Abstract
Trisomic and monosomic (aneuploid) embryos account for at least 10% of human pregnancies and, for women nearing the end of their reproductive lifespan, the incidence may exceed 50%. The errors that lead to aneuploidy almost always occur in the oocyte but, despite intensive investigation, the underlying molecular basis has remained elusive. Recent studies of humans and model organisms have shed new light on the complexity of meiotic defects, providing evidence that the age-related increase in errors in the human female is not attributable to a single factor but to an interplay between unique features of oogenesis and a host of endogenous and exogenous factors.
It has been over 50 years since trisomy 21 was identified as the cause of Down’s syndrome, providing the first link between a clinical disorder and a chromosome abnormality1,2. In the intervening half-century, the importance of numerical chromosome abnormalities to human disease pathology has been well-documented (reviewed in Ref. 3). Studies of live births conducted during the 1960s and 1970s demonstrated that approximately 0.3% of newborn infants were trisomic or monosomic, whereas subsequent studies of spontaneous abortions identified a much higher incidence: approximately 35% (Table 1). Taken together, these studies established aneuploidy as the leading known cause of congenital birth defects and miscarriage and demonstrated that most aneuploid conceptuses perish in utero. More recently, investigations of gametes and preimplantation embryos conceived using assisted reproductive technology (ART; Box 1) have identified aneuploidy as the leading impediment to successful pregnancies in this setting. As discussed below, advances in preimplantation genetic diagnosis in ART provide powerful new approaches to the study of aneuploidy (Box 2), allowing us to re-examine the levels of aneuploidy in human embryos and to address questions about the influence of environmental factors on human female meiosis.
Table 1.
Aneuploidy in humans: estimated levels at different stages
| Population | Methodology* | Timeframe of studies |
Incidence of aneuploidy‡ |
Most common aneuploidies | Refs |
|---|---|---|---|---|---|
| Newborns | Karyotyping | 1960s–1970s | 0.3% | +13; +18; +21; XXX; XXY; XYY | 125 |
| Stillbirths | Karyotyping | 1970s–1980s | 4% | 45,X; +13; +18; +21; XXX; XXY | 125 |
| Spontaneous abortions | Karyotyping | 1970s–1980s | >35% | 45,X; +15: +16; +21; +22 | 125 |
| Preimplantation embryos | Karyotyping | 1990s | 20–40% | +16; +17; +18 | 6,126 |
| FISH | 1990s–present | 25–>70% | Various | 127–130 | |
| CGH, SNP array, CGH array | 2000–present | 30–60% | +15; +16; +21; +22 | 9,12–14 | |
| Eggs or polar bodies | Karyotyping | 1990s | 10–35% | +16; +17; +18; +21; +22 | 7,8 |
| FISH | 1990s–present | 20–>70% | Various | 7,8 | |
| CGH, SNP array, CGH array | 2000–present | 30–70% | +15; +16; +21; +22 | 10,11,15,131 | |
| Sperm | Karyotyping | 1980s–1990s | 1–4% | XY disomy; +21; +22 | 132,133 |
| FISH | 1990s–present | 1–3% | XY disomy; +13; +21; +22 | 134 |
For sperm, karyotyping involved analyses of human sperm that had penetrated hamster oocytes; for eggs, karyotyping involved analyses of meiosis II oocytes. Fluorescence in situ hybridization (FISH) assays varied widely among studies, with different numbers of chromosome-specific FISH probes used per experiment.
Levels of aneuploidy have been estimated across all maternal age groups. Almost all FISH analyses of embryos and eggs or polar bodies involved only a subset of human chromosomes, typically between 3–12 chromosomes per assay. Thus, the overall rates of aneuploidy would presumably be much higher than the values cited here. CGH, comparative genomic hybridization.
Box 1 | Techniques used in assisted reproductive technology.
Ovarian stimulation
Various stimulation protocols are used to promote follicle growth and thereby to increase the number of eggs for fertilization. Most protocols involve using exogenous hormones to modulate gonadotropin-releasing hormone (GnRH) and gonadotropins. As discussed in the main text, accumulating data suggest that some stimulation protocols may increase aneuploidy levels107. Accordingly, some clinics have implemented milder stimulation protocols or the use of natural cycles, and initial reports suggest improvement in embryo quality108,109. Importantly, the introduction of array-based technologies (Box 2) provides the first practical means of comparing established methods and assessing new ones to improve the quality of the resultant eggs and embryos.
Fertilization and in vitro culture
Currently, two approaches are routinely used to achieve fertilization in vitro: standard in vitro fertilization (IVF), which involves the mixing of eggs and sperm in a culture dish, and intracytoplasmic sperm injection (ICSI), which is the injection of a single sperm directly into the egg cytoplasm. Following fertilization by either procedure, embryos may be cultured in vitro for several days before being transferred or frozen.
