Abstract
The migration of neurons along glial fibers from a germinal zone (GZ) to their final laminar positions is essential for morphogenesis of the developing brain, aberrations in this process are linked to profound neurodevelopmental and cognitive disorders. During this critical morphogenic movement, neurons must navigate complex migration paths, propelling their cell bodies through the dense cellular environment of the developing nervous system to their final destinations. It is not understood how neurons can successfully migrate along their glial guides through the myriad processes and cell bodies of neighboring neurons. Although much progress has been made in understanding the substrates (1–4), guidance mechanisms (5–7), cytoskeletal elements (8–10), and post-translational modifications (11–13) required for neuronal migration, we have yet to elucidate how neurons regulate their cellular interactions and adhesive specificity to follow the appropriate migratory pathways. Here I will examine recent developments in our understanding of the mechanisms controlling neuronal cell adhesion and how these mechanisms interact with crucial neurodevelopmental events, such as GZ exit, migration pathway selection, multipolar-to-radial transition, and final lamination.
Keywords: neuronal migration, glial-guided migration, endocytosis, Rap1, N-Cadherin, PAR complex
INTRODUCTION
Until the mid-1990s, adhesive mechanisms were the focus of efforts to understand the recognition and pathway-specific migration of neurons in the developing brain (14–16). Early classical ultrastructural electron microscopy (17) and correlated high-resolution time-lapse electron microscopy (18) provided tantalizing clues to the exquisite specificity of neuronal adhesion to migration substrates and the potential dynamic remodeling of neuron-glial junctions. The molecular cloning of adhesion receptors identified molecules that putatively mediate neuron-neuron or neuron-glial adhesive events (19–24). Moreover, gene targeting in the mouse showed that many of these molecules play key roles in migratory events in vivo during assembly of the brain’s cortical regions (3, 25– 29). However, it proved difficult to elucidate the molecular mechanisms that control neuronal adhesive affinity or avidity by altering levels or types of adhesion molecules expressed at the cell surface. For example, the mechanisms proposed to alter cell surface adhesion receptor strength, such as carbohydrate modification of N-CAM (30–33) or transcription of ASTN1 mRNA during cerebellar granule neuron (CGN) differentiation (16, 20), take much longer than the seconds to minutes needed to remodel neuronal junctions demonstrated by time-lapse/electron microscopy for neuron migrating along glial fibers (18). Moreover, few tools other than antibodies were available to observe the molecular components of junctions and manipulate adhesion receptor function.
Great progress in our ability to examine the molecular mechanisms controlling cell adhesion during neuronal migration have resulted from striking advances in ex vivo and in vivo manipulation of neurons, cell biology tools to alter receptor function, small-molecule inhibitors, and advanced time-lapse imaging. These tools have not only confirmed some early ultrastructure-based predictions about vesicle recycling and exocytosis but also implicated conserved polarity signaling pathways in adhesion receptor trafficking, linked conserved adhesion pathways to extrinsic signaling molecules like Reelin, and, for the first time, allowed direct visualization of adhesion receptor trafficking at the neuronal cell surface.
ENDOCYTOSIS AND NEURONAL ADHESION
Adhesion receptor trafficking has long been implicated in migration of motile cells, such as fibroblasts and leukocytes (34–38). Insertion of new adhesion receptors forward of the cell body is thought to generate traction that pulls cell components forward, while removal of adhesive elements in the rear may facilitate forward translocation. Thus, the balance between exocytosis and endocytosis and the site of adhesion receptor insertion and retrieval are major factors in the motility of non-neuronal cells.
Adhesion receptor trafficking was postulated as a general control mechanism of the substrate specificity of migrating neurons, as classical EM studies revealed clathrin-coated pits closely proximal to neuron-glial junctions of both radially and tangentially migrating neurons (17, 18, 39, 40). Not surprisingly, recent studies found that many molecules involved in neuron-neuron or neuron-glial adhesion, such as ASTN1, N-Cadherin, or integrins, are localized to the endocytic compartment near cell contact sites (41–43). Importantly, mechanistic studies clearly implicate endocytosis as a key regulator of neuronal migration (See Figure 1A and B). Segal and coworkers reported that TrkB-enriched signaling endosomes were required to orient CGN migration to the IGL in response to brain-derived neurotrophic factor (BDNF) (6, 44). Hatten and colleagues showed that the newly identified astrotactin 2 (ASTN2) protein is not a neuron-glial adhesion molecule like its homolog ASTN1; instead, it functions in CGN-glial junction formation by forming a complex with ASTN1 to regulate ASTN1 cell surface recruitment (41). Shieh et al. reported that multiple endocytic adaptor proteins are located primarily in the portion of the leading process just proximal to the neuronal cell body and that endocytic recycling of activated integrin receptors is required for the tangential migration of subventricular zone (SVZ)-a neurons (42). Interestingly, both of the latter studies found that small-molecule endocytosis inhibitors block neuronal migration, both in vitro and in ex vivo slice cultures; these results mesh nicely with findings that nonspecific inhibition of endocytosis via overexpression of dominant-negative Dynamin constructs inhibits migration. In addition, Shieh et. al. found that inhibition of endocytosis led to a acculation of adhesion receptors at the rear of the migrating neuron. Finally, Kawauchi and coworkers reported that endocytic trafficking pathways involving the Rab5, Rab7 and Rab11 proteins (small Ras-related GTPases regulating discrete stages of endocytosis) control neocortical radial migration. A Rab5/11 pathway by precisely regulating N-cadherin surface expression and Rab7 appears to controls the final somal translocation of pyramidal neurons within the cortical plate. (43). Thus, as in general cell motility models, endocytic trafficking of adhesion receptors dynamically orchestrates adhesive interactions required for neuronal migration.
