ABSTRACT
The human appendix has historically been considered a vestige of evolutionary development with an unknown function. While limited data are available on the microbial composition of the appendix, it has been postulated that this organ could serve as a microbial reservoir for repopulating the gastrointestinal tract in times of necessity. We aimed to explore the microbial composition of the human appendix, using high-throughput sequencing of the 16S rRNA gene V4 region. Seven patients, 5 to 25 years of age, presenting with symptoms of acute appendicitis were included in this study. Results showed considerable diversity and interindividual variability among the microbial composition of the appendix samples. In general, however, Firmicutes was the dominant phylum, with the majority of additional sequences being assigned at various levels to Proteobacteria, Bacteroidetes, Actinobacteria, and Fusobacteria. Despite the large diversity in the microbiota found within the appendix, however, a few major families and genera were found to comprise the majority of the sequences present. Interestingly, also, certain taxa not generally associated with the human intestine, including the oral pathogens Gemella, Parvimonas, and Fusobacterium, were identified among the appendix samples. The prevalence of genera such as Fusobacterium could also be linked to the severity of inflammation of the organ. We conclude that the human appendix contains a robust and varied microbiota distinct from the microbiotas in other niches within the human microbiome. The microbial composition of the human appendix is subject to extreme variability and comprises a diversity of biota that may play an important, as-yet-unknown role in human health.
IMPORTANCE
There are currently limited data available on the microbial composition of the human appendix. It has been suggested, however, that it may serve as a “safe house” for commensal bacteria that can reinoculate the gut at need. The present study is the first comprehensive view of the microbial composition of the appendix as determined by high-throughput sequencing. We have determined that the human appendix contains a wealth of microbes, including members of 15 phyla. Important information regarding the associated bacterial diversity of the appendix which will help determine the role, if any, the appendix microbiota has in human health is presented.
Introduction
It has recently been hypothesized that the human appendix functions as a reservoir of beneficial microbes that can be used for recovery following events of pathogen colonization, diarrheal disease, or antibiotic treatment (1, 2). Bollinger et al. theorized that the vermiform appendix serves as a microbial reservoir or “safe house” for beneficial bacteria capable of repopulating the gut. The associated lymphoid tissue of the appendix has been recognized to provide an ideal environment for bacterial growth in biofilms acting as an enteric reservoir (3, 4). Furthermore, the presence of the appendix may reduce the risk of Clostridium difficile recurrence (5), the protective effect being attributed to the presence of beneficial microbial biofilms and/or to an immune defense (6). Conversely, however, another recent study suggested that the appendix may actually promote C. difficile acquisition, carriage, and disease (7).
Acute appendicitis is one of the most common causes of abdominal pain, with surgical appendectomy being the standard choice of treatment, and is still considered a clinical emergency. There is now evidence that obstructions in the organ are unlikely to be the primary cause of appendicitis (8), and bacterial infection is believed to be central to appendix inflammation (9). Despite this, however, there are limited data on the causal agents of acute appendicitis and of the microbial composition of the human appendix. Culturing-dependent studies have documented the dominance of Bacteroides species in both healthy and inflamed appendices (10, 11), in addition to Escherichia coli and Streptococcus spp. being recovered from the tissue (11). Recent studies using fluorescence in situ hybridization (FISH) reported that local invasion with species of Fusobacterium is the cause in the majority of cases of suppurative appendicitis (9, 12). The presence of Fusobacterium spp. in the mucosal lesions correlated positively with the severity of acute appendicitis, and the presence of other fecal organisms, including members of Bacteroides, Eubacterium rectale, Faecalibacterium prausnitzii, and Akkermansia muciniphila inversely correlated with the inflammation of the organ (9).
In this study, we determined the microbial composition of the human appendix, using next-generation sequencing technologies. The data presented constitute, to our knowledge, the first account of the entire microbiota of this organ and provide insight into the diversity of its associated taxa. Important information regarding the bacterial diversity of inflamed appendices is presented, and although full conclusions cannot yet be made, it is a step toward assigning a role for the microbiota of the appendix in human health.
RESULTS
Seven patients from whom appendices were surgically removed had been fasting prior to surgery and received preoperative antibiotics (Table 1). Both the macroscopic appearance of the appendices and microscopic histopathology were used to determine the severity of the disease of the organs. Samples A, B, and D were deemed by macroscopic examination to be the most inflamed, with the histopathology indicating a diagnosis of acute appendicitis, whereas samples C, E, F, and G appeared less inflamed and therefore were considered healthier samples.
TABLE 1 .