Preimplantation genetic diagnosis for aneuploidy
To assess the genetic quality of eggs and embryos, the chromosome constitution of biopsied polar bodies and/or blastomeres can be determined by several different techniques (Box 2). This typically involves dissection through the zona pellucida and removal of the first and second polar body or removal of a single blastomere from an early cleavage embryo. A determination of the chromosomal constitution of these biopsied products provides a means of inferring the chromosomal status of the egg or embryo and hence a means of choosing those with the greatest likelihood of normal development.
Embryo transfer
Embryos obtained following IVF or ICSI are transferred to the uterus. To obtain a viable pregnancy, embryo transfer must be timed carefully to coincide with the period of uterine receptivity. If an excess number of embryos are produced or transfer is delayed (for example, owing to preimplantation genetic diagnosis), embryos may be frozen for transfer during a subsequent cycle.
Embryo freezing
In lieu of direct transfer, embryos can be frozen for subsequent transfer months or even years later. Currently, two different techniques of storing embryos are used: either a standard stepwise embryo freezing method using cryoprotectants, or vitrification, which is an ultra-rapid freezing technique that is gaining popularity because it prevents formation of intracellular ice crystals.
Box 2 | Aneuploidy detection in assisted reproductive technology.
Beginning in the 1990s, preimplantation genetic diagnosis (PGD) protocols were developed to identify embryos with the greatest likelihood of producing a chromosomally normal pregnancy. In these assays, the first and/or second polar body or 1–2 cells from preimplantation embryos are biopsied, and they are tested for trisomy or monosomy using one of the following approaches.
Fluorescence in situ hybridization (FISH)
This was the first technique developed for PGD of aneuploidy. Typically, chromosome-specific FISH probes for a subset of chromosomes involved in clinically relevant trisomies (for example, chromosomes 13, 18, 21 and the sex chromosomes) are hybridized to biopsied polar bodies or blastomeres, and the FISH signals are counted to infer the chromosome constitution of the embryo. Although this technique was the mainstay of aneuploidy detection in assisted reproductive technology for over 10 years, its use has diminished for two reasons. First, many FISH-based studies reported extremely high, biologically implausible levels of aneuploidy, calling into question the accuracy of the technique. Second, clinical trials comparing successful pregnancy rates with and without FISH-based PGD have found little or no benefit of FISH120,121. This remains a contentious issue, with some practitioners suggesting that the approach does indeed improve pregnancy success rates122 but only in laboratories with sufficient skill in the requisite techniques (for example, in embryo biopsies and FISH). Nevertheless, an ‘anti-FISH’ consensus has been building for the past few years, and FISH is being replaced by new genome-based techniques.
Comparative genomic hybridization (CGH)
DNA from individual cells is subjected to whole-genome amplification, and this ‘test’ DNA and chromosomally normal ‘reference’ DNA are differentially labelled with fluorochromes and hybridized to normal metaphase chromosomes, and fluorescence ratios of test/reference signals are calculated to detect additional or missing chromosomes. Unlike typical FISH assays, CGH yields information on all chromosomes; however, it is time-consuming and can interfere with timely embryo transfer to the mother. Consequently, CGH is gradually being supplanted by the array-based approaches outlined below.
Array comparative genomic hybridization (aCGH)
This is a variation on CGH that uses hybridization to microarray chips decorated with thousands of probes that cover the entire genome. Like CGH, the analysis can reveal chromosome gains or losses, but analytical automation of microarray chips provides swift data generation (that is, typically within 24 hours), allowing embryo screening without compromising embryo transfer.
SNP arrays
The approach is similar to aCGH, except that the microarray chip contains SNP-detecting probes. This not only allows for the detection of chromosome gains and losses but can also provide information on the parental origin of aneuploidy and data on crossovers123. Like aCGH, the analysis can be completed in a timely manner for embryo transfer.
The results from early studies demonstrated that most aneuploidies are due to errors in maternal meiosis and that increasing maternal age is a powerful contributor to the occurrence of aneuploidy3. However, studies during the past 10–15 years have also implicated events that occur at the onset of female meiosis in the fetal ovary and during the protracted dictyate arrest (Fig. 1). The duration of the division (10 to 50 years and beyond) (Fig. 1) provides ample opportunity for errors to occur and to accumulate, which is a feature that has been the basis of a number of hypotheses to explain the maternal age effect (for example, Ref. 4). Indeed, the emerging picture indicates that aneuploidy is not due to a single causal factor but involves a complex constellation of effects that begins in utero, continues throughout the reproductive lifespan of the woman, is exacerbated by age and is facilitated by the unique features of cell cycle control in the oocyte.