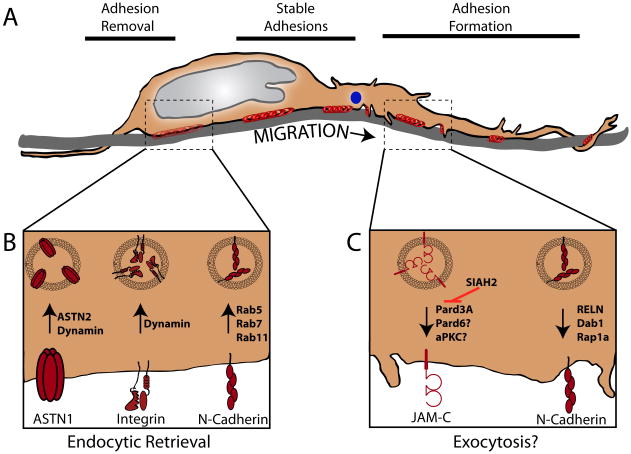
Figure 1.
A. During glial-guided of migration of CNS neurons in the developing mammalian brain neurons are polarized in the direction of migration (arrow). A leading process extends in the direction of migration, cytoplasmic organelles like the centrosome (blue circle) are often found where the leading process tapers into the neuronal cell body and the nucleus (grey oval) is located in rear the cell body. Similar to what is observed in other migrating cells, a gradient of adhesive contacts exist where neurons contact a migration substrate (glial fiber is shaded grey, adhesions are depicted in red): new adhesions are primarily located in the leading process, stable adhesions near the soma and adhesions are removed near the rear of the soma. B. Summary of recently reported signaling pathways that control adhesion receptor endocytosis: ASTN1 endocytosis is controlled by ASTN2 and dynamin in CGNs, Integrin receptor endocytosis occurs at the rear of migrating SVZa neurons via dynamin and N-Cadherin surface levels is regulated via endocytic recycling controlled by the Rab5, 7 and 11 GTPases. C. Summary of recently reported signaling pathways that control adhesion trafficking to the neuronal cell surface. JAM-C trafficking is facilitated by the Pard3A component of the PAR complex and is negatively regulated by the Siah E3 ubiquitin ligase. A RELN, Dab1 and Rap1A dependent pathway controls multipolar transition and final cortical plate positioning by regulating N-Cadherin. The precise mechanisms controlling receptor exocytosis are currently unclear.
POLARITY SIGNALING AND NEURONAL ADHESION
While the endocytosis studies described above provide some mechanistic insight into retrieval of adhesion receptors from the neuron surface, new reports suggest that conserved polarity signaling molecules, such as the partitioning defective (PAR) complex or the Rap1 GTPase regulate the extent of cell-surface adhesion receptor recruitment (Figure 1C). Moreover, fine-tuning of adhesion receptor function through polarity signaling molecules provides a new mechanism for the control of GZ exit, multipolar-to-radial migration, and final laminar positioning.
Reelin, Rap1, and N-Cadherin
The Reelin signaling pathway has long been a model for studies of the mechanisms controlling inside-out lamination of the vertebrate cerebral cortex (45, 46). Reelin is a large extracellular glycoprotein secreted by early-born cortical neurons that allows later-born neurons to migrate past them. Interestingly, although neurons can migrate along glial fibers in the absence of Reelin, its receptors (VLDLR/ApoER2) (47), or Dab1 (48, 49) (an essential signaling adaptor of the pathway), cortical layering appears to become inverted because of defective positioning within the cortical plate target area.
Despite tremendous progress in elucidating the genetics and biochemistry of Reelin pathway signaling, the precise cellular mechanism controlling selection of a final cortical plate position remains controversial. Appropriate positioning may be impeded when Reelin-deficient neurons fail to detach from glial fibers, via either a mechanism involving the α3 integrin receptor (50) or altered interactions between migrating neurons and those already positioned in the cortical plate (51, 52). Reports from the Cooper and Mueller laboratories shed new light on how Reelin signaling regulates cellular interactions (53, 54). Cortical pyramidal neurons normally migrate toward the cortical plate in three phases: multipolar migration, radial translocation along glial fibers, and somal translocation (55–57). Jossin et al. and Franco et al. used in utero electroporation, conditional mouse models, and time-lapse microscopy to elegantly show that deficiency in Reelin signaling perturbs the multipolar-to-radial migration transition and the terminal somal translocation phase within the cortical plate, without affecting migration along glial fibers. How does Reelin signaling mechanistically control these two events? Building on earlier studies from the Cooper laboratory suggesting that Reelin signaling activates the Rap1 small GTPase, Franco et al. and Yossin et al. argued compellingly that Rap1 is an essential downstream component of Reelin signaling during cerebral cortical development. First, Reelin stimulates Rap1 activation in cultured cortical neurons in both a Dab1- and VLDLR-dependent manner. Second, overexpression of Rap1GAP (a specific Rap1 inhibitor) and Rap1 silencing demonstrate that Rap1 activity is required for proper cortical plate targeting but not for migration along glial fibers; this phenotype closely resembles the deficits seen with Reeler and Dab1 loss of function. Finally, Rap1 gain of function partially rescues migration phenotypes induced by overexpression of a dominant-negative VLDR construct with altered Dab1 function and Reelin-induced Rap1 activation.