Description of patients and appendix samples
| Sample | Patient |
Macroscopic appearance | Microscopic analysisa | Antibiotic(s) | |
|---|---|---|---|---|---|
| Gender | Age (yr) | ||||
| A | M | 14 | Congested appendix | Acute appendicitis | Amoxicillin-clavulanate |
| Bb | M | 17 | Mucosal hemorrhagic, red inflamed appearance |
Acute suppurative appendicitis | Amoxicillin-clavulanate + gentamicin |
| C | F | 20 | No perforation | No evidence of active inflammation |
Amoxicillin-clavulanate |
| D | M | 13 | Congested appendix, red appearance | Acute appendicitis | Amoxicillin-clavulanate + gentamicin + metronidazole |
| E | M | 25 | No perforation, congested surface | Serositis | Amoxicillin-clavulanate |
| F | M | 5 | No perforation | No evidence of inflammation | Amoxicillin-clavulanate |
| G | F | 14 | No perforation | No pathological changes | Amoxicillin-clavulanate |
Result of histopathological analysis of the appendix.
Stool sample also obtained from this patient.
qPCR analysis of appendix samples.
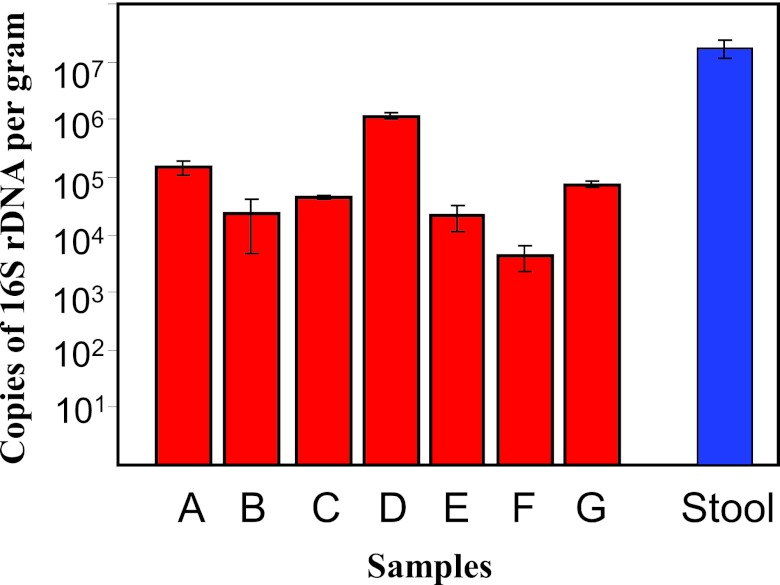
Shifts in phyla may be due to a depletion of or increase in bacteria which may alter the final bacterial numbers. Quantitative PCR (qPCR), therefore, was performed to determine total bacterial numbers in the 7 appendices. Absolute quantification revealed that all appendix samples harbored between 104 and 106 copies of 16S rRNA/g appendix (Fig. 1). Statistical analysis using the Kruskal-Wallis test revealed a significant difference in total 16S rRNA gene copies across the 7 appendix samples (P < 0.01), with sample D having the highest total numbers (Fig. 1). The microbial loads in the appendices were compared to the load in the fecal sample from patient B. The stool sample from patient B had 107 copies of 16S rRNA/g wet stool, whereas the comparative appendix sample harbored only 104 copies. This value for the stool sample is comparable to results obtained by similar methods for fecal samples (13), despite the use of antibiotics in the present study.
FIG 1 .
Numbers of bacteria determined by real-time qPCR. Error bars represent standard errors of the means (n = 3). Red bars represent the appendix samples A to G, blue bar represents the stool sample.
Compositional high-throughput sequencing.
The microbial content of the appendices and stool sample were investigated by high-throughput sequencing (Roche-454 GS-FLX Titanium) of 16S rRNA (V4) amplicons generated from extracted DNA. The sequence reads averaged 9,934 reads per appendix sample, and 11,010 reads represented the stool sample. Diversity, richness, and coverage estimations were calculated for each data set at 97% similarity levels (Table 2). The Chao1 estimator of species richness indicated a sufficient level of overall phylotype diversity (Table 2). The Shannon index is reflective of both species numbers and evenness of their abundance, and therefore the value increases with the number of unique species or the evenness of species abundance. The Shannon indices indicated a high level of overall diversity within samples, with all values exceeding 4.2. Good’s coverage, a measure of sampling completeness, ranged from 85.7 to 93.8% for all samples, indicating sufficient overall sampling.
TABLE 2 .
