Abstract
BACKGROUND
The use of tyrosine kinase inhibitors to target the epidermal growth factor receptor gene (EGFR) in patients with non–small-cell lung cancer is effective but limited by the emergence of drug-resistance mutations. Molecular characterization of circulating tumor cells may provide a strategy for noninvasive serial monitoring of tumor genotypes during treatment.
METHODS
We captured highly purified circulating tumor cells from the blood of patients with non–small-cell lung cancer using a microfluidic device containing microposts coated with antibodies against epithelial cells. We performed EGFR mutational analysis on DNA recovered from circulating tumor cells using allele-specific polymerase-chain-reaction amplification and compared the results with those from concurrently isolated free plasma DNA and from the original tumor-biopsy specimens.
RESULTS
We isolated circulating tumor cells from 27 patients with metastatic non–small-cell lung cancer (median number, 74 cells per milliliter). We identified the expected EGFR activating mutation in circulating tumor cells from 11 of 12 patients (92%) and in matched free plasma DNA from 4 of 12 patients (33%) (P = 0.009). We detected the T790M mutation, which confers drug resistance, in circulating tumor cells collected from patients with EGFR mutations who had received tyrosine kinase inhibitors. When T790M was detectable in pretreatment tumor-biopsy specimens, the presence of the mutation correlated with reduced progression-free survival (7.7 months vs. 16.5 months, P<0.001). Serial analysis of circulating tumor cells showed that a reduction in the number of captured cells was associated with a radiographic tumor response; an increase in the number of cells was associated with tumor progression, with the emergence of additional EGFR mutations in some cases.
CONCLUSIONS
Molecular analysis of circulating tumor cells from the blood of patients with lung cancer offers the possibility of monitoring changes in epithelial tumor genotypes during the course of treatment.
Increasing knowledge of molecular abnormalities that drive human cancers offers the promise of therapies targeted at specific genetic lesions.1,2 Genetic abnormalities may define a cancer at diagnosis, but mutations, some of which lead to acquired drug resistance, may emerge during treatment. For many epithelial cancers, minimally invasive biopsies provide insufficient material for molecular analysis at diagnosis, and tumors typically are not sampled repeatedly during treatment to monitor changes in genetic abnormalities. Although tumor cells are known to circulate in the blood of patients with metastatic cancer,3 their use in monitoring of tumor genotypes has been limited by relatively insensitive detection strategies.4,5 The detection of circulating tumor cells in some patients with the use of magnetic bead–conjugated antibodies against epithelial-cell adhesion molecule (EpCAM) may be useful as a prognostic marker.6–9 However, the small number of circulating tumor cells isolated by this method is below the dynamic range required for measuring treatment response, and the low purity of such cells prevents reliable molecular analyses.10
We recently developed a microfluidic-based device (called the CTC-chip) that can isolate, quantify, and analyze circulating tumor cells from a blood sample. In the CTC-chip, blood flows past 78,000 EpCAM-coated microposts under controlled conditions that optimize the capture of circulating tumor cells.11 An average of 132 circulating tumor cells per milliliter (median, 67 cells per milliliter) are isolated at high purity from virtually all tested patients with metastatic cancers — including non–small-cell lung cancer and prostate, pancreas, breast, and colorectal cancers — but not from healthy controls.11 The prevalence and quantity of circulating tumor cells that are isolated from patients with advanced cancer may thus provide a measure of tumor response, whereas the high purity of such cells allows repeated analysis of molecular markers.
Tumor-associated activating mutations in the epidermal growth factor receptor (EGFR) gene identify patients with non–small-cell lung cancer who have a dramatic response to EGFR tyrosine kinase inhibitors, including gefitinib (Iressa) and erlotinib (Tarceva).12–14 However, most patients have a relapse within 1 year after the initiation of therapy.15 Studies of tumors at relapse show the acquisition of a secondary EGFR mutation, in which methionine is substituted for threonine at position 790 (T790M). This mutation hinders drug binding but may be susceptible to second-generation, “irreversible” tyrosine kinase inhibitors, which form covalent cross-links with the receptors.16–18 Other mechanisms of resistance to tyrosine kinase inhibitors have also been reported.19,20 We tested the ability of microfluidic techniques to isolate a sufficient number of circulating tumor cells from patients with non–small-cell lung cancer to permit mutational analysis of EGFR.
METHODS
PATIENTS AND CLINICAL SPECIMENS
Patients with advanced non–small-cell lung cancer were recruited according to one of two protocols that were approved by the institutional review board. A total of 31 patients in Group A (Patients 1 to 27 and 43 to 46), who were treated at the Massachusetts General Hospital Cancer Center, donated 10 ml of blood on one or more occasions for CTC-chip analysis. Blood samples were analyzed for the quantity of circulating tumor cells11 (for details, see the Methods section in the Supplementary Appendix, available with the full text of this article at www.nejm.org). We analyzed circulating tumor cells, free plasma DNA, archived paraffin-embedded tumor tissue, or all three specimens for EGFR mutations using the Scorpion Amplification Refractory Mutation System (SARMS) technology (DxS), standard nucleotide sequencing, or both.
The number of tumor-biopsy specimens that were available for comparison of EGFR sequencing and SARMS analysis was extended by the inclusion of 15 patients in Group B (Patients 28 to 42) who had participated in a multicenter clinical trial of gefitinib21 but were not available for the analysis of circulating tumor cells. We reviewed the medical charts of all patients, and an independent radiologist quantified the tumor burden at various times as the sum of the unidimensional size of all measurable tumor sites, according to the Response Evaluation Criteria in Solid Tumors (RECIST).22 Patients who had been treated with an EGFR tyrosine kinase inhibitor (gefitinib or erlotinib) were assessed for the best response to therapy with the use of RECIST.