Rap1 is an ancient cell polarity signaling molecule with diverse activities. It regulates polarized cdc42 activation (in budding yeast) and cell-cell and cell-matrix interactions (in mammals) by modulating the adhesive activity of integrin or cadherin receptors (58, 59). The Cooper and Mueller laboratories report that N-Cadherin is likely a downstream functional target of the Rap1 arm of the Reelin signaling cascade. N-Cadherin, like Rap1, is required for cortical plate targeting, as N-Cadherin silencing or overexpression of a dominant-negative cadherin construct arrests migration deep within the cortical plate. Moreover, N-Cadherin gain of function rescues migration defects induced by Rap1 deficiency. How does Rap1 activity downstream of Reelin control N-Cadherin function? Jossin et al. report that Rap1 is localized to transport vesicles in cortical neurons as well as other types of migrating cells. Both Rap1 loss of function (via overexpression of Rap1GAP) and perturbation of Reelin signaling (via overexpression of a dominant negative VLDR construct) significantly reduced N-Cadherin plasma membrane levels in cultured cortical neurons; Rap1GAP overexpression also inhibited binding of cortical neurons to N-Cadherin substrates. Thus, Rap1 signaling appears to control surface N-Cadherin levels as neurons transition between migratory phases during neocortical development.
Several intriguing questions about the Reelin/Rap1/N-Cadherin signaling pathway remain. First, while both the Cooper and Mueller groups agree that terminal translocation is defective when Reelin or Rap1 is perturbed, it is unknown how multipolar-radial transition ultimately affects cortical plate targeting. Second, Franco et al. propose that terminal translocation requires the attachment of cortical neurons to elements within the marginal zone, but it is unclear what these elements are and whether attachment is N-Cadherin-dependent. Finally, it is unknown how the presumed increase in N-Cadherin adhesion is integrated with 1) Reelin activity to decrease α3 integrin adhesion during cortical plate targeting and 2) Rab GTPase–dependent endocytic retrieval of N-Cadherin, which is important for glial-guided migration.
Siah, Pard3A, and JAM-C
The PAR complex, comprising the Pard3 and Par6 adaptor proteins, atypical PKCζ, and the CDC42 Rho GTPase, is perhaps the best-characterized evolutionarily conserved cell polarity signaling entity and is crucial for tight junction formation, mitotic spindle orientation, cell migration, and axon specification in various cell polarity models (60–62). While previous studies showed that Par6α is required to organize the cytoskeletal components that coordinate CGN nucleokinesis during migration along Bergman glial fibers (10, 63), the role of other PAR complex components and the identity of upstream regulators of polarity during migration are relatively unexplored. Recent examination of the regulation of PAR complex during neuronal differentiation revealed a surprising signaling pathway that controls adhesion receptor trafficking as CGNs exit their germinal zone niche (64). Developing CGNs are an excellent model for analyzing the mechanisms regulating GZ exit and for elucidating migration pathway selection, as they undergo two migration phases (65–67): tangential migration near the cerebellar surface followed by radial migration away from the EGL, during which CGNs cross the molecular layer (ML) and traverse to a final site within the internal granule layer (IGL). Interestingly, Pard3A expression is low in immature CGNs and increases as CGNs terminally differentiate, suggesting that it plays a role in GZ exit or the switch from tangential to radial migration. Indeed, systematic gain- or loss-of-function analyses in ex vivo cerebellar slices confirmed that Pard3A activity is necessary and sufficient for CGN GZ exit.
What controls Pard3A levels in differentiating CGNs? Famulski et al. used a two-hybrid screen to identify PAR complex–binding proteins and showed the seven in absentia homolog (Siah) family of E3 ubiquitin ligases to be key regulators of Pard3A. The interaction of Pard3A and Siah ubiquitin ligases requires Siah degron sequences within Pard3A and the Sina substrate-binding domain of Siah. Moreover, Siah ligases appear to antagonize Pard3A function, as Siah overexpression can induce proteosomal degradation of Pard3A, while levels of Pard6 or aPKCζ are unaffected. During cerebellar development, Siah expression is complementary to Pard3A expression: high in undifferentiated CGNs and extinguished during terminal differentiation. Systematic gain- or loss-of-function analyses in ex vivo cerebellar slices show that Siah activity is necessary and sufficient to maintain immature CGNs within the EGL and that Siah inhibition of GZ exit is dependent on its targeting of Pard3A for degradation, as Pard3A overexpression rescues any Siah-dependent inhibition of migration. Interestingly, long-term time-lapse imaging in cerebellar slices revealed that excess Siah activity does not deter the motility of CGNs but restricts their movement to the EGL, blocking radial migration to the IGL. Thus, the apparent antagonistic relationship between Siah and Pard3A controls migration initiation and pathway selection in developing CGNs.
Regulation of cell adhesion through junctional adhesion molecule (JAM)-C is a major downstream function of Siah and Pard3A. JAM-C, a member of the immunoglobulin superfamily, is an essential component of epithelial tight junctions and is reported to interact directly with Pard3A via a JAM-C cytoplasmic PDZ binding motif (68–72). In epithelial cells, this interaction is essential for formation of tight junctions and-JAM-C recruitment to tight junctions. Interestingly, not only is JAM-C expressed in differentiating CGNs but JAM-C-mediated adhesion is also necessary and sufficient for CGN GZ exit. Moreover, Pard3A activity is essential to recruit JAM-C to neuron-neuron or neuron-glial cell contacts, and disruption of Pard3A/JAM-C interaction by overexpression of the Pard3A binding site in JAM-C blocks CGN movement to the IGL. Thus, Pard3A’s activity in promoting GZ exit of differentiated CGNs through JAM-C recruitment to the plasma membrane facilitates the polarity-dependent integration of new neurons into the cerebellar cortex.