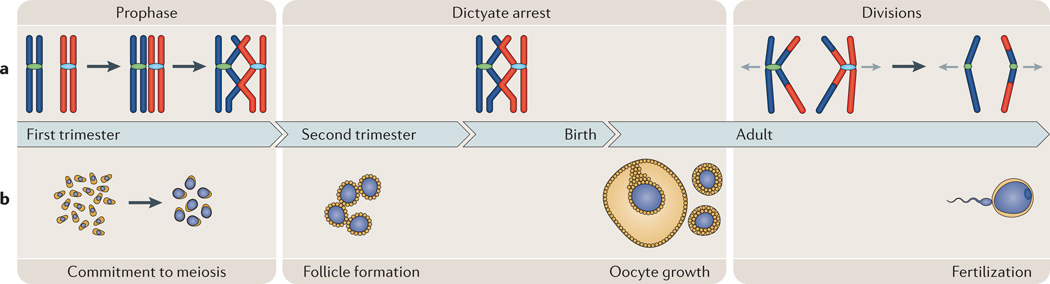
Figure 1. Oogenesis and the female meiotic cycle.
a | Meiosis. Female meiosis can be divided into three temporally distinct phases. Prophase: after DNA replication, homologous chromosomes (shown in red and blue) undergo pairing, synapsis and recombination, and arrest at the diplotene (dictyate) stage. Dictyate arrest: oocytes remain in meiotic arrest until the female reaches maturity and the oocyte has completed an extensive period of growth following follicle formation. The divisions: the luteinizing hormone surge that triggers ovulation also causes resumption and completion of the first meiotic division in the periovulatory oocyte. The ovulated egg is arrested at second meiotic metaphase, and anaphase onset and completion of meiosis II only occur if the egg is fertilized. b | Oogenesis. The process of making an egg is complex and involves four distinct developmental phases. First, commitment to meiosis and meiotic initiation, which occurs at 8–10 weeks of gestation in humans. Second, follicle formation, which occurs during the second trimester in humans. Third, oocyte growth, which occurs in the sexually mature female under the control of paracrine and endocrine signals. Oocyte growth is thought to take approximately 85 days in humans and typically culminates in the ovulation of a single egg. Last, fertilization of the ovulated egg results in the completion of the second meiotic division.
In the sections that follow, we discuss the evidence that has led to this view. Initially, we summarize recent observations on the incidence and aetiology of human aneuploidy from studies of eggs and embryos. In subsequent sections, we discuss the emerging evidence for sex-specific differences in the stringency of meiotic cell cycle controls and the types of errors that evade these checkpoints in the female. We conclude by considering possible environmental agents that may influence the rates of aneuploidy in humans.
The frequency of aneuploidy
Strong in utero selection against chromosomally abnormal conceptions means that the ‘true’ incidence of human aneuploidy can only be determined from studies of fertilized eggs. Clearly, such data will never be obtained from naturally occurring human pregnancies. However, the introduction of ART to treat infertility brought not only a means of assessing aneuploidy levels in gametes or early embryos but also a drive to use aneuploidy assessment to optimize the chances of reproductive success for infertile couples (Box 2).
Aneuploidy in ART-derived pregnancies
Although the initial cytogenetic surveys of live births and miscarriages reported surprisingly high levels of aneuploidy (Table 1), many thought this represented only the tip of the iceberg. It was not possible to study pregnancy losses that occurred before about 6 weeks of gestation, and it was assumed that many aneuploid conceptions would be eliminated during the earliest stages of pregnancy. The first karyotypic studies of human gametes and preimplantation embryos from infertility clinics were consistent with this idea, implying levels of aneuploidy of at least 10–40% at the time of conception (for example, Refs 5,6) (Table 1).
Although these initial results were consistent with expectation, subsequent results from the use of preimplantation genetic diagnosis for ART pregnancies raised eyebrows; that is, in the 1990s traditional karyotypic analysis was replaced by fluorescence in situ hybridization (FISH)-based analyses of eggs and preimplantation embryos (Box 2), and the estimated rates of aneuploidy skyrocketed. Typically, only 3–6 chromosomes were analysed per gamete or embryo; thus, the expectation was that aneuploidy rates would be lower than those detected by conventional chromosome analysis. In fact, a number of studies reported remarkably high rates of aneuploidy of 50% or more (reviewed in Refs 7,8) (Table 1). Given the limited number of chromosomes studied, these data imply biologically implausible total aneuploidy levels, suggesting that FISH-based assays are unable to provide reliable estimates of aneuploidy in human conceptions.