Estimation of diversity at the 97% similarity level within each data set
| Samplea | Chao1 richness | Shannon index | Good’s coverage |
|---|---|---|---|
| A | 790 | 5.5 | 86.8 |
| B | 697 | 5.3 | 86.7 |
| C | 466 | 4.7 | 90.8 |
| D | 705 | 4.8 | 93.8 |
| E | 358 | 4.2 | 85.7 |
| F | 834 | 5.3 | 92.4 |
| G | 727 | 5.3 | 89.4 |
| Stool | 694 | 5 | 93.9 |
Appendix samples A to G and comparative stool sample B.
Individual variation of the microbiota of the human appendix.
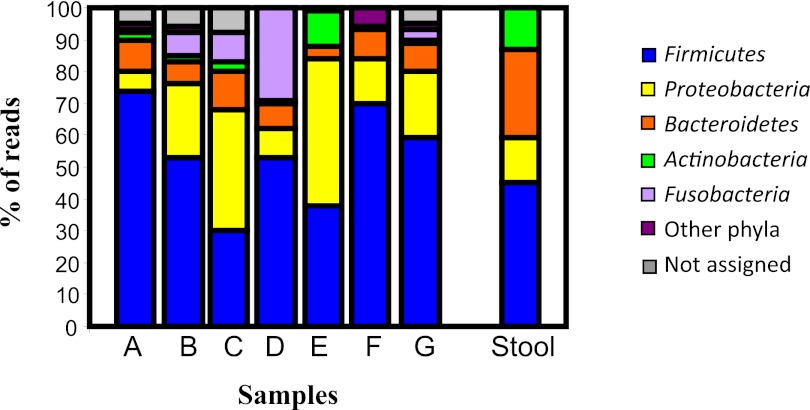
The overall phylum distribution of the appendix samples is shown in Fig. 2 and indicates that the individual composition data sets show much variation with respect to each other. Across the 7 appendix samples, there was considerable diversity, with 15 phyla being represented. In general, however, the appendices were dominated by 5 major phyla—Firmicutes, Proteobacteria, Bacteroidetes, Fusobacteria, and Actinobacteria—with others being detected at low levels (<1 to 2%) in certain samples, including Deferribacteres, Verrucomicrobia, Deinococcus-Thermus, Chloroflexi, Lentisphaera, Viridiplantae, Spirochaetes, candidate division TM7, and candidate phyla OP10 and OP11.
FIG 2 .
Phylum distribution among appendix samples (A to G) and the stool sample.
Firmicutes was generally the dominant phylum, comprising 30% to 75% of the total assignable sequences among the 7 appendix samples. Proteobacteria was also well represented, however, and in sample C (37%) and E (46%) was the predominant phylum. Although Actinobacteria represented just 3% of the sequences on average, sample E harbored relatively high levels (11%) of this phylum. Bacteroidetes levels varied between samples (4% to 12%) but generally were not high (Fig. 2). The levels of the phylum Fusobacteria varied considerably between samples, ranging from <1% to 29% of assignable reads. Fusobacteria was most highly represented in sample D, from an inflamed appendix.
Subpopulations of the microbiota of the appendix.
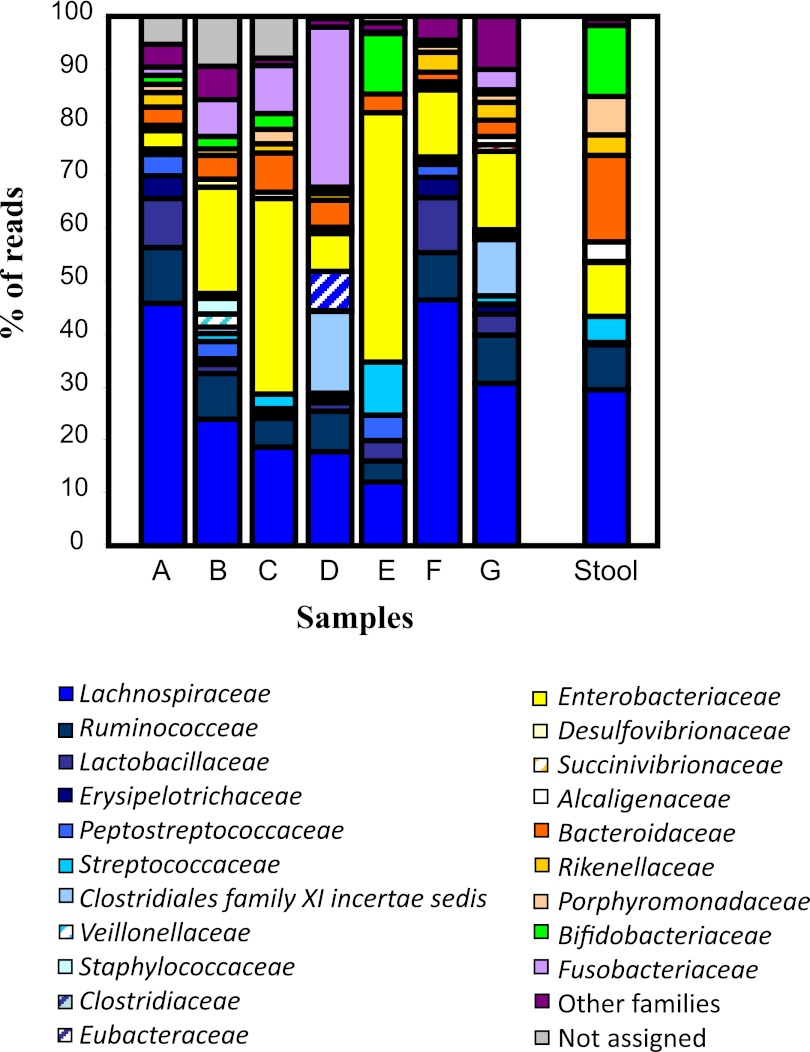
At the family level, it appeared that the subpopulations of the assigned phyla also differed somewhat between samples (Fig. 3). In general, Lachnospiraceae was the dominant subgroup of Firmicutes and corresponded to 46% of total assignable sequences in sample A. However, >12 subpopulations of Firmicutes were identified within the 7 appendices, with the majority of the remaining reads being assigned to the families Ruminococcaceae, Lactobacillaceae, and Streptococcaceae (Fig. 3). Of the subpopulations of Proteobacteria identified, a large proportion of reads from each sample was assigned to the family Enterobacteriaceae. The families Bacteroidaceae, Fusobacteriaceae, and Bifidobacteriaceae comprised the dominant or only families of the phyla Bacteroidetes, Fusobacteria, and Actinobacteria, respectively, in all samples (Fig. 3).
FIG 3 .