Several intriguing questions about the Siah/Pard3A/JAM-C pathway remain. First, it is currently unclear what upstream signals impinge on the Siah E3 ubiqutin ligase controls Pard3A turnover during neuronal differentiation. Interestingly, studies from other model systems suggest that Siah may be a downstream component of the Ras signaling pathway and a target of hypoxia signaling (73), suggesting that these pathways may intersect with polarity signaling during CGN development. Second, it is also unclear how Pard3A controls JAM-C surface recruitment. Recent studies in C. elegans and cultured fibroblasts implicate PAR3, aPKC and the NUMB polarity protein in surface receptor trafficking (74, 75). It will be interesting to determine if similar endocytosic mechanisms or perhaps exocytosis controls the PAR complex dependent recruitment of JAM-C during tangential to radial migration switch.
NEURONAL ADHESION DYNAMICS
In general models of cell motility, adhesion formation and disassembly drive migration by regulating how a cell binds to actomyosin and uses it to pull against migration substrates (76, 77). While various adhesion molecules are known to mediate binding of neurons to their glial or nonglial guides (1–3), we lack the tools to examine the basic features of neuronal adhesion in living cells in a dynamic, high-throughput fashion.
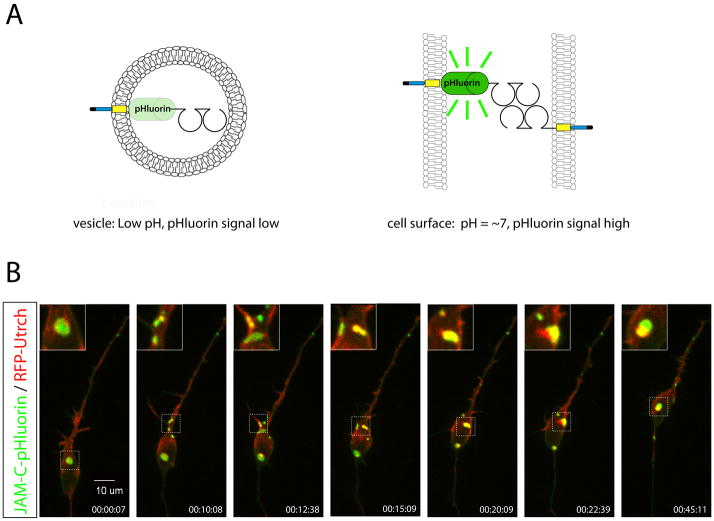
Two recent studies using fluorescence-labeled adhesion molecules and live cell imaging to examine CGN radial migration demonstrated that adhesion dynamics are linked to the saltatory movement of neurons along their glial guide. In the first study, Wilson et al. tracked neuron-glial attachment sites with an ASTN1-Venus fusion protein(41). During early CGN migration, when the neuron is presumably attached to the glial fiber, ASTN1-Venus was located in the leading aspect of the neuronal soma, where a specialized “interstitial junction” had previously been observed in correlated time-lapse microscopy and EM studies. As CGNs translocated along the glial fiber and the somal junction was released, ASTN1-Venus flowed from the soma to the proximal domain of the leading process, where a new adhesion site would form. In the second study, Famulski et al. developed a novel neuronal adhesion live imaging probe by fusing pHluorin (78) to the extracellular domain of JAM-C, a PAR complex–dependent adhesion molecule required for CGN radial migration (64). A similar approach has been to track SYNCAM1 trafficking at putative synaptic connections (79). pHluorin pH sensitivity ensures that JAM-C-pHluorin fluoresces only as the JAM-C extracellular domain traffics to cell contacts, allowing direct examination of cell surface adhesion dynamics (Figure 2A). Surface JAM-C was observed at punctate junctions within the leading process and in larger adhesion plaques in the neuronal soma (Figure 2B). During the movement cycle, JAM-C puncta appeared in the f-actin–rich region at the base of the leading process, where actomyosin activity helps to power nucleokinesis. As the soma translocated forward, these leading-process junctions coalesced into the larger plaques seen in the leading portion of the soma. Surprisingly, the soma translocated past large adhesion plaques originally located in the leading somal pole, and the plaques subsequently disassembled within the trailing process. Interestingly, f-actin was heavily enriched at sites of surface JAM-C recruitment at two points of the cycle: after JAM-C puncta appeared in the leading process and at the time of their disassembly in the trailing process. The recruitment of f-actin to newly formed JAM-C puncta provides strong evidence that these locations are bona fide cell-cell adhesion sites and that the leading process is the locus of strong traction forces during migration. In future studies, it will be important to determine whether these adhesion dynamics observed in CGNs are applicable to other neuronal cell types. It will also be important to examine the precise relationship of actomyosin activity to neuron-glial adhesion sites in the leading process, as recent evidence suggests that actomyosin plays a direct role in the leading portion of the cell (10), where myosin II–based contractions generated forward of the nucleus are required for efficient adhesion and cell motility.
Figure 2.
A. Rational of JAM-C-pHluorin time lapse imaging probe. B. Time lapse spinning disk confocal imaging of CGNs nucleofected with JAM-C-pHluorin (green) and RFP-UTRCH (red), an F-actin probe. During CGN migration JAM-C-pHluorin puncta co-localize with UTRCH-RFP (yellow signal within enlarged insets). RFP-UTRCH quickly co-localized with newly formed JAM-C-pHluorin puncta, indicating that JAM-C-pHluorin puncta are sites of F-actin recruitment and therefore are bona fide cell-cell adhesion sites. Both panels reproduced with permission of Famulski et al.
CONCLUSIONS
Over the past two decades, much progress has been made in identifying molecules that mediate the interaction of migrating neurons with glial or neuronal substrates on their journey to a final laminar position. Clearly, regulation of the affinity or avidity of the array of cell-surface adhesion receptors is crucial to guide a neuron’s migration path. An important challenge facing this field is to identify how cell-extrinsic cues cooperate with both the genetic programs controlling neuronal maturation and cell-intrinsic adhesion receptor trafficking mechanisms to regulate how immature neurons initiate, execute, and terminate this essential nervous system morphogenic event.