MOLECULAR ANALYSIS
DNA that was extracted from captured circulating tumor cells with the use of a PicoPure DNA Extraction Kit (Molecular Devices) was subjected to two rounds of linear amplification with a TransPlex amplification kit (Rubicon Genomics). DNA from plasma was isolated with the use of plasma preparation tubes (Vacutainer PPT) and the QIAmp DNA Blood Midi Kit (Fisher Scientific) and a standard method using proteinase K. For identification of EGFR mutations with the SARMS assay, 1.5 ng of DNA was analyzed with the use of ABI 7500 Real-Time PCR System (Applied Biosystems). The assay detects grouped deletions within exon 19, insertions within exon 20, and mutations affecting codon 719 (G719X), as well as the individual mutations T790M, L858R, L861Q, and S768I. The rate of amplification of these mutant alleles was compared with that of EGFR exon 2 as an internal control. Standard bidirectional nucleotide sequencing was performed with the use of dye terminator chemistry and a Capillary ABI 3100 sequencer (Applied Biosystems).
STATISTICAL ANALYSIS
The relationship between the quantity of circulating tumor cells and tumor burden was analyzed with the use of Spearman’s correlation coefficient. Fisher’s exact test was used to compare mutations that were identified in different populations. The relationship between patients’ baseline T790M status and progression-free survival (the time between the initiation of therapy with a tyrosine kinase inhibitor and either tumor progression or death) was analyzed with a multivariate Cox model and the Kaplan–Meier method with a log-rank test (for details, see the Supplementary Appendix).
RESULTS
IDENTIFICATION OF CIRCULATING TUMOR CELLS
Blood samples were obtained from 23 patients with EGFR mutant tumors, including 5 patients who had undergone no previous treatment, 10 patients who had previously been treated with erlotinib or gefitinib, and 8 patients who had previously been treated with another chemotherapy agent or multiple regimens, including both tyrosine kinase inhibitors and chemotherapy. Four patients whose tumors had wild-type EGFR were also analyzed. (A schematic depiction of the strategy for microfluidic isolation of circulating tumor cells and representative images of captured cells are shown in Figure 1 in the Supplementary Appendix.)
Circulating tumor cells were identified in all patients, with a median of 74 cells per milliliter (mean, 133; range, 5 to 771), with a similar number in patients with or without EGFR mutant tumors (Table 1). The number of circulating tumor cells that were isolated from this series of patients with lung cancers enriched for EGFR mutations was similar to that from patients with other cancers.11 The tumor burden on matched radiographic measurements that were performed close to the time of analysis of circulating tumor cells (median, 8 days; range, 0 to 38) showed that the quantity of circulating tumor cells at a single time point was not well correlated with simple tumor volume (Spearman’s correlation coefficient, −0.028; P = 0.88). This finding suggested that additional tumor characteristics, such as invasiveness and vascularity, probably influenced the number of circulating tumor cells.
Table 1.
Detection of Circulating Tumor Cells in Patients with Non–Small-Cell Lung Cancer.*
| Patient No. and EGFR Mutation Status | Sex | Age | Histologic Features | Time since Diagnosis | Previous Systemic Therapy† | Tumor Burden‡ | Circulating Tumor Cells§ |
|---|---|---|---|---|---|---|---|
| yr | mo | cm | no. per ml | ||||
| EGFR mutation present | |||||||
|
| |||||||
| 1 | M | 58 | Adeno | 3.2 | None | 19.8 | 156 |
|
| |||||||
| 2 | M | 55 | Adeno | 14.4 | C, E | 2.4 | 50 |
|
| |||||||
| 3 | F | 66 | Adeno | 18.2 | G, C | 19.5 | 9 |
|
| |||||||
| 4 | M | 59 | Adeno/BAC | 20.7 | G | 2.0 | 771 |
|
| |||||||
| 5 | M | 57 | Adeno | 10.8 | C | 1.5 | 152 |
|
| |||||||
| 6 | F | 74 | Adeno | 18.3 | E | 9.8 | 5 |
|
| |||||||
| 7 | M | 64 | NSCLC | 13.1 | G, E, C | 4.6 | 196 |
|
| |||||||
| 8 | F | 70 | Adeno | 1.4 | None | 5.8 | 175 |
|
| |||||||
| 9 | F | 63 | Adeno | 0.9 | None | 28.6 | 143 |
|
| |||||||
| 10 | F | 66 | Adeno/BAC | 1.3 | None | 8.2 | 112 |
|
| |||||||
| 11 | F | 74 | Adeno/BAC | 4.8 | G | 4.5 | 74 |
|
| |||||||
| 12 | F | 62 | Adeno/BAC | 56.8 | G, E, O | 7.2 | 9 |
|
| |||||||
| 13 | M | 27 | Adeno | 9.9 | G | 4.1 | 47 |
|
| |||||||
| 14 | F | 55 | Adeno | 11 | G | 5.2 | 241 |
|
| |||||||
| 15 | F | 70 | Adeno/BAC | 54.7 | G, C | 30.2 | 31 |
|
| |||||||
| 16 | M | 53 | Adeno/BAC | 9.2 | E | 7.0 | 49 |
|
| |||||||
| 17 | F | 60 | Adeno/BAC | 97.0 | G | 4.3 | 103 |
|
| |||||||
| 18 | F | 37 | Adeno | 29.4 | G | 13.1 | 70 |
|
| |||||||
| 19 | M | 66 | Adeno/BAC | 3.0 | None | 8.8 | 20 |
|
| |||||||
| 20 | M | 63 | Adeno | 40.7 | G, E, C | 7.3 | 64 |
|
| |||||||
| 21 | F | 58 | Adeno | 19.7 | C, E | 2.1 | 107 |
|
| |||||||
| 22 | M | 86 | Adeno | 6.4 | E | 1.4 | 62 |
|
| |||||||
| 23 | M | 65 | NSCLC | 5.0 | E | 3.8 | 62 |
|
| |||||||
| EGFR mutation absent | |||||||
|
| |||||||
| 24 | F | 65 | Adeno | 1.1 | None | 22.1 | 538 |
|
| |||||||
| 25 | F | 71 | Adeno | 4.6 | C | 6.7 | 219 |
|
| |||||||
| 26 | M | 47 | Adeno | 0.1 | None | 26.6 | 84 |
|
| |||||||
| 27 | F | 70 | Adeno | 15.4 | C, E | 3.6 | 43 |
Adeno denotes adenocarcinoma, Adeno/BAC adenocarcinoma with bronchoalveolar features, C chemotherapy, E erlotinib, G gefitinib, NSCLC non–small-cell lung cancer not otherwise specified, and O other or experimental agent.