Family-level comparison of appendix samples (A to G) and the stool sample. Similar bar colors correspond to samples within a particular phylum. Families of lower abundance are grouped.
At the genus level, despite the large diversity of bacteria represented, a small number of taxa comprised the majority of the bacteria in the samples (Table 3). Although a large number of phyla (>15), families (>40), and their subpopulations are represented within the 7 samples tested indicating a high level of microbial diversity, it is evident that in certain samples a few major families and genera comprise the majority of the sequences (Table 3). It should be noted, however, that percentages are based on the sequences that can be assigned at this level, and assignments to genus level are difficult, owing to limited taxonomic assignments within certain families and phyla. Members of the families Enterobacteriaceae and Lachnospiraceae and the genera Fusobacterium, Lactobacillus, Bacteroides, and Bifidobacterium are represented in the majority of samples at various levels (Table 3). Fusobacterium spp., in particular Fusobacterium nucleatum, are generally regarded as oral pathogens but are also commonly found in the lining of the gut and have been identified in the stomach (14) and most notably as the likely etiological agents of acute appendicitis (9). In addition, the genus Gemella, usually associated with mucous membranes of oral cavities (15), is represented in samples A and B. Parvimonas, which is also generally associated with the oral microbiome and considered a periodontal pathogen (16), was identified in high numbers in samples D and G (Table 3). Parvimonas micros, however, has been previously identified from appendix tissue of patients with acute appendicitis (17).
TABLE 3 .
Comparison of the dominant subpopulations of the appendix samples A to G and the stool sample (S)
| Genus | Abundance (%) ina: |
|||||||
|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | S | |
| Enterobacteriaceae members | 9 | 25 | 36 | 9 | 49 | 27 | 25 | 11 |
| Lachnospiraceae incertae sedis | 10 | 6 | 11 | 16 | ||||
| Ruminococcaceae incertae sedis | 8 | 4 | 3 | 7 | 4 | 6 | ||
| Fusobacterium | 3 | 8 | 9 | 41 | 7 | |||
| Lactobacillus | 16 | 4 | 4 | 23 | 7 | |||
| Bacteroides | 7 | 9 | 7 | 7 | 4 | 4 | 5 | 18 |
| Bifidobacterium | 4 | 3 | 3 | 12 | 15 | |||
| Eubacterium | 10 | |||||||
| Streptococcus | 10 | 5 | ||||||
| Parvimonas | 21 | 18 | ||||||
| Parabacteroides | 8 | |||||||
| Oscillibacter | 3 | |||||||
| Gemella | 5 | 3 | ||||||
| Faecalibacterium | 3 | 4 | ||||||
| Alistipes | 5 | |||||||
| Mucispirillum | 5 | 5 | ||||||
| Allobaculum | 9 | |||||||
| Blautia | 7 | |||||||
| Other genera | 23 | 34 | 28 | 9 | 10 | 27 | 29 | 14 |
| Not assigned | 11 | 8 | ||||||
Only genera representing >3% of assignable sequences have values listed here. Values are representative of the relative abundances of total sequences assignable at genus level. All genera representing <3% of assignable sequences are grouped together as “Other genera.”