Acknowledgments
I thank Niraj Trivedi and Danielle Howell for critically reading the manuscript and helpful discussion. Sharon Naron provided expert editorial support. Funded by the American Lebanese Syrian Associated Charities (ALSAC) and by Grant Number 1R01NS066936 from the National Institute Of Neurological Disorders (NINDS). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NINDS or the NIH.
References
- 1.Fishell G, Hatten ME. Astrotactin provides a receptor system for CNS neuronal migration. Development. 1991 Nov;113:755. doi: 10.1242/dev.113.3.755. [DOI] [PubMed] [Google Scholar]
- 2.Elias LA, Wang DD, Kriegstein AR. Gap junction adhesion is necessary for radial migration in the neocortex. Nature. 2007 Aug 23;448:901. doi: 10.1038/nature06063. [DOI] [PubMed] [Google Scholar]
- 3.Anton ES, Kreidberg JA, Rakic P. Distinct functions of alpha3 and alpha(v) integrin receptors in neuronal migration and laminar organization of the cerebral cortex. Neuron. 1999 Feb;22:277. doi: 10.1016/s0896-6273(00)81089-2. [DOI] [PubMed] [Google Scholar]
- 4.Anton ES, Marchionni MA, Lee KF, Rakic P. Role of GGF/neuregulin signaling in interactions between migrating neurons and radial glia in the developing cerebral cortex. Development. 1997 Sep;124:3501. doi: 10.1242/dev.124.18.3501. [DOI] [PubMed] [Google Scholar]
- 5.Polleux F, Whitford KL, Dijkhuizen PA, Vitalis T, Ghosh A. Control of cortical interneuron migration by neurotrophins and PI3-kinase signaling. Development. 2002 Jul;129:3147. doi: 10.1242/dev.129.13.3147. [DOI] [PubMed] [Google Scholar]
- 6.Zhou P, et al. Polarized signaling endosomes coordinate BDNF-induced chemotaxis of cerebellar precursors. Neuron. 2007 Jul 5;55:53. doi: 10.1016/j.neuron.2007.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Renaud J, et al. Plexin-A2 and its ligand, Sema6A, control nucleus-centrosome coupling in migrating granule cells. Nat Neurosci. 2008 Apr;11:440. doi: 10.1038/nn2064. [DOI] [PubMed] [Google Scholar]
- 8.Schaar BT, McConnell SK. Cytoskeletal coordination during neuronal migration. Proc Natl Acad Sci U S A. 2005 Sep 20;102:13652. doi: 10.1073/pnas.0506008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci. 2007 Jul 8;10:970. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- 10.Solecki DJ, et al. Myosin II motors and F-actin dynamics drive the coordinated movement of the centrosome and soma during CNS glial-guided neuronal migration. Neuron. 2009 Jul 16;63:63. doi: 10.1016/j.neuron.2009.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patrick GN, Zhou P, Kwon YT, Howley PM, Tsai LH. p35, the neuronal-specific activator of cyclin-dependent kinase 5 (Cdk5) is degraded by the ubiquitin-proteasome pathway. J Biol Chem. 1998 Sep 11;273:24057. doi: 10.1074/jbc.273.37.24057. [DOI] [PubMed] [Google Scholar]
- 12.Suetsugu S, et al. Regulation of actin cytoskeleton by mDab1 through N-WASP and ubiquitination of mDab1. Biochem J. 2004 Nov 15;384:1. doi: 10.1042/BJ20041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karakuzu O, Wang DP, Cameron S. MIG-32 and SPAT-3A are PRC1 homologs that control neuronal migration in Caenorhabditis elegans. Development. 2009 Mar;136:943. doi: 10.1242/dev.029363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rakic P, Cameron RS, Komuro H. Recognition, adhesion, transmembrane signaling and cell motility in guided neuronal migration. Curr Opin Neurobiol. 1994 Feb;4:63. doi: 10.1016/0959-4388(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 15.Rakic P. Contact Regulation of Neuronal Migration. In: Edelman GM, Thiery JP, editors. The Cell in Contact: Adhesion and Junctions of Morphogenetic Determinants. Wiley and Sons; New York: 1985. [Google Scholar]
- 16.Hatten ME, Mason CA. Mechanisms of glial-guided neuronal migration in vitro and in vivo. Experientia. 1990 Sep 15;46:907. doi: 10.1007/BF01939383. [DOI] [PubMed] [Google Scholar]
- 17.Rakic P. Neuron-glia relationship during granule cell migration in developing cerebellar cortex. A Golgi and electronmicroscopic study in Macacus Rhesus. J Comp Neurol. 1971 Mar;141:283. doi: 10.1002/cne.901410303. [DOI] [PubMed] [Google Scholar]
- 18.Gregory WA, Edmondson JC, Hatten ME, Mason CA. Cytology and neuron-glial apposition of migrating cerebellar granule cells in vitro. J Neurosci. 1988 May;8:1728. doi: 10.1523/JNEUROSCI.08-05-01728.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cunningham BA, et al. Neural cell adhesion molecule: structure, immunoglobulin-like domains, cell surface modulation, and alternative RNA splicing. Science. 1987 May 15;236:799. doi: 10.1126/science.3576199. [DOI] [PubMed] [Google Scholar]
- 20.Zheng C, Heintz N, Hatten ME. CNS gene encoding Astrotactin, which supports neuronal migration along glial fibers. Science. 1996;272:417. doi: 10.1126/science.272.5260.417. [DOI] [PubMed] [Google Scholar]
- 21.Grumet M, Mauro V, Burgoon MP, Edelman GM, Cunningham BA. Structure of a new nervous system glycoprotein, Nr-CAM, and its relationship to subgroups of neural cell adhesion molecules. The Journal of cell biology. 1991 Jun;113:1399. doi: 10.1083/jcb.113.6.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prince JT, Alberti L, Healy PA, Nauman SJ, Stallcup WB. Molecular cloning of NILE glycoprotein and evidence for its continued expression in mature rat CNS. Journal of neuroscience research. 1991 Nov;30:567. doi: 10.1002/jnr.490300315. [DOI] [PubMed] [Google Scholar]
