [DOI] [PubMed] [Google Scholar]
- Otaegui D, Baranzini SE, Armananzas R, Calvo B, Munoz-Culla M, Khankhanian P, et al. Differential micro RNA expression in PBMC from multiple sclerosis patients. PLoS One. 2009;4(7):e6309. doi: 10.1371/journal.pone.0006309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang RT, Leung CO, Ye TM, Liu W, Chiu PC, Lam KK, et al. MicroRNA-34a suppresses invasion through downregulation of Notch1 and Jagged1 in cervical carcinoma and choriocarcinoma cells. Carcinogenesis. 2010;31(6):1037–44. doi: 10.1093/carcin/bgq066. [DOI] [PubMed] [Google Scholar]
- Parkinson DB, Bhaskaran A, Arthur-Farraj P, Noon LA, Woodhoo A, Lloyd AC, et al. c-Jun is a negative regulator of myelination. J Cell Biol. 2008;181(4):625–37. doi: 10.1083/jcb.200803013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira JA, Baumann R, Norrmen C, Somandin C, Miehe M, Jacob C, et al. Dicer in Schwann cells is required for myelination and axonal integrity. J Neurosci. 2010;30(19):6763–75. doi: 10.1523/JNEUROSCI.0801-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peru RL, Mandrycky N, Nait-Oumesmar B, Lu QR. Paving the axonal highway: from stem cells to myelin repair. Stem Cell Rev. 2008;4(4):304–18. doi: 10.1007/s12015-008-9043-z. [DOI] [PubMed] [Google Scholar]
- Potzner MR, Tsarovina K, Binder E, Penzo-Mendez A, Lefebvre V, Rohrer H, et al. Sequential requirement of Sox4 and Sox11 during development of the sympathetic nervous system. Development. 2010;137(5):775–84. doi: 10.1242/dev.042101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson WD, Kessaris N, Pringle N. Oligodendrocyte wars. Nat Rev Neurosci. 2006;7(1):11–8. doi: 10.1038/nrn1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santosh PS, Arora N, Sarma P, Pal-Bhadra M, Bhadra U. Interaction map and selection of microRNA targets in Parkinson’s disease-related genes. J Biomed Biotechnol. 2009;2009:363145. doi: 10.1155/2009/363145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer SS, Wrabetz L. Molecular mechanisms of inherited demyelinating neuropathies. Glia. 2008;56(14):1578–89. doi: 10.1002/glia.20751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, et al. Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol. 2008;15(8):842–8. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Shin JY, McManus MT, Ptacek LJ, Fu YH. Dicer ablation in oligodendrocytes provokes neuronal impairment in mice. Ann Neurol. 2009;66(6):843–57. doi: 10.1002/ana.21927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124–8. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Topilko P, Schneider-Maunoury S, Levi G, Baron-Van Evercooren A, Chennoufi AB, Seitanidou T, et al. Krox-20 controls myelination in the peripheral nervous system. Nature. 1994;371(6500):796–9. doi: 10.1038/371796a0. [DOI] [PubMed] [Google Scholar]
- Trapp BD, Nave KA. Multiple sclerosis: an immune or neurodegenerative disorder? Annu Rev Neurosci. 2008;31:247–69. doi: 10.1146/annurev.neuro.30.051606.094313. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132(5):875–86. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrier JD, Lau P, Hudson L, Murashov AK, Renne R, Notterpek L. Peripheral myelin protein 22 is regulated post-transcriptionally by miRNA-29a. Glia. 2009;57(12):1265–79. doi: 10.1002/glia.20846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrier JD, Semple-Rowland S, Madorsky I, Papin JE, Notterpek L. Reduction of Dicer impairs Schwann cell differentiation and myelination. J Neurosci Res. 2010;88(12):2558–68. doi: 10.1002/jnr.22418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo NK, Cambronne XA, Goodman RH. MicroRNA pathways in neural development and plasticity. Curr Opin Neurobiol. 2010;20(4):457–65. doi: 10.1016/j.conb.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Wegner M. A matter of identity: transcriptional control in oligodendrocytes. J Mol Neurosci. 2008;35(1):3–12. doi: 10.1007/s12031-007-9008-8. [DOI] [PubMed] [Google Scholar]
- Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137(4):647–58. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- Yu Y, Casaccia P, Lu QR. Shaping the oligodendrocyte identity by epigenetic control. Epigenetics. 2010;5(2):124–8. doi: 10.4161/epi.5.2.11160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun B, Anderegg A, Menichella D, Wrabetz L, Feltri ML, Awatramani R. MicroRNA-deficient Schwann cells display congenital hypomyelination. J Neurosci. 2010;30(22):7722–8. doi: 10.1523/JNEUROSCI.0876-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, He X, Han X, Yu Y, Ye F, Chen Y, et al. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron. 2010;65(5):612–26. doi: 10.1016/j.neuron.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, Li H, Zhu Y, Zhu Q, Qiu M. MicroRNAs are essential for the developmental switch from neurogenesis to gliogenesis in the developing spinal cord. J Neurosci. 2010;30(24):8245–50. doi: 10.1523/JNEUROSCI.1169-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]