Comparative analysis of the microbial composition of the appendix with the microbiota of the gut.
The microbiota of the healthy adult gut is generally thought to be dominated by a small number of phyla, with considerable interindividual variation (18–21). Studies on the gut of young adults have indicated that the majority of bacteria belong to the phylum Firmicutes or Bacteroidetes, followed by Proteobacteria and Actinobacteria (18, 19). The results from the appendix samples in this study indicate, in addition to the above phyla, the substantial presence of Fusobacteria at the phylum level and a more diverse biota at the family and genus levels compared to data previously presented (20) (Fig. 2 and 3; Table 3).
For further comparison, we used one fecal sample (patient B), and the microbial composition was compared to that of its corresponding appendix sample at the phylum (Fig. 2), family (Fig. 3), and genus (Table 3) levels. At phylum level, the stool sample harbored Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria (Fig. 2). However, the appendix sample from the same patient had, in addition to the above 4 phyla, a significant proportion of sequences from the phylum Fusobacteria (7%), and a low percentage of reads were also assigned to the phyla Deferribacteres, Verrucomicrobia, and Viridiplantae and the candidate division OP11. Examination of the subpopulations (Fig. 3; Table 3) clearly shows that there is more diversity in the appendix sample than the fecal sample. Of the subpopulations of the phylum Firmicutes, only Lachnospiraceae, Ruminococcaceae, and Streptococcaceae were represented in the stool, whereas the subpopulation of Firmicutes in the appendix sample comprised 11 families, with the most reads being assigned to Lachnospiraceae (Fig. 3). Taken together, these data suggest that there is a more diverse microbial composition in the human appendix than in the human intestine. It is also evident, however, that despite some apparent differences, the microbiota in both samples share some of the dominant biota represented at the phylum, family, and genus levels (Fig. 2 and 3; Table 3).
DISCUSSION
This study represents a first look at the bacterial composition of the human appendix by next-generation sequencing technologies. Overall, this work reveals that the appendix harbors a wealth of microbiota distinct from other niches within the human microbiome and shows considerable interindividual variation. However, comparison of the differences between samples may be limited, given that the patients in this study varied in age, gender, and clinical presentation (Table 1). Also, antibiotic use has been widely documented to disturb the gut microbiota (for a review, see reference 22), and due to the nature of the clinical admittance of the patients participating in this study, it was unavoidable that antibiotics were taken prior to surgery. However, it should be emphasized that the appendix may “protect” the microbiota from full antibiotic exposure, given that it is outside the main flow of intestinal contents. The possibility that the composition of the stool sample used in this study was not affected by antibiotic use cannot be ruled out; however, the microbiota of the stool sample in this study is not dramatically different from those examined in previous studies (18–20).
qPCR analysis of the total bacterial numbers suggests that the appendix contains fewer bacteria than the comparative fecal sample (Fig. 1). Culturing techniques also indicated a lower overall bacterial count in the appendix than in the comparative stool sample (data not shown). This is expected, as the bacterial density increases from the upper (ileum and jejunum) to lower (cecum) colonic sites (23, 24). Compositional sequencing revealed a diverse microbiota, with at least 15 phyla being identified in the human appendix. In addition to the 4 major phyla associated with the gut, Fusobacteria were represented extensively among the samples (Fig. 2). The presence of this phylum and the proportionate reads assigned to the genus Fusobacterium is interesting, as species of Fusobacterium have been suggested to be causative agents of acute appendicitis (9, 12). Appendicitis is a clinical diagnosis, and therefore its uniformity cannot be assumed. The severity of the disease of the appendix samples in this study was based on a macroscopic and histopathological analysis (Table 1). The levels of Firmicutes, Proteobacteria, and Bacteroidetes varied among samples, regardless of disease state (Fig. 2), whereas the levels of Actinobacteria were highest in the nonperforated sample E (11%). The phylum Fusobacteria and the genus Fusobacterium were most represented in sample D (Table 3). Appendix sample D was obtained from a red and inflamed appendix, and a clinical diagnosis of acute appendicitis was made. Fusobacterium spp. were also found at various levels among the other appendix samples except samples E and F, which were deemed to be healthier samples in this study (Tables 1 and 3).