- 23.Prince JT, Milona N, Stallcup WB. Characterization of a partial cDNA clone for the NILE glycoprotein and identification of the encoded polypeptide domain. The Journal of neuroscience: the official journal of the Society for Neuroscience. 1989 May;9:1825. doi: 10.1523/JNEUROSCI.09-05-01825.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furley AJ, et al. The axonal glycoprotein TAG-1 is an immunoglobulin superfamily member with neurite outgrowth-promoting activity. Cell. 1990 Apr 6;61:157. doi: 10.1016/0092-8674(90)90223-2. [DOI] [PubMed] [Google Scholar]
- 25.Adams NC, Tomoda T, Cooper M, Dietz G, HME Mice that lack astrotactin have slowed neuronal migration. Development. 2002;129:965. doi: 10.1242/dev.129.4.965. [DOI] [PubMed] [Google Scholar]
- 26.Sakurai T, et al. Overlapping functions of the cell adhesion molecules Nr-CAM and L1 in cerebellar granule cell development. The Journal of cell biology. 2001 Sep 17;154:1259. doi: 10.1083/jcb.200104122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Demyanenko GP, et al. Close homolog of L1 modulates area-specific neuronal positioning and dendrite orientation in the cerebral cortex. Neuron. 2004 Oct 28;44:423. doi: 10.1016/j.neuron.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 28.Belvindrah R, Graus-Porta D, Goebbels S, Nave KA, Muller U. Beta1 integrins in radial glia but not in migrating neurons are essential for the formation of cell layers in the cerebral cortex. J Neurosci. 2007 Dec 12;27:13854. doi: 10.1523/JNEUROSCI.4494-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belvindrah R, Hankel S, Walker J, Patton BL, Muller U. Beta1 integrins control the formation of cell chains in the adult rostral migratory stream. J Neurosci. 2007 Mar 7;27:2704. doi: 10.1523/JNEUROSCI.2991-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorkin BC, Hoffman S, Edelman GM, Cunningham BA. Sulfation and phosphorylation of the neural cell adhesion molecule, N-CAM. Science. 1984 Sep 28;225:1476. doi: 10.1126/science.6474186. [DOI] [PubMed] [Google Scholar]
- 31.Chuong CM, Edelman GM. Alterations in neural cell adhesion molecules during development of different regions of the nervous system. The Journal of neuroscience: the official journal of the Society for Neuroscience. 1984 Sep;4:2354. doi: 10.1523/JNEUROSCI.04-09-02354.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rutishauser U. NCAM and its polysialic acid moiety: a mechanism for pull/push regulation of cell interactions during development? Development. 1992;99 [PubMed] [Google Scholar]
- 33.Rutishauser U, Acheson A, Hall AK, Mann DM, Sunshine J. The neural cell adhesion molecule (NCAM) as a regulator of cell-cell interactions. Science. 1988 Apr 1;240:53. doi: 10.1126/science.3281256. [DOI] [PubMed] [Google Scholar]
- 34.Bretscher MS. Distribution of receptors for transferrin and low density lipoprotein on the surface of giant HeLa cells. Proceedings of the National Academy of Sciences of the United States of America. 1983 Jan;80:454. doi: 10.1073/pnas.80.2.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bretscher MS. Getting membrane flow and the cytoskeleton to cooperate in moving cells. Cell. 1996 Nov 15;87:601. doi: 10.1016/s0092-8674(00)81380-x. [DOI] [PubMed] [Google Scholar]
- 36.Bretscher MS. Moving membrane up to the front of migrating cells. Cell. 1996 May 17;85:465. doi: 10.1016/s0092-8674(00)81246-5. [DOI] [PubMed] [Google Scholar]
- 37.Bretscher MS. Endocytosis: relation to capping and cell locomotion. Science. 1984 May 18;224:681. doi: 10.1126/science.6719108. [DOI] [PubMed] [Google Scholar]
- 38.Lawson MA, Maxfield FR. Ca(2+)- and calcineurin-dependent recycling of an integrin to the front of migrating neutrophils. Nature. 1995 Sep 7;377:75. doi: 10.1038/377075a0. [DOI] [PubMed] [Google Scholar]
- 39.O’Rourke NA, Sullivan DP, Kaznowski CE, Jacobs AA, McConnell SK. Tangential migration of neurons in the developing cerebral cortex. Development. 1995 Jul;121:2165. doi: 10.1242/dev.121.7.2165. [DOI] [PubMed] [Google Scholar]
- 40.Rakic P. Mode of cell migration to the superficial layers of fetal monkey neocortex. J Comp Neurol. 1972 May;145:61. doi: 10.1002/cne.901450105. [DOI] [PubMed] [Google Scholar]
- ••41.Wilson PM, Fryer RH, Fang Y, Hatten ME. Astn2, a novel member of the astrotactin gene family, regulates the trafficking of ASTN1 during glial-guided neuronal migration. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2010 Jun 23;30:8529. doi: 10.1523/JNEUROSCI.0032-10.2010. This study critically demonstrates not only that endocytosis regulates surface retrieval of the neuron-glial adhesion molecule ASTN1, but also reveals a surprising role for the closely related ASTN2 molecule in controling ASTN1 endocytosis. Small molecule inhibition of dyanmin blocks CGN migration to the IGL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••42.Shieh JC, Schaar BT, Srinivasan K, Brodsky FM, McConnell SK. Endocytosis regulates cell soma translocation and the distribution of adhesion proteins in migrating neurons. PloS one. 2011;6:e17802. doi: 10.1371/journal.pone.0017802. This study demonstrates that endocytic adaptors; like a-adaptin and ap-2, as well as clathrin coated vesicles containing activated integrin adhesion receptors are enriched in the SVZa neuronal leading process. Small molecule inhbition of dyanmin blocks SVZa migration and leads to an erichment of clathrin coated vesicles, endocytic adaptors and integrin receptors in the trailing aspect of the SVZa neuron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••43.Kawauchi T, et al. Rab GTPases-dependent endocytic pathways regulate neuronal migration and maturation through N-cadherin trafficking. Neuron. 