Previous systemic therapies are listed in the order they were administered.
The tumor burden was measured by unidimensional diameter, according to the Response Evaluation Criteria in Solid Tumors (RECIST).
The number of circulating tumor cells per milliliter was calculated on the basis of the analysis of 1 to 5 ml of whole blood per patient. Patients are listed in the order of specimen collection. Each blood sample was processed once through the CTC-chip. There was no correlation between the tumor burden and the number of circulating tumor cells (Spearman’s correlation coefficient, −0.02; P = 0.88).
DETECTION OF EGFR MUTATIONS IN TUMORS
We tested the suitability of the allele-specific SARMS assay23 for detecting EGFR mutations in rare cell populations. This test is designed to detect multiple drug sensitivity-associated types of EGFR mutation, including the multiple in-frame exon 19 deletions (collectively analyzed as “Del” mutations) and the L858R missense mutation, which together comprise 90% of EGFR mutations. The test also detects the T790M mutation associated with resistance to tyrosine kinase inhibitors.16–18 To validate the results of the SARMS assay, we first analyzed 26 paraffin-embedded tumors of non–small-cell lung cancers previously identified as having the EGFR mutation and 8 specimens reported as having wild-type alleles by sequencing (Table 2). The SARMS assay and sequencing identified the same mutation in 25 tumor specimens, whereas all 8 wild-type tumors were confirmed as being negative, yielding a sensitivity of 96% and a specificity of 100%. The single discrepancy was due to a rare deletion mutation that was not within the detection capacity of the SARMS assay.
Table 2.
Allele-Specific SARMS Analysis of EGFR Mutations in Tumor Samples and Best Clinical Response.*
| Patient No. and EGFR Mutation Status | EGFR Mutation by Sequencing | EGFR Mutation by SARMS Assay† | Duration of Therapy with Tyrosine Kinase Inhibitor‡ | Best Clinical Response | ||
|---|---|---|---|---|---|---|
| Primary Mutation | T790M | Other | mo | |||
| EGFR mutation present | ||||||
|
| ||||||
| 2 | Del T751_I759 insS | None detected§ | No | 18.2¶ | SD | |
|
| ||||||
| 3 | G719S | G719X | Yes | 3.9 | PR | |
|
| ||||||
| 4 | L858R | L858R | No | >36.9 | PR | |
|
| ||||||
| 5 | Del E746_A750 | Del | Yes | 8.6¶ | PR | |
|
| ||||||
| 8 | L861Q | L861Q | Yes | 4.0 | SD | |
|
| ||||||
| 9 | Del E746_A750 | Del | Yes | 6.2 | PR | |
|
| ||||||
| 10 | L858R | L858R | Yes | 8.3 | PR | |
|
| ||||||
| 11 | Del E746_A750 | Del | No | >14.0 | PR | |
|
| ||||||
| 13 | Del E746_A750 | Del | Yes | 8.3 | PR | |
|
| ||||||
| 14 | Del L747_P753 insS | Del | No | 13.5 | PR | |
|
| ||||||
| 15 | Del E746_S752 insV | Del | No | 30.8 | CR | |
|
| ||||||
| 22 | Del E746_A750 | Del | No | >9.2 | PR | |
|
| ||||||
| 23 | L858R | L858R | No | >7.7 | SD | |
|
| ||||||
| 28 | Del E746_A750 | Del | No | 14.9 | PR | |
|
| ||||||
| 29 | L858R | L858R | No | 11.8 | PR | |
|
| ||||||
| 30 | L858R | L858R | No | Del§ | 3.6 | PR |
|
| ||||||
| 31 | G719A | G719X | Yes | Del§ | 3.9 | SD |
|
| ||||||
| 32 | Del E746_A750 | Del | No | >16.7 | SD | |
|
| ||||||
| 33 | Del E746_A750 | Del | No | 11.4 | PR | |
|
| ||||||
| 34 | Del E746_A750 | Del | No | 17.5 | CR | |
|
| ||||||
| 35 | Del E746_A750 | Del | No | 7.6 | SD | |
|
| ||||||
| 36 | L858R | L858R | No | 9.5 | PR | |
|
| ||||||
| 37 | Del L747_T751 | Del | Yes | 9.2 | PR | |
|
| ||||||
| 38 | L858R | L858R | Yes | 6.9 | PR | |
|
| ||||||
| 39 | Del E746_A750 | Del | No | 14.3 | PR | |
|
| ||||||
| 40 | Del E746_A750 insIP | Del | Yes | 11.3 | SD | |
|
| ||||||
| EGFR mutation absent | ||||||
|
| ||||||
| 24 | None (wild type) | None detected | No | 0.7¶ | PD | |
|
| ||||||
| 25 | None (wild type) | None detected | No | None | NA | |
|
| ||||||
| 26 | None (wild type) | None detected | No | 1.2¶ | PD | |
|
| ||||||
| 27 | None (wild type) | None detected | No | 7.5¶ | SD | |
|
| ||||||
| 41 | None (wild type) | None detected | No | >4.4¶ | SD | |
|
| ||||||
| 42 | None (wild type) | None detected | No | None | NA | |
|
| ||||||
| 43 | None (wild type) | None detected | No | 0.7 | PD | |
|
| ||||||
| 44 | None (wild type) | None detected | No | 2.6 | SD | |
The best clinical response was defined according to the Response Evaluation Criteria in Solid Tumors (RECIST). CR denotes complete response, Del deletions in exon 19, NA not applicable, PD progressive disease, PR partial response, SARMS Scorpion Amplification Refractory Mutation System, and SD stable disease.