Is there a role for the microbiota of the human appendix in gut health? The function of the appendix as a microbial reservoir for the gut has recently received much attention (1, 2). Based on this study, the appendix appears to harbors a large diversity of gut microbes, including significant amounts of “beneficial bacteria” or indicators of gut health, including the genera Bacteroides, Lactobacillus, and Bifidobacterium (Table 3). It is plausible that these bacteria are present in biofilms on the epithelial layer of the appendix and may serve as a reservoir for replenishing populations that have been eradicated from the gut. The genus Clostridium was not largely represented in any appendix sample (Table 3), and therefore these results do not indicate that the appendix serves as an organ to carry this organism, as previously suggested (7).
The composition of the appendix microbiota differed somewhat from that of the stool sample microbiota in this study (Fig. 2 and 3; Table 3). This is not surprising, however, based on their differing ecological conditions. The appendix is an extension of the cecum, which has a lower pH and higher fatty acid content than the gut and in consequence has been found to harbor bacteria different from those found in fecal samples (23). However, despite some variation, the profiles were not radically different, and the appendix does contain a high proportion of intestinal microbes. In addition to these, however, there were also a number of phyla that are not generally regarded as gut microbes, including Deinococcus-Thermus, Spirochaetes, and Chloroflexi. Deinococcus-related organisms are largely associated with extreme environments but have recently been identified in the human stomach (14) and in endodontic infections (25). The presence of these phyla indicates that environmentally resistant organisms reside within the appendix. At the genus level, Parvimonas and Gemella, which are generally considered pathogenic organisms of the oral cavity (15, 16), were represented. The high incidence of oral pathogens in the appendix samples is interesting, as microbial community analysis previously indicated that the oral cavity and gastrointestinal tract share few bacterial species (26).
This study presents the first comprehensive view of the microbiota of the human appendix. It would appear that this organ harbors a diverse microbiota and that although it shares a substantial amount of microbes with the intestinal tract, it has its own defined microbiome. The study presented here provides some insights into the composition of the human appendix and information on pathogens present that may possibly contribute to appendicitis. The microbial diversity may be shaped through the coevolution of the microbial communities and specific ecological factors of the appendix tissue. It may also be the case that the microbial dynamics of surrounding intestinal niches influence the microbiota of this organ. Further studies of this diverse biota may be fruitful in ultimately assigning it a role in gut health.
MATERIALS AND METHODS
Participants.
Approval for the study protocol was obtained from the Clinical Research Ethics Committee of the Cork Teaching Hospitals, Cork, Ireland. Informed consent was obtained from all subjects or from parents of children, in accordance with the local Clinical Research Ethics Committee guidelines. The subject details, preoperative antibiotics administered and the macro- and microscopic details of the appendices are outlined in Table 1. Material from 7 appendices removed during laparoscopic emergent appendectomy was used in this study. One stool sample (patient B) was also taken for comparative analysis. All patients had been fasting for >15 h prior to surgery. Samples were transported under anaerobic conditions and kept at 4°C until processing.
Generation of 16S rRNA amplicons for 454 sequencing.
Total bacterial metagenomic DNA was extracted from appendix and stool samples (triplicate samples for each subject) using the QIAamp DNA stool minikit (Qiagen) (27). 16S rRNA bacterial gene amplicons (V4) were generated with a view to sequencing using the Roche Genome Sequencer FLX platform. Amplicons of 239 bp were generated using a combination of one forward primer, i.e., F1 (5′ AYTGGGYDTAAAGNG), and a combination of 4 reverse primers, R1 (5′ TACCRGGGTHTCTAATCC), R2 (5′ TACCAGAGTATCTAATTC), R3 (5′ CTACDSRGGTMTCTAATC), and R4 (5′ TACNVGGGTATCTAATC). These primers also contained an A (F primer) or B (R primers) adapter and different versions of the F primer, each containing a distinct multiple identifier (MID), were employed for each sample. PCRs were performed using Biomix Red (MyBio), and conditions were as follows: 94°C for 2 min followed by 35 cycles of 94°C for 1 min, 52°C for 1 min and 72°C for 1 min followed by a temperature step of 72°C for 2 min. All samples were completed in duplicate. PCR products were cleaned using Agentcourt AMPure kit (Beckman Coulter Genomics) as per manufacturer’s instructions. Samples were quantified using Quant-iT Picogreen quantification kit (Biosciences, Ireland) and the Nanodrop 3300 (Thermo Scientific). Equimolar solutions of samples were pooled for sequencing, cleaned, and requantified (as described above). Emulsion based clonal amplification was completed as part of the 454 pyrosequencing process. Sequencing was performed at the Teagasc 454 sequencing facility on a Genome Sequencer FLX platform (Roche Diagnostics Ltd.) according to the manufacturer’s protocols.