2010 Aug 26;67:588. doi: 10.1016/j.neuron.2010.07.007. This study demonstrates a critical role for the Rab5, 7, and 11 small GTPases in endocytosis control and recycling during coritcal pyramidal neuron migration. A Rab5 and Rab11 pathway regulates surface expression of N-Cadherin in migrating neurons such that inhbition of enocytosis leads to increased surface N-Cadherin, excessive adhesion and perturbation in migration. Rab7 appears to regulate final somal translocation within the cortical plate. [DOI] [PubMed] [Google Scholar]
- ••44.Zhou P, et al. Numb links extracellular cues to intracellular polarity machinery to promote chemotaxis. Developmental cell. 2011 May 17;20:610. doi: 10.1016/j.devcel.2011.04.006. This study demonstrates that Numb and the aPKC component of the PAR complex cooperatively regulate polarized TrkB receptor endocytosis during CGN glial guided migration. Genetic deletion of Numb slows CGN movement to the IGL by perturbing chemotaxis to a BDNF gradient previously characterized by the Segal laboratory. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rice DS, Curran T. Role of the reelin signaling pathway in central nervous system development. Annual review of neuroscience. 2001;24:1005. doi: 10.1146/annurev.neuro.24.1.1005. [DOI] [PubMed] [Google Scholar]
- 46.Tissir F, Goffinet AM. Reelin and brain development. Nature reviews. Neuroscience. 2003 Jun;4:496. doi: 10.1038/nrn1113. [DOI] [PubMed] [Google Scholar]
- 47.Trommsdorff M, et al. Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell. 1999 Jun 11;97:689. doi: 10.1016/s0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- 48.Howell BW, Hawkes R, Soriano P, Cooper JA. Neuronal position in the developing brain is regulated by mouse disabled-1. Nature. 1997 Oct 16;389:733. doi: 10.1038/39607. [DOI] [PubMed] [Google Scholar]
- 49.Sheldon M, et al. Scrambler and yotari disrupt the disabled gene and produce a reeler-like phenotype in mice. Nature. 1997 Oct 16;389:730. doi: 10.1038/39601. [DOI] [PubMed] [Google Scholar]
- 50.Dulabon L, et al. Reelin binds alpha3beta1 integrin and inhibits neuronal migration. Neuron. 2000 Jul;27:33. doi: 10.1016/s0896-6273(00)00007-6. [DOI] [PubMed] [Google Scholar]
- 51.Pinto Lord MC, Caviness VS., Jr Determinants of cell shape and orientation: a comparative Golgi analysis of cell-axon interrelationships in the developing neocortex of normal and reeler mice. The Journal of comparative neurology. 1979 Sep 1;187:49. doi: 10.1002/cne.901870104. [DOI] [PubMed] [Google Scholar]
- 52.Tabata H, Nakajima K. Neurons tend to stop migration and differentiate along the cortical internal plexiform zones in the Reelin signal-deficient mice. Journal of neuroscience research. 2002 Sep 15;69:723. doi: 10.1002/jnr.10345. [DOI] [PubMed] [Google Scholar]
- ••53.Franco SJ, Martinez-Garay I, Gil-Sanz C, Harkins-Perry SR, Muller U. Reelin regulates cadherin function via Dab1/Rap1 to control neuronal migration and lamination in the neocortex. Neuron. 2011 Feb 10;69:482. doi: 10.1016/j.neuron.2011.01.003. This study utilized an elegant mix of conditional mouse genetics and time-lapse imaging to show that the Dab1 component of the RELN signaling cascade is required for the terminal somal translocation of both early and late born cortical pyramidal neurons. Additionally, epistasis analysis showed that the Rap1A small GTPase and N-Cadherin are functionally downstream of Dab1, suggesting that RELN signaling controls N-Cadherin function during the final somal translocation phase of migration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••54.Jossin Y, Cooper JA. Reelin, Rap1 and N-cadherin orient the migration of multipolar neurons in the developing neocortex. Nature neuroscience. 2011 Jun;14:697. doi: 10.1038/nn.2816. This study utilized an elegant mix of in utero electroporation, systmatic necessity/sufficiency testing, time-lapse imaging and cell biological techniques to show that Rap1A activity, downstream of RELN, controls surface expression and adhesion by N-Cadherin during cortical pyramidal neuron migration to cortical plate. The authors propose that N-Cadherin surface levels control the transition out multipolar state and regulates the final position of neurons within the cortical plate. While reference 53 and 54 agree that Rap1 and N-Cadherin are downstream of RELN, the role of multipolar transition and terminal translocation has not been fully reconciled. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tabata H, Nakajima K. Multipolar migration: the third mode of radial neuronal migration in the developing cerebral cortex. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2003 Nov 5;23:9996. doi: 10.1523/JNEUROSCI.23-31-09996.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004 Feb;7:136. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 57.Nadarajah B, Brunstrom JE, Grutzendler J, Wong RO, Pearlman AL. Two modes of radial migration in early development of the cerebral cortex. Nat Neurosci. 2001 Feb;4:143. doi: 10.1038/83967. [DOI] [PubMed] [Google Scholar]