The SARMS assay groups all variant breakpoints of the in-frame (exon 19) EGFR deletion mutations as a single Del mutation and all mutations at codon 719 as G719X. The presence or absence of T790M is indicated. Other mutations that were identified with the use of the SARMS assay are listed only when they were present.
When the number of months is preceded by “>,” therapy was ongoing.
The mutation identified by sequencing in Patient 2 was not within the detection capacity of the SARMS assay. In Patients 30 and 31, a second activating EGFR allele was detected at low frequency (other), in addition to the prevalent activating mutation (primary mutation).
In these patients, either gefitinib or erlotinib was administered as second-line or third-line therapy. In all other patients, the drugs were administered as first-line therapy for advanced cancers.
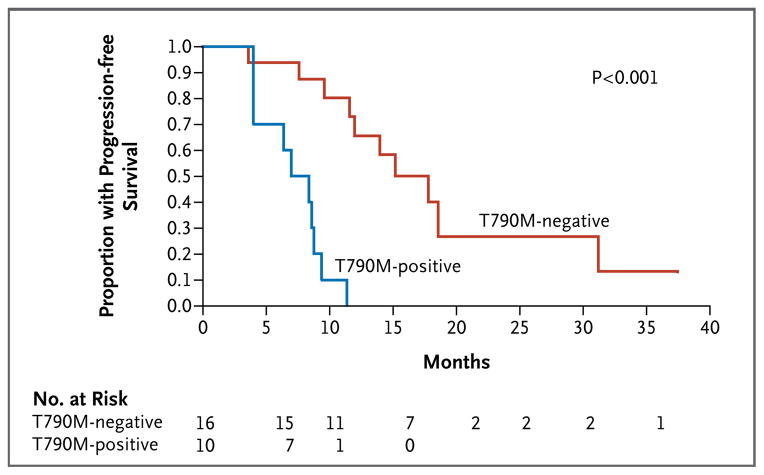
Using the SARMS test, we were able to identify rare EGFR mutant alleles below the detection limit of standard sequencing. In addition to the known primary EGFR mutation, low levels of T790M were detected in pretreatment tumor samples from 10 of 26 patients (38%). The relatively high number of amplification cycles that were required to detect T790M suggests that the mutation is present in only a small number of cells. Indeed, the sequencing of cloned polymerase-chain-reaction (PCR) products from one of these tumors identified only one T790M mutation in 500 EGFR alleles. The presence of the drug-resistance mutation at such a low frequency did not preclude significant responses to therapy with tyrosine kinase inhibitors among patients with EGFR mutant tumors, but it was associated with a striking difference in progression-free survival, with a median of 7.7 months in patients with a detectable T790M allele, as compared with 16.5 months in those without a detectable allele (hazard ratio for progression for the T790M allele, 11.5; 95% confidence interval, 2.94 to 45.1; P<0.001) (Fig. 1). It seems likely that therapy with tyrosine kinase inhibitors results in the selection of the pre-existing T790M resistance allele. Such selection would be predicted to contribute to variation in the duration of response to therapy with tyrosine kinase inhibitors in patients with sensitizing EGFR mutations.
Figure 1. Correlation between the Presence of T790M Mutations in Tumor-Biopsy Specimens and Decreased Progression-free Survival.
In patients with non–small-cell lung cancer with EGFR mutations who were receiving therapy with gefitinib or erlotinib, the presence of the drug-resistance mutation T790M before initiation of treatment was associated with decreased progression-free survival.
DETECTION OF EGFR MUTATIONS IN CIRCULATING TUMOR CELLS
Having established the reliability of the SARMS assay, we applied it to the analysis of circulating tumor cells isolated from peripheral blood. We first compared the EGFR mutations detected in circulating tumor cells by SARMS with those reported for the tumor specimen using either standard sequencing or SARMS. Among specimens from 20 patients that were available for molecular analysis of circulating tumor cells, SARMS identified EGFR mutations in 19 patients (95%) (Table 3). In addition to the primary activating mutation, T790M was detected in circulating tumor cells from 2 of 6 patients (33%) who had a response to tyrosine kinase inhibitors and from 9 of 14 patients (64%) who had clinical progression (P = 0.34). This finding was consistent with the reported prevalence of T790M (about 50%) in patients with the EGFR mutation who were undergoing repeat tumor biopsy after the development of resistance to tyrosine kinase inhibitors.18
Table 3.