However, other genome-based methods of aneuploidy detection — conventional comparative genomic hybridization (CGH), its array-based derivative (array CGH) and SNP array analysis (Box 2) — have recently been developed and provide optimism for future ART studies. The clinical use of these technologies in preimplantation genetic diagnosis is in its infancy. However, initial results9–13 show aneuploidy rates that are more in line with the 20–40% estimates from conventional cytogenetic studies of preimplantation embryos (Table 1) and, as in clinically recognized pregnancies, abnormalities involving small and/or acrocentric chromosomes are overrepresented.
Natural conceptions versus ART-derived conceptions
Not all observations from natural conceptions, however, are replicated by the ART studies. In at least some studies, the proportion of aneuploidy due to maternal meiosis II errors exceeds that attributable to meiosis I errors (for example, Ref. 10), and chromosomally abnormal cells that include not just one but multiple trisomies and/or monosomies are common occurrences (for example, Refs 11,12,14,15). These observations contrast sharply with data from naturally occurring pregnancies, where most aneuploid abnormalities involve a single chromosome and are attributable to errors at maternal meiosis I.
Methodological differences are likely to explain some of these discrepancies. Studies of the origin of trisomies in naturally occurring pregnancies rely on retrospective analyses of DNA polymorphisms in parents and trisomic offspring, whereas the genome-based ART approaches directly assay chromosome content in polar bodies or embryos. Thus, there is variation in the way that interpretations are made. For example, studies of naturally occurring pregnancies use centromeric heterozygosity or homozygosity to infer the meiotic stage of error16. By contrast, ART approaches provide data on the stage at which segregation mistakes become evident, but this does not necessarily reflect the point at which the precipitating event occurs (for example, errors in segregation that occur at meiosis II may have their genesis at meiosis I; see Box 3).
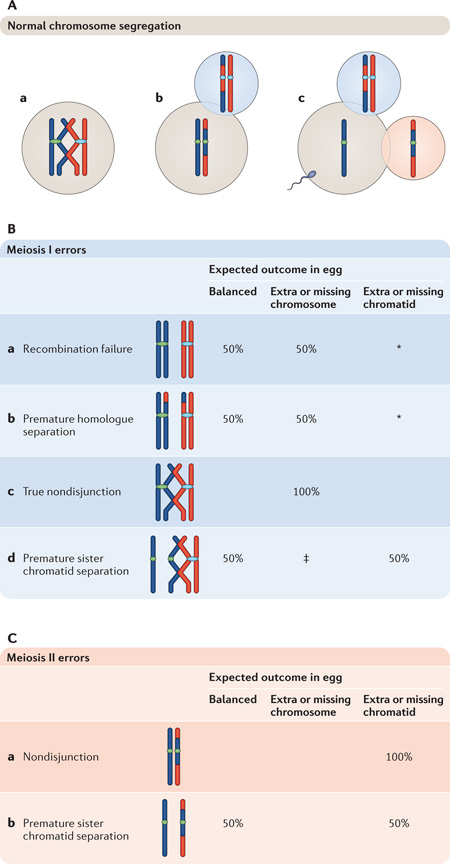
Box 3 | Normal and abnormal meiotic chromosome segregation.
Normal chromosome segregation
Meiosis I oocyte
Homologues are physically tethered at the sites of recombination, facilitating their attachment to opposite poles of the meiosis I spindle (see panel Aa of the figure).
Meiosis II egg
Homologues separate at anaphase I, with one remaining in the egg and the other segregating to the first polar body. The ovulated egg is arrested at metaphase II (Ab).
Fertilized egg
Fertilization triggers the second meiotic division, which results in the separation of sister chromatids, one remaining in the egg and the other segregating to the second polar body (Ac).
Meiosis I errors
The major categories of first meiotic division errors and their effects on the chromosome constitution of the ovulated egg are depicted in panel B of the figure. Predicted frequencies assume that unassociated homologues or sister chromatids segregate randomly. Errors may involve misdivision of whole chromosomes or sister chromatids, but the zygotic outcomes will be the same: that is, monosomic or trisomic conceptions.
Recombination failure
Failure to recombine results in two unpaired univalents at meiosis I. Assuming random segregation of univalents, production of an egg with a balanced chromosome composition or with a missing or extra chromosome are equally likely (Ba). (*Because univalent chromosomes may form attachments to opposite spindle poles and segregate chromatids at meiosis I68–70, eggs with a missing or extra chromatid may also be produced.)
Premature homologue separation
As in recombination failure, premature resolution of connections between homologues produces two unpaired univalents at meiosis I, with the same segregation outcomes (Bb).
True nondisjunction
Failure to resolve connections between homologues results in segregation of both to the same pole, producing eggs with missing or additional whole chromosomes (Bc).