Bioinformatic analysis.
16S rRNA sequencing reads were quality trimmed using a locally installed version of the Ribosomal Database Project (RDP) Pyrosequencing Pipeline and applying the criteria as previously described (28). Trimmed FASTA sequences were searched with BLAST (29) against a locally installed version of the SILVA 16S rRNA database (30). The resulting BLAST output was parsed using MEGAN (version 4.6) (31) using modified accession look-up tables for mapping the SILVA assignments to NCBI taxonomy. MEGAN assigns reads to NCBI taxonomies by employing the Lowest Common Ancestor algorithm. Bit scores (bit-score of 86) as previously described for 16S rRNA data (32) were used from within MEGAN to filter the results prior to tree construction and summarization of taxa. Phylum, family and genera counts for each subject were extracted from MEGAN. Clustering and diversity analysis of the sequence data was performed using the MOTHUR software package (33, 34).
qPCR-based analysis.
Absolute quantification of total bacterial numbers was performed by qPCR using the Roche 480 LightCycler platform. To quantify total 16S rRNA bacterial counts, a standard curve was established using 109 to 102 copies of 16S rRNA/µl. Values were then converted to copies of 16S rRNA/g appendix or wet stool using the previously published formula (35). Samples were made up of 2 µl of PCR grade water, 1 µl of forward primer F1 (5’ AYTGGGYDTAAAGNG) (0.15 µM), 1 µl of the reverse primer R1 (5’ TACCRGGGTHTCTAATCC) (0.15 µM), 1 µl DNA, and 5 µl of SYBR green (Roche Diagnostics Ltd.), giving a final reaction volume of 10 µl. All samples, negative controls (where template DNA was replaced with PCR-grade water), and standards were run in triplicate.
Statistical analyses.
Minitab release 15.1.1.0 (Minitab Inc.; 2007) was used to run nonparametric statistics (Kruskal-Wallis tests) to determine differences between the microbial numbers from the appendices as determined by PCR. Statistical significance was accepted at a P value of <0.05.
ACKNOWLEDGMENTS
We thank Fiona Crispie and Eva Rosberg-Cody for high-throughput DNA sequencing services. We are grateful to the people who kindly donated their samples to this study.
Fiona Fouhy is in receipt of an Irish Research Council for Science, Engineering and Technology EMBARK scholarship and is a Teagasc Walsh fellow. The Alimentary Pharmabiotic Centre is a research centre funded by Science Foundation Ireland (SFI), through the Irish Government’s National Development Plan. The authors and their work were supported by SFI (grant no. 02/CE/B124 and 07/CE/B1368).
Footnotes
Citation Guinane CM, et al. 2013. Microbial composition of human appendices from patients following appendectomy. mBio 4(1):e00366-12. doi:10.1128/mBio.00366-12.