Analysis of EGFR Mutations in Circulating Tumor Cells and Free Plasma DNA and the Concordance with Tumor Mutation.*
| Patient No., EGFR Mutation Status, and Response to Therapy | SARMS Assay Results† | Concordance with Tumor Mutation‡ | |||||
|---|---|---|---|---|---|---|---|
| Circulating Tumor Cells | Free Plasma | ||||||
| primary mutation | T790M | other | primary mutation | T790M | other | ||
|
|
|
||||||
| EGFR mutation present and response during therapy§ | |||||||
|
| |||||||
| 1 | Del | No | UN | UN | C¶|| | ||
|
| |||||||
| 9 | Del | Yes | None detected | Yes | C | ||
|
| |||||||
| 11 | Del | No | None detected | No | C | ||
|
| |||||||
| 21 | Del | No | Del | Yes | NA | ||
|
| |||||||
| 22 | Del | No | G719X | None detected | Yes | C | |
|
| |||||||
| 23 | Del | Yes | L858R | Del | Yes | L858R | C,P |
|
| |||||||
| EGFR mutation present and disease progression during therapy§ | |||||||
|
| |||||||
| 3 | Del | No | G719X | UN | UN | C¶ | |
|
| |||||||
| 6 | Del | Yes | Del | No | C,P | ||
|
| |||||||
| 7 | None detected | Yes | L858R | Yes | P | ||
|
| |||||||
| 10 | L858R | Yes | Del | None detected | No | C | |
|
| |||||||
| 12 | Del | Yes | None detected | No | C | ||
|
| |||||||
| 13 | Del | Yes | Del | Yes | C,P | ||
|
| |||||||
| 14 | Del | No | None detected | No | C | ||
|
| |||||||
| 15 | Del | No | None detected | No | C | ||
|
| |||||||
| 16 | Del | Yes | None detected | No | C | ||
|
| |||||||
| 17 | Del | Yes | Del | Yes | NA | ||
|
| |||||||
| 18 | Del | No | G719X | None detected | No | NA | |
|
| |||||||
| 20 | Del | No | None detected | No | NA** | ||
|
| |||||||
| 45 | Del | Yes | None detected | No | NA | ||
|
| |||||||
| 46 | Del | Yes | Del | Yes | NA | ||
|
| |||||||
| EGFR mutation absent | |||||||
|
| |||||||
| 26 | None detected | No | None detected | No | C,P | ||
|
| |||||||
| 43 | None detected | No | None detected | No | C,P | ||
|
| |||||||
| 44 | None detected | No | None detected | No | C,P | ||
Listed are patients in Group A, for whom genotypes were available from at least two of the following: circulating tumor cells, plasma, and tumor-biopsy specimens. Molecular analyses of circulating tumor cells and plasma were performed within 6 months after the initial quantitation of circulating tumor cells. Del denotes deletions in exon 19, NA not applicable due to unavailable tumor specimen, SARMS Scorpion Amplification Refractory Mutation System, and UN sample unavailable for analysis.
The primary activating EGFR mutations are listed when present, including the presence or absence of the specific drug-resistance allele T790M. In all cases, other activating mutations that are listed as “other” were present at a lower allele frequency than that of the primary mutation.
Listed is the concordance between the presence of the primary mutation in the tumor specimen and that in circulating tumor cells (C) and free plasma DNA (P).
Patients who were receiving EGFR tyrosine kinase inhibitors were classified as having either a response or disease progression at the time of the analysis of circulating tumor cells.
A plasma specimen was not available for this patient.
The mutation in the primary tumor was detected by high-performance liquid chromatography but was below the level of detection by standard sequencing analysis.
The presence of a mutation of unknown significance (S885L) that was not included in the SARMS assay was reported in the primary tumor. An additional tumor specimen was not available for SARMS analysis.
A recent study reported the detection of EGFR mutations with the use of the SARMS assay in free plasma DNA from patients with metastatic non–small-cell lung cancer.24 Therefore, we studied the accuracy of mutational analysis in purified circulating tumor cells and free plasma DNA using blood samples from 18 patients with EGFR mutant tumors and 3 controls with wild-type tumors (Table 3). The SARMS assay identified EGFR mutations in 17 of 18 specimens of circulating tumor cells (94%) and in 7 of 18 plasma samples (39%). Among 12 patients for whom specimens of the primary tumor, circulating tumor cells, and plasma were all available for analysis, genotyping of circulating tumor cells had a sensitivity of 92% (in 11 of 12 patients), whereas plasma genotyping had a sensitivity of 33% (4 of 12 patients) (P = 0.009).
The sensitive SARMS assay also identified rare secondary EGFR activating mutations in a subgroup of tumor samples, circulating tumor cells, and plasma (Tables 2 and 3 and Fig. 2A and 3, and Table 1 in the Supplementary Appendix). To assess the significance of these mutations, we monitored genotypes of circulating tumor cells and quantity over time in a subgroup of patients.