Premature sister chromatid separation
Premature loss of cohesion between sister centromeres results in their independent segregation at meiosis I, producing eggs with a balanced chromosome constitution and with extra or missing chromatids in equal frequency (Bd). (‡Several other outcomes are also possible, depending on whether one or both homologues exhibit premature separation between sisters.)
Meiosis II errors
Meiosis II errors are depicted in panel C of the figure.
Nondisjunction
Failure to resolve connections between sister centromeres results in nondisjunction, producing fertilized eggs with missing or extra chromatids (Ca).
Premature sister chromatid separation
Premature loss of cohesion between sister centromeres results in their independent segregation at meiosis II, producing eggs with a balanced chromosome constitution and with extra or missing chromatids in equal frequency (Cb).
Biological differences between the populations, however, may also be important. Couples attending infertility clinics are not necessarily representative of the general population, and some nondisjunction events may be increased in, or restricted to, infertile individuals. Further, selection pressures would result in the early demise of embryos with multiple errors, effectively restricting them to the ART setting. Finally, as detailed in the final section, there is growing evidence that some of the procedures used in ART may increase the likelihood of aneuploidy.
Sex-specific differences in meiosis
As discussed above, studies of clinically recognized pregnancies demonstrate that most human aneuploidies are maternally derived (reviewed in Ref. 3). This begs the question: why is female meiosis so error-prone? In this section, we review oocyte development, summarizing recent evidence that errors that predispose to chromosome missegregation are increased in the oocyte and that sex-specific differences in meiotic cell cycle checkpoints allow oocytes with these errors to develop into mature eggs. The general conclusion from these studies is straightforward: consistent with previous studies of human trisomies (Box 4), there are many ways in which chromosome dynamics can be disturbed in oogenesis and, consequently, there are many routes to human aneuploidy.
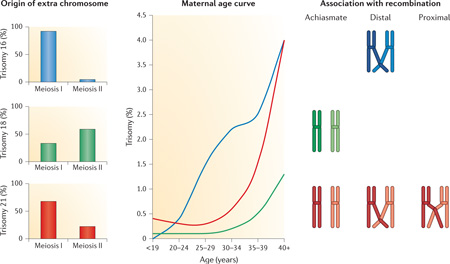
Box 4 | The complexity of human nondisjunction: chromosome-specific variation in the genesis of trisomies.
Studies of clinically recognized pregnancies indicate extraordinary variation in the origin of trisomies and in the importance of the two known risk factors for nondisjunction, altered recombination and increasing maternal age. Shown in the figure are relevant observations for three common human trisomies that involve small chromosomes: +16 (in blue on the figure), +18 (in green) and +21 (in red). Data for these and other individual trisomies are taken from Refs 16,58–61,124.
The origin of trisomy
Inheritance of DNA polymorphisms has been used to determine the parent and meiotic stage of origin of trisomies. Most trisomies are maternally derived, but as shown here, the relative contribution of maternal meiosis I and meiosis II errors varies widely among chromosomes.
The effect of maternal age
Most trisomies increase in frequency with advancing maternal age, but there is variation in the slopes of the curves; for example, for trisomy 16 the increase is roughly linear, whereas both trisomies 18 and 21 are characterized by exponential increases.
Association with altered recombination
Three abnormal crossover configurations have been linked to human trisomies: an absence of crossovers (known as ‘achiasmate’ bivalents), distal-only crossovers and proximal crossovers. However, the importance of the configurations varies among trisomies: each of the three has been reported for trisomy 21, but for trisomies 16 and 18 only a single configuration appears to be contributory.
A bad start: recombination and aneuploidy
In mammalian females, meiotic recombination occurs in the fetal ovary, and the importance of the resultant physical connections for chromosome segregation is well-documented: studies in the 1990s identified failure to recombine and/or suboptimally located crossovers as prominent contributors to human trisomy (Box 4; reviewed in Ref. 3). The importance of altered recombination pertains to paternally as well as maternally derived trisomies but, as most aneuploidy arises during oogenesis, the female is clearly at greater risk. Therefore, either more recombination errors are made in the female or these errors are more efficiently culled in the male. Immunofluorescence methodology has made it possible to examine crossover associated proteins in pachytene spermatocytes and oocytes and thereby to test these alternatives. Strikingly, in the male, almost all chromosomes are joined by at least one crossover17, but the same does not apply to the female. Indeed, it appears that over 10% of all human oocytes contain at least one ‘crossover-less’ bivalent18. Because half of all such bivalents are expected to result in aneuploidy (Box 3), the stage appears to be set for meiotic errors from the beginning of oogenesis.