REFERENCES
- 1. Laurin M, Everett ML, Parker W. 2011. The cecal appendix: one more immune component with a function disturbed by post-industrial culture. Anat. Rec. (Hoboken) 294:567–579 [DOI] [PubMed] [Google Scholar]
- 2. Smith HF, et al. 2009. Comparative anatomy and phylogenetic distribution of the mammalian cecal appendix. J. Evol. Biol. 22:1984–1999 [DOI] [PubMed] [Google Scholar]
- 3. Bollinger RR, et al. 2003. Human secretory immunoglobulin A may contribute to biofilm formation in the gut. Immunology 109:580–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bollinger RR, Barbas AS, Bush EL, Lin SS, Parker W. 2007. Biofilms in the normal human large bowel: fact rather than fiction. Gut 56:1481–1482 [PMC free article] [PubMed] [Google Scholar]
- 5. Im GY, et al. 2011. The appendix may protect against Clostridium difficile recurrence. Clin. Gastroenterol. Hepatol. 9:1072–1077 [DOI] [PubMed] [Google Scholar]
- 6. Na X, Kelly C. 2011. The vermiform appendix and recurrent Clostridium difficile infection: a curious connection. Clin. Gastroenterol. Hepatol. 9:1017–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Merchant R, et al. 2012. Association between appendectomy and Clostridium difficile infection. J. Clin. Med. Res. 4:17–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carr NJ. 2000. The pathology of acute appendicitis. Ann. Diagn. Pathol. 4:46–58 [DOI] [PubMed] [Google Scholar]
- 9. Swidsinski A, et al. 2011. Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut 60:34–40 [DOI] [PubMed] [Google Scholar]
- 10. Elhag KM, Alwan MH, Al-Adnani MS, Sherif RA. 1986. Bacteroides fragilis is a silent pathogen in acute appendicitis. J. Med. Microbiol. 21:245–249 [DOI] [PubMed] [Google Scholar]
- 11. Roberts JP. 1988. Quantitative bacterial flora of acute appendicitis. Arch. Dis. Child. 63:536–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Swidsinski A, et al. 2012. Mucosal invasion by fusobacteria is a common feature of acute appendicitis in Germany, Russia, and China. Saudi J. Gastroenterol. 18:55–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fouhy F, et al. 2012. High-throughput sequencing reveals the incomplete, short-term recovery of the infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamycin. Antimicrob. Agents Chemother. 56:5811–5820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bik EM, et al. 2006. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. U. S. A. 103:732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Diaz PI, et al. 2012. Using high throughput sequencing to explore the biodiversity in oral bacterial communities. Mol. Oral Microbiol. 27:182–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ota-Tsuzuki C, Alves Mayer MP. 2010. Collagenase production and hemolytic activity related to 16S rRNA variability among Parvimonas micra oral isolates. Anaerobe 16:38–42 [DOI] [PubMed] [Google Scholar]
- 17. Rautio M, Saxén H, Siitonen A, Nikku R, Jousimies-Somer H. 2000. Bacteriology of histopathologically defined appendicitis in children. Pediatr. Infect. Dis. J. 19:1078–1083 [DOI] [PubMed] [Google Scholar]
- 18. Eckburg PB, et al. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tap J, et al. 2009. Towards the human intestinal microbiota phylogenetic core. Environ. Microbiol. 11:2574–2584 [DOI] [PubMed] [Google Scholar]
- 20. Claesson MJ, et al. 2011. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl 1):4586–4591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Claesson MJ, et al. 2012. Gut microbiota composition correlates with diet and health in the elderly. Nature 488:178–184 [DOI] [PubMed] [Google Scholar]
- 22. Cotter PD, Stanton C, Ross RP, Hill C. 2012. The impact of antibiotics on the gut microbiota as revealed by high throughput DNA sequencing. Discov. Med. 13:193–199 [PubMed] [Google Scholar]
- 23. Marteau P, et al. 2001. Comparative study of bacterial groups within the human cecal and fecal microbiota. Appl. Environ. Microbiol. 67:4939–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Simon GL, Gorbach SL. 1995. Normal alimentary tract microflora. In Blaser MJ, Smith PD, Rafdin JI, Greenberg HB, Guerrant RL, Infections of the gastrointestinal tract. Raven, New York, NY. [Google Scholar]
- 25. Li L, et al. 2010. Analyzing endodontic infections by deep coverage pyrosequencing. J. Dent. Res. 89:980–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. He X, et al. 2010. In vitro communities derived from oral and gut microbial floras inhibit the growth of bacteria of foreign origins. Microb. Ecol. 60:665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hussey S, et al. 2011. Parenteral antibiotics reduce bifidobacteria colonization and diversity in neonates. Int. J. Microbiol. 2011:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. O’Sullivan Ó, et al. 2011. Correlation of rRNA gene amplicon pyrosequencing and bacterial culture for microbial compositional analysis of faecal samples from elderly Irish subjects. J. Appl. Microbiol. 111:467–473 [DOI] [PubMed] [Google Scholar]
- 29. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 30. Pruesse E, et al. 2007. Silva: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huson DH, Auch AF, Qi J, Schuster SC. 2007. Megan analysis of metagenomic data. Genome Res. 17:377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Urich T, et al. 2008. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One 3:e2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schloss PD, Handelsman J. 2008. A statistical toolbox for metagenomics: assessing functional diversity in microbial communities. BMC Bioinformatics 9:34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schloss PD, et al. 2009. Introducing MOTHUR: pen-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang H, et al. 2009. Human gut microbiota in obesity and after gastric bypass. Proc. Natl. Acad. Sci. U. S. A. 106:2365–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]