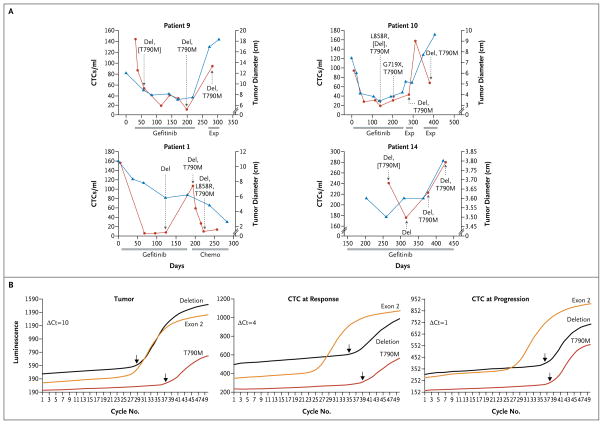
Figure 2. Serial Analyses of Circulating Tumor Cells during Therapy.
Panel A shows serial analyses of the numbers of circulating tumor cells (CTCs) per milliliter (red curve) and the radiographic tumor burden in centimeters (blue curve) in four patients with non–small-cell lung cancer with EGFR mutations, as measured at multiple time points during the course of treatment with gefitinib, another chemotherapy agent (chemo), or an experimental agent (exp). The duration of each therapy is indicated by the gray bars. The genotypes of circulating tumor cells are shown for various time points. Mutations in brackets are those that were present at low allele frequencies. In Panel B, SARMS analysis of EGFR genotypes in Patient 9 shows an increased abundance of the T790M drug-resistance allele during disease progression. Arrows denote the cycle of threshold for amplification cycles (Ct) required for detection of the primary mutation (Del or Deletion, referring to the grouped exon 19 deletions) and the T790M mutation, as compared with the exon 2 control. ΔCt reflects the difference in allele frequency between the primary mutation and T790M in the tumor-biopsy specimen, the circulating tumor cells that were isolated at the time of response to gefitinib therapy, and the circulating tumor cells that were isolated at the time of disease progression. Luminescence was measured quantitatively in relative light units.
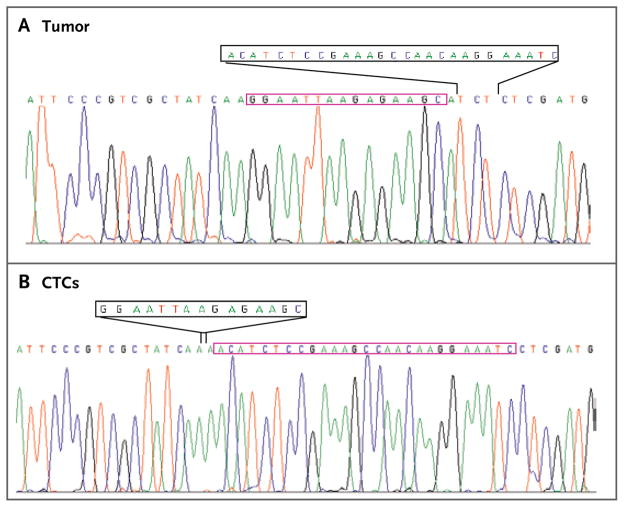
Figure 3. Tracings of EGFR Nucleotide Sequencing from the Tumor and Circulating Tumor Cells of Patient 2.<.
br>Analysis of a tumor-biopsy specimen from Patient 2 shows a T751_I759delinsS mutation that is distinct from the Del 746_750 mutation present in the analysis of circulating tumor cells (CTCs). The 27 nucleotides that were deleted in the tumor (black box) are present in the DNA in the CTCs (red box), whereas the 15-nucleotide deletion in DNA in the CTCs (black box) is present in the tumor DNA (red box). The tracing of CTCs represents direct nucleotide sequencing of DNA lysed from cells captured on the CTC-chip, indicating a high degree of captured tumor-cell purity.
SERIAL MEASUREMENTS
Detailed serial analyses of the quantity and genotype of circulating tumor cells were available for four patients with EGFR mutations after the initiation of gefitinib therapy (Fig. 2A). In all four patients, gefitinib treatment was associated with a profound decline in the number of circulating tumor cells. The interval between blood-sample analyses was dictated by previously scheduled clinical visits, and therefore we could not ascertain the precise timing in the decline in the number of circulating tumor cells. However, in one case (Patient 9), a 50% decline in the quantity of circulating tumor cells was evident within a week after the initiation of therapy, with the nadir at 3 months. Clinical progression was associated with an increase in the number of circulating tumor cells, and in one case (Patient 1), a second decline was evident as the tumor responded to subsequent chemotherapy. In contrast to the lack of a relationship between absolute values for tumor burden and the quantity of circulating tumor cells, as measured among different patients at the time of study enrollment, a close concordance was observed between radiographic assessment of tumor volume and the relative number of circulating tumor cells in patients who were followed serially during the course of therapy.
Genotypes of circulating tumor cells evolved during treatment, with the consistent presence of the primary EGFR activating mutation and the emergence of the T790M drug-resistance mutation. T790M was present at a very low allele frequency in initial tumor specimens, as determined by the relative number of cycles required for amplification, and serial analyses indicated the increased prevalence of the resistance allele within circulating tumor cells over time, a finding that was consistent with the acquisition of clinical drug resistance. Remarkably, seven patients showed the emergence of additional EGFR activating mutations (Table 3 and Fig. 2A and 3, and Table 1 in the Supplementary Appendix). Although these secondary mutant alleles were typically less prevalent than the primary mutation, in at least one case (Patient 2) there was a clear demonstration of the potential for evolution in the predominant genotype within a patient (Fig. 3). In this case, sufficient DNA was isolated from captured circulating tumor cells to allow direct sequencing of EGFR, which confirmed that the predominant mutation in circulating tumor cells differed from that in the original tumor specimen.