The differing susceptibility of the sexes to disruptions in recombination poses another question: how are recombination events determined in males and females? Recent studies have shed light on molecular players that mediate recombination hotspots in mammals19 and on at least one recombination gene that acts differently in males than it does in females20. However, we still have little understanding of when recombination levels are ‘set’, and we still do not know why the chromosomal locations of exchanges vary between the sexes21. Given the crucial role of crossovers in setting the stage for normal chromosome segregation, an understanding of the sex-specific signals that control patterns of exchanges is essential.
Sex differences in prophase checkpoint control
In somatic cells, a G2/M checkpoint functions to prevent the onset of metaphase in the presence of DNA damage (reviewed in Ref. 22). Evidence from both yeast23 and mammals24 suggests that an analogous checkpoint mechanism functions in meiotic cells and results in the demise of meiocytes when the repair of programmed double-stand breaks (DSBs) is perturbed. However, meiocytes are more complex than somatic cells, and synapsis between homologous chromosomes appears to have imposed an additional level of control.
Synaptic defects during meiotic prophase have been studied extensively in the mouse, and in the male they almost always result in spermatocyte death, either at the pachytene stage or at first meiotic metaphase. By contrast, females retain fertility in the face of many mutations that cause complete meiotic arrest and sterility in males, although their reproductive lifespan may be substantially shortened (reviewed in Ref. 25, and see Refs 26–29). Our understanding of this sex-specific difference has deepened with the recognition that synaptic failure leads to transcriptional silencing of unsynapsed chromosomal regions. In the male, synapsis between the sex chromosomes is limited to the small pseudoautosomal region, and transcriptional silencing of the remaining unsynapsed regions of the X and Y chromosomes occurs in the pachytene spermatocyte (reviewed in Refs 30–32). Although the mechanisms by which this meiotic sex chromosome inactivation (MSCI) is accomplished vary, sex chromosome inactivation in the heterogametic sex is highly conserved33. In mammals, silencing appears to involve a host of players, including components of the synaptonemal complex, the DNA repair machinery, and histone modifiers31.
MSCI is essential for male fertility32 and, as it occurs only in males, it provides a satisfying explanation for the difference in sensitivity in the sexes to the presence of unsynapsed chromatin. However, the real situation is likely to be more complex because the mechanisms that silence the large asynaptic regions of the X and Y chromosomes also act on unsynapsed autosomal chromosomes. This process is known as meiotic silencing of unsynapsed chromatin (MSUC) and occurs in both males and females34,35. In the male, unsynapsed autosomes are apparently transcriptionally silenced before the X and Y chromosomes, and this interferes with MSCI36. Accordingly, it has been hypothesized that failure to inactivate the sex chromosomes is the main cause of asynapsis-related male sterility35–38.
In females, the data indicate that transcriptional silencing of unsynapsed autosomal regions is detrimental but, in the absence of a requirement for sex chromosome silencing, the consequences are milder. Studies of mice with chromosome translocations or meiotic mutations that impede synapsis indicate that synaptic defects result in elimination of some, but not all, oocytes29,39–41. Indeed, in many situations, female fertility is maintained, whereas the male is sterile (reviewed in Ref. 25 and see Refs 26–29). The conclusion from the available data is that pachytene checkpoint mechanisms are less stringent in the female and that differences in sex chromosome activity during meiosis are likely to underlie the differences between the sexes, although a more complete understanding is needed.
Increasing nondisjunction with maternal age: is cohesin the reason?
As important as the above early sex-specific differences may be, the most obvious difference between spermatogenesis and oogenesis occurs after pachytene: male gametes proceed quickly through the rest of meiosis, but oocytes arrest in a late stage of prophase for weeks to months (in mice) or years, if not decades (in humans).
In addition to this long resting phase, human female meiosis is complicated by an intriguing and complex relationship between maternal age and the genetic quality of the egg. The maternal age curve for the incidence of trisomies among naturally occurring pregnancies is J-shaped, with a slight increase at the youngest maternal ages and an exponential increase in the decade preceding menopause42. The presence of maternal age effects at both extremes of reproductive life, coupled with the observation that the age curves are variable for individual human chromosomes (Box 4; reviewed in Ref. 3), points towards multiple mechanisms by which ageing affects chromosome segregation43. In this section, we briefly summarize the evidence for one mechanism: loss of sister chromatid cohesion.