- 58.Bos JL. Linking Rap to cell adhesion. Current opinion in cell biology. 2005 Apr;17:123. doi: 10.1016/j.ceb.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 59.Bos JL, de Rooij J, Reedquist KA. Rap1 signalling: adhering to new models. Nature reviews. Molecular cell biology. 2001 May;2:369. doi: 10.1038/35073073. [DOI] [PubMed] [Google Scholar]
- 60.Munro EM. PAR proteins and the cytoskeleton: a marriage of equals. Curr Opin Cell Biol. 2006 Feb;18:86. doi: 10.1016/j.ceb.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 61.Barnes AP, Polleux F. Establishment of axon-dendrite polarity in developing neurons. Annu Rev Neurosci. 2009;32:347. doi: 10.1146/annurev.neuro.31.060407.125536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nance J, Zallen JA. Elaborating polarity: PAR proteins and the cytoskeleton. Development. 2011 Mar;138:799. doi: 10.1242/dev.053538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Solecki DJ, Model L, Gaetz J, Kapoor TM, Hatten ME. Par6alpha signaling controls glial-guided neuronal migration. Nat Neurosci. 2004 Nov;7:1195. doi: 10.1038/nn1332. [DOI] [PubMed] [Google Scholar]
- ••64.Famulski JK, et al. Siah Regulation of Pard3A Controls Neuronal Cell Adhesion During Germinal Zone Exit. Science. 2010 Nov 25; doi: 10.1126/science.1198480. This study used ex vivo cerebellar slices, systematic necessity-sufficiency testing and quantitative time-lapse imaging to reveal a key neuronal polarity-signaling cascade converging on the Pard3A polarity protein including both upstream regulatory (i.e. Siah2 ubiquitin ligase) and downstream adhesive effector (i.e. JAM-C) components. The authors propose that an antagonistic relationship between Siah and Pard3A regulates the amount of JAM-C adhesion receptor at the CGN cell surface, ultimately controlling germinal zone exit and tangential to radial migration of CGNs as they move to the IGL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edmondson JC, Hatten ME. Glial-guided granule neuron migration in vitro: a high-resolution time-lapse video microscopic study. J Neurosci. 1987 Jun;7:1928. doi: 10.1523/JNEUROSCI.07-06-01928.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ryder EF, Cepko CL. Migration patterns of clonally related granule cells and their progenitors in the developing chick cerebellum. Neuron. 1994 May;12:1011. doi: 10.1016/0896-6273(94)90310-7. [DOI] [PubMed] [Google Scholar]
- 67.Komuro H, Yacubova E, Rakic P. Mode and tempo of tangential cell migration in the cerebellar external granular layer. J Neurosci. 2001 Jan 15;21:527. doi: 10.1523/JNEUROSCI.21-02-00527.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ebnet K, et al. The cell polarity protein ASIP/PAR-3 directly associates with junctional adhesion molecule (JAM) EMBO J. 2001 Jul 16;20:3738. doi: 10.1093/emboj/20.14.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ebnet K, Suzuki A, Ohno S, Vestweber D. Junctional adhesion molecules (JAMs): more molecules with dual functions? J Cell Sci. 2004 Jan 1;117:19. doi: 10.1242/jcs.00930. [DOI] [PubMed] [Google Scholar]
- 70.Ooshio T, et al. Cooperative roles of Par-3 and afadin in the formation of adherens and tight junctions. Journal of cell science. 2007 Jul 15;120:2352. doi: 10.1242/jcs.03470. [DOI] [PubMed] [Google Scholar]
- 71.Fukuhara A, et al. Involvement of nectin in the localization of junctional adhesion molecule at tight junctions. Oncogene. 2002 Oct 31;21:7642. doi: 10.1038/sj.onc.1205875. [DOI] [PubMed] [Google Scholar]
- 72.Kuramitsu K, Ikeda W, Inoue N, Tamaru Y, Takai Y. Novel role of nectin: implication in the co-localization of JAM-A and claudin-1 at the same cell-cell adhesion membrane domain. Genes to cells: devoted to molecular & cellular mechanisms. 2008 Aug;13:797. doi: 10.1111/j.1365-2443.2008.01206.x. [DOI] [PubMed] [Google Scholar]
- 73.House CM, Moller A, Bowtell DD. Siah proteins: novel drug targets in the Ras and hypoxia pathways. Cancer Res. 2009 Dec 1;69:8835. doi: 10.1158/0008-5472.CAN-09-1676. [DOI] [PubMed] [Google Scholar]
- 74.Balklava Z, Pant S, Fares H, Grant BD. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nature cell biology. 2007 Sep;9:1066. doi: 10.1038/ncb1627. [DOI] [PubMed] [Google Scholar]
- 75.Nishimura T, Kaibuchi K. Numb controls integrin endocytosis for directional cell migration with aPKC and PAR-3. Developmental cell. 2007 Jul;13:15. doi: 10.1016/j.devcel.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 76.Ridley AJ, et al. Cell migration: integrating signals from front to back. Science. 2003 Dec 5;302:1704. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 77.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009 Nov;10:778. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miesenbock G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998 Jul 9;394:192. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- ••79.Stagi M, Fogel AI, Biederer T. SynCAM 1 participates in axo-dendritic contact assembly and shapes neuronal growth cones. Proceedings of the National Academy of Sciences of the United States of America. 2010 Apr 20;107:7568. doi: 10.1073/pnas.0911798107. This study was the first to use a pHluorin fusioin protein to quantitatively examine cell surface recruitment of the neuronal adhesion receptor Syncam1 in living neurons. [DOI] [PMC free article] [PubMed] [Google Scholar]