DISCUSSION
We have shown that the CTC-chip reproducibly isolated circulating tumor cells in sufficient quantity and with sufficient purity to allow molecular analyses. Circulating tumor cells were readily identified in all patients in numbers that were higher than those identified with previously available methods by a factor of approximately 100.6–9
The use of the allele-specific SARMS assay to identify EGFR mutations in rare cell populations was made possible by the recurrent nature of these mutations, with only 2 of 31 patients carrying mutations identified by sequencing that were absent from the assay panel. Together with the CTC-chip, the SARMS assay may allow for non-invasive genotyping in patients with non–small-cell lung cancer, which could be repeated at therapeutic decision-making points during a patient’s course of therapy. Genotyping of circulating tumor cells appeared to be more sensitive than analysis of free plasma DNA (P = 0.009), and the concomitant quantification of circulating tumor cells provided an important context in which to interpret genotyping results.
The analysis of circulating tumor cells commonly identified the T790M drug-resistance mutation in a majority of patients who had clinical tumor progression while receiving therapy with tyrosine kinase inhibitors. Unexpectedly, use of the highly sensitive allele-specific assay showed that a subgroup of patients with the EGFR mutation harbor rare T790M alleles both before exposure to tyrosine kinase inhibitors and during clinical response. T790M is thought to emerge through selective pressure during therapy, although it has been reported in rare cases in patients without previous drug exposure,25,26 and the mutation confers additional transforming properties when combined in cis with the more common EGFR activating mutations.27 Thus, T790M may initially arise by virtue of its oncogenicity and may rapidly emerge as a dominant allele after treatment. The presence of rare T790M alleles in pretreatment tumor specimens did not preclude dramatic responses to tyrosine kinase inhibitors but did have a significantly adverse effect on progression-free survival. We speculate that this molecular marker may be a way of distinguishing patients who are likely to have a prolonged response to erlotinib or gefitinib from those whose response is short-lived and who may be appropriate candidates for second-generation, irreversible tyrosine kinase inhibitors or combination targeted-therapy regimens. Although amplification of the c-met proto-oncogene (MET) has recently been reported as a second mechanism of acquired resistance to tyrosine kinase inhibitors,19,20 we did not detect it in pretreatment tumor-biopsy specimens or in circulating tumor cells collected during therapy using a quantitative PCR assay (data not shown).
The most unexpected observation in our study was the emergence of additional activating EGFR mutations in circulating tumor cells during therapy. Although the SARMS assay cannot determine whether two different mutations are present on the same or on separate alleles, the patient for whom we had sufficient DNA for direct sequencing had mutually exclusive mutations in the original tumor and in circulating tumor cells (Fig. 3). We therefore presume that the identification of different EGFR activating mutations represents the emergence of different tumor clones. In some patients, additional mutations emerged during tumor progression after chemotherapy. Therefore, such mutations may be associated with drug-induced shifts in tumor-cell populations, reflecting clonal selection during treatment.
The mutation of EGFR is thought to be an early molecular event in the genesis of non–small-cell lung cancer in nonsmokers on the basis of the transforming potential of these mutations in vitro and in mouse models.27–30 Although most non–small-cell lung cancers appear to have a single clonal EGFR mutation that is evident at multiple sites of metastatic disease, rare cases of bronchoalveolar cancer may present with multifocal tumors, each harboring different EGFR mutations.17,25 Circulating tumor cells may be derived from multiple disease sites with different responses to therapy and be associated with an evolution in tumor genotypes that cannot be appreciated by a single tissue biopsy performed at the time of presentation. The absence of treatment-induced genetic change may explain why recent clinical trials of first-line tyrosine kinase inhibitors in patients with EGFR mutations have shown a high concordance between tumor genotype and clinical response,21,31,32 as compared with earlier second-line and third-line studies, in which multiple courses of chemotherapy separated the diagnostic tumor specimen from the clinical evaluation of responsiveness to tyrosine kinase inhibitors.33,34 Therefore, genotype-directed clinical trials of molecularly targeted agents may benefit from “real time” tumor genotyping, either in the form of coincident tumor biopsy or the analysis of circulating tumor cells.
Direct sequencing of an EGFR mutation indicates that the captured circulating tumor cells represent a largely pure tumor-derived cell population. Further characterization of such precursors of metastasis35 may provide important opportunities for diagnostic and therapeutic interventions. However, optimization and automation of the device for capturing circulating tumor cells for high-throughput processing will be required to allow large-scale clinical trials that use this novel technology.
Supplementary Material
Acknowledgments
Supported by grants from the National Institutes of Health; the Doris Duke, Ellison, and Monell Foundations; the ESSCO–MGH Research Fund; the National Foundation for Cancer Research; and the Howard Hughes Medical Institute. SARMS assay reagents were provided by DxS.
Dr. Sequist reports receiving grant support from AstraZeneca and Genentech; Drs. Iafrate, Settleman, and Haber, receiving research support from AstraZeneca; Drs. Bell, Settleman, Lynch, and Haber, receiving royalties as coinventors on a patent awarded for the discovery of EGFR mutations, licensed to Genzyme Genetics, which was not involved in this study; Drs. Tompkins and Toner, being coinventors of the CTC-chip technology, licensed to CellPoint Diagnostics, which was not involved in this study; Drs. Tompkins, Toner, and Haber, receiving consulting fees from CellPoint Diagnostics; and Dr. Lynch, receiving consulting fees from AstraZeneca, Genentech, OSI Pharma, Chugai, and Roche.
We thank the patients for their participation in the study.
Footnotes
No other potential conflict of interest relevant to this article was reported.