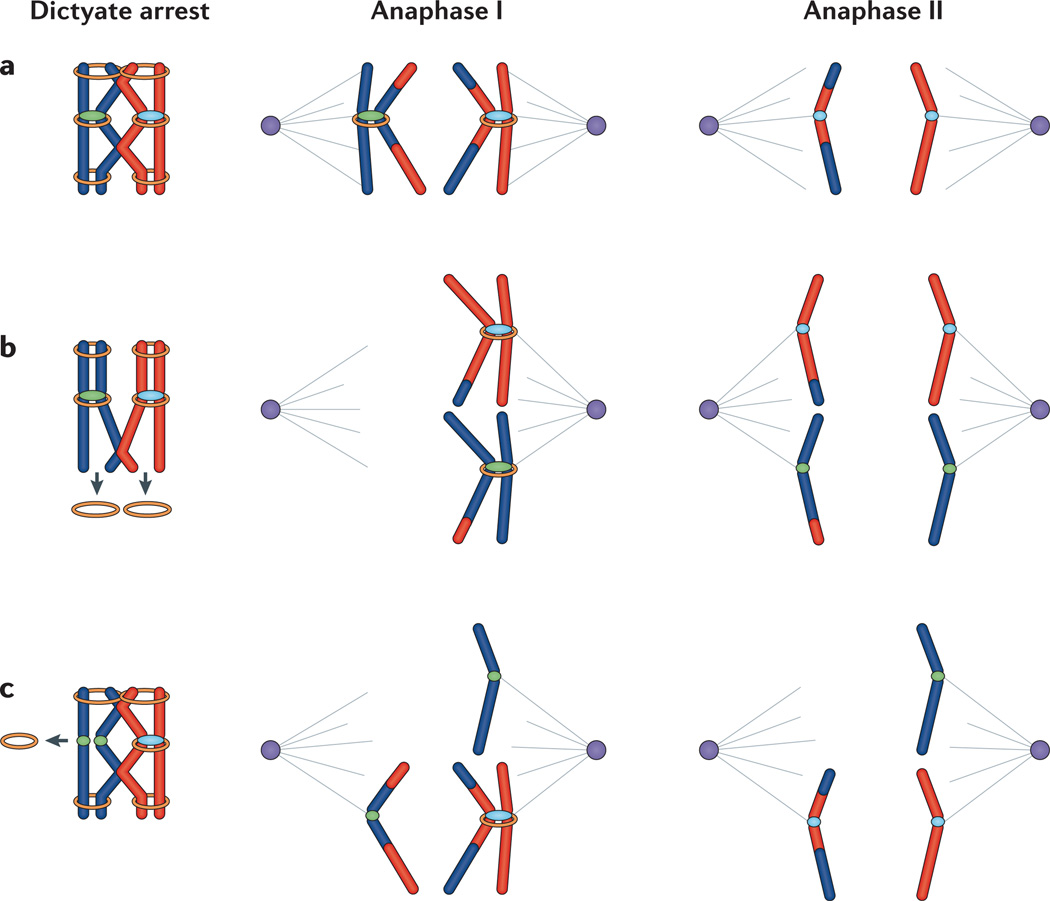
The sequential loss of sister chromatid cohesion from chromosome arms at anaphase I and from sister centromeres at anaphase II is essential to orchestrate the complex chromosome segregation events necessary to produce haploid gametes (Fig. 2). Failure to establish connections between homologues is one of the oldest postulated mechanisms of human aneuploidy44 and, as detailed above, studies of human trisomies suggest that recombination failure is, indeed, an important mechanism of human nondisjunction. However, on the basis of data from humans and model organisms, premature loss of connections between homologues is also an important contributor and could be due to loss of sister chromatid cohesion; for example, if homologues are only joined by a distally located crossover, loss of cohesion past the point of exchange could uncouple the homologues16,45,46(Fig. 2b; Box 3). Similarly, premature loss of cohesion between sister centromeres can lead to segregation errors at either the first or second meiotic division (Fig. 2c; Box 3). Early studies of eggs from women undergoing assisted reproductive procedures suggested that premature separation of sister centromeres is a major mechanism of human aneuploidy47 and, as discussed below, recent experimental findings strongly support this hypothesis.
Figure 2. Releasing sisters: normal and premature loss of cohesion.
a | The normal situation. Cohesion between sister chromatids (shown by the orange rings) is established during pre-meiotic S phase. Following recombination, cohesion distal to the sites of exchanges tethers homologues throughout dictyate arrest. During the first meiotic division, release of cohesion along chromosome arms but retention at sister centromeres allows homologues to segregate while retaining a centromeric connection between sister chromatids. During the second meiotic division, cleavage of the remaining centromeric cohesion allows sister chromatids to segregate. (Note that in this panel, we have followed segregation of only one of the two homologues; that is, the homologue on the right at anaphase I. Similarly, in the following panels only one of the two possible meiosis II configurations is shown.) b | Premature loss of arm cohesion. Loss of cohesion distal to sites of exchange before anaphase I may result in premature homologue separation into two unpaired univalents, which will then segregate independently of one another at meiosis I. If both homologues travel together, the production of unbalanced gametes is almost certain. For example, as shown here, the sisters of each homologue separate at meiosis II, yielding an oocyte (and second polar body) with an extra chromatid. c | Premature loss of centromeric cohesion. Loss of cohesion between sister centromeres can occur at meiosis I (as shown here) or meiosis II, leading to random segregation of sister centromeres.
Because cohesion is established during pre-meiotic S phase in the fetal ovary but chromosome segregation occurs years later in the adult, the idea that degradation of cohesion during the protracted meiotic arrest is the basis of the human maternal age effect is attractive. Studies in female Drosophila melanogaster yielded the first report that weakened cohesion leads to an age-related increase in meiotic nondisjunction48. Subsequent studies of mice with a mutation in the meiosis-specific cohesin structural maintenance of chromosomes 1B (Smc1b) demonstrated premature separation of homologues and of sister centromeres and suggested an age-related loss of cohesion49. Evidence that cohesins are lost from meiotic chromosomes in an age-related fashion has now been found in various mouse models, and increasing aneuploidy levels have been attributed to this loss49–53.
The hypothesis that loss of cohesion is the basis of the maternal age effect makes an important assumption. Specifically, it presupposes no turnover of the proteins in the cohesin complex that is loaded on meiotic chromosomes during fetal development; that is, chromosome segregation in the oocyte of a 50-year-old woman presumably relies on a complex of 50-year-old cohesin proteins. However, meiosis-specific cohesin transcripts are detected in growing oocytes49,54, suggesting that cohesin proteins may be replenished during oogenesis. Two recent complementary mouse studies have elegantly addressed this possibility. In the first study, a stage-specific knockout of Smc1b in the oocyte allowed for normal synthesis of this meiosis-specific cohesin in the fetal ovary but did not allow for any new protein synthesis during oocyte growth in the adult ovary54. The second study analysed the meiosis-specific cohesin REC8, testing whether REC8 synthesized in the growing oocyte could replenish protein that had been loaded during fetal development but then destroyed experimentally55. The results of the two studies were in agreement, suggesting that cohesins loaded onto chromosomes during fetal development are necessary and sufficient to mediate cohesion in the fully mature oocyte.
Thus, the combined data from studies using mouse models49–52,54,55 and studies of human oocytes10,47,56,57 suggest that loss of cohesin is an important mechanism of meiotic nondisjunction. However, several lines of evidence indicate that it is not the sole basis for the age effect. Perhaps most importantly, studies of human trisomies indicate that no single nondisjunctional mechanism applies to all chromosomes; that is, both the mechanisms of nondisjunction and the influence of age vary remarkably among chromosomes (Box 4). However, the differences are not simply dependent on chromosome size, as might be expected if cohesin loss were the only mechanism involved. The relationship between the two known aetiological agents — altered recombination and maternal age — is entirely dependent on chromosomal context: recombination failure has been linked to some58,59 but not other trisomies60 involving older women, and the relationship between altered location of crossovers and age is similarly chromosome-specific61. Age-related loss of cohesion is an attractive candidate mechanism for some situations (for example, small chromosomes held together by a single crossover) but is less so for others (for example, recombination failure, chromosomes held together by multiple crossovers and chromosomes held together by proximal crossovers). Thus, the evidence from humans indicates that there are multiple mechanisms that contribute to the maternal age effect.
Further, given the differences in chromosome abnormality rates and reproductive lifespans of mice and humans, caution must be exercised when transferring ideas across species. For example, the baseline level of aneuploidy in fertilized mouse eggs is an order of magnitude lower than it is in humans, and the effect of maternal age pales by comparison62. Thus, any attempt to extrapolate from mice to humans must take into account these differences in the nondisjunctional ‘phenotypes’ of the two species. Similarly, in the recent studies of cohesion loss in ageing mice, increases in nondisjunction were apparent only in reproductively senescent females51,52. Because age-related increases in human trisomies begin at least a decade before the onset of menopause, the relevance of the mouse data to the human situation is uncertain. Finally, in the only human study to date, no obvious differences in levels of meiotic cohesins were detected in oocytes from women of different ages63. This does not mean that a marked reduction in cohesins does not occur in humans — loss may indeed occur during the many years of prophase arrest. Nevertheless, given the duration of the arrest and the complexity of both nondisjunctional patterns and the influence of age, loss of cohesin seems unlikely to be the only force driving the precipitous increase in nondisjunction observed in the decade preceding menopause in humans.
The spindle assembly checkpoint: the final gatekeeper
The inability to maintain associations between homologous chromosomes — owing either to recombination failure or to impaired sister chromatid cohesion — results in the presence of unpaired univalents at the first meiotic division (Fig. 2