References
- 1.Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papadopoulos N, Kinzler KW, Vogelstein B. The role of companion diagnostics in the development and use of mutation-targeted cancer therapies. Nat Biotechnol. 2006;24:985–95. doi: 10.1038/nbt1234. [DOI] [PubMed] [Google Scholar]
- 3.Ashworth TR. A case of cancer in which cells similar to those in the tumors were seen in the blood after death. Aust Med J. 1869;14:146. [Google Scholar]
- 4.Zieglschmid V, Hollmann C, Böcher O. Detection of disseminated tumor cells in peripheral blood. Crit Rev Clin Lab Sci. 2005;42:155–96. doi: 10.1080/10408360590913696. [DOI] [PubMed] [Google Scholar]
- 5.Allard WJ, Matera J, Miller MC, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897–904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 6.Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–91. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 7.Braun S, Marth C. Circulating tumor cells in metastatic breast cancer — toward individualized treatment? N Engl J Med. 2004;351:824–6. doi: 10.1056/NEJMe048163. [DOI] [PubMed] [Google Scholar]
- 8.Cristofanilli M, Hayes DF, Budd GT, et al. Circulating tumor cells: a novel prognostic factor for newly diagnosed meta-static breast cancer. J Clin Oncol. 2005;23:1420–30. doi: 10.1200/JCO.2005.08.140. [Erratum, J Clin Oncol 2005; 23:4808.] [DOI] [PubMed] [Google Scholar]
- 9.Smerage JB, Hayes DF. The measurement and therapeutic implications of circulating tumour cells in breast cancer. Br J Cancer. 2006;94:8–12. doi: 10.1038/sj.bjc.6602871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smirnov DA, Zweitzig DR, Foulk BW, et al. Global gene expression profiling of circulating tumor cells. Cancer Res. 2005;65:4993–7. doi: 10.1158/0008-5472.CAN-04-4330. [DOI] [PubMed] [Google Scholar]
- 11.Nagrath S, Sequist LV, Maheswaran S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–9. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 13.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 14.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sequist LV, Bell DW, Lynch TJ, Haber DA. Molecular predictors of response to epidermal growth factor receptor antagonists in non-small-cell lung cancer. J Clin Oncol. 2007;25:587–95. doi: 10.1200/JCO.2006.07.3585. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non–small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–92. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 17.Kwak EL, Sordella R, Bell DW, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc Natl Acad Sci U S A. 2005;102:7665–70. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2(3):e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 20.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–7. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sequist LV, Martins RG, Spigel D, et al. First-line gefitinib in advanced non-small-cell lung cancer patients harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–9. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 22.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors: European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–16. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 23.Whitcombe D, Theaker J, Guy SP, Brown T, Little S. Detection of PCR products using self-probing amplicons and fluorescence. Nat Biotechnol. 1999;17:804–7. doi: 10.1038/11751. [DOI] [PubMed] [Google Scholar]
- 24.Kimura H, Kasahara K, Kawaishi M, et al. Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non-small-cell lung cancer. Clin Cancer Res. 2006;12:3915–21. doi: 10.1158/1078-0432.CCR-05-2324. [DOI] [PubMed] [Google Scholar]
- 25.Bell DW, Gore I, Okimoto RA, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet. 2005;37:1315–6. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- 26.Inukai M, Toyooka S, Ito S, et al. Presence of epidermal growth factor receptor gene T790M mutation as a minor clone in non-small cell lung cancer. Cancer Res. 2006;66:7854–8. doi: 10.1158/0008-5472.CAN-06-1951. [DOI] [PubMed] [Google Scholar]
- 27.Godin-Heymann N, Bryant I, Rivera MN, et al. Oncogenic activity of epidermal growth factor receptor kinase mutant alleles is enhanced by the T790M drug resistance mutation. Cancer Res. 2007;67:7319–26. doi: 10.1158/0008-5472.CAN-06-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greulich H, Chen TH, Feng W, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2005;2(11):e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Politi K, Zakowski MF, Fan PD, Schonfeld EA, Pao W, Varmus HE. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev. 2006;20:1496–510. doi: 10.1101/gad.1417406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji H, Li D, Chen L, et al. The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer Cell. 2006;9:485–95. doi: 10.1016/j.ccr.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 31.Asahina H, Yamazaki K, Kinoshita I, et al. A phase II trial of gefitinib as first-line therapy for advanced non-small cell lung cancer with epidermal growth factor receptor mutations. Br J Cancer. 2006;95:998–1004. doi: 10.1038/sj.bjc.6603393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inoue A, Suzuki T, Fukuhara T, et al. Prospective phase II study of gefitinib for chemotherapy-naive patients with advanced non-small-cell lung cancer with epidermal growth factor receptor gene mutations. J Clin Oncol. 2006;24:3340–6. doi: 10.1200/JCO.2005.05.4692. [DOI] [PubMed] [Google Scholar]
- 33.Bell DW, Lynch TJ, Haserlat SM, et al. Epidermal growth factor receptor mutations and gene amplification in non-small-cell lung cancer: molecular analysis of the IDEAL/INTACT gefitinib trials. J Clin Oncol. 2005;23:8081–92. doi: 10.1200/JCO.2005.02.7078. [DOI] [PubMed] [Google Scholar]
- 34.Tsao MS, Sakurada A, Cutz JC, et al. Erlotinib in lung cancer — molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–44. doi: 10.1056/NEJMoa050736. [Erratum, N Engl J Med 2005;355:1746.] [DOI] [PubMed] [Google Scholar]
- 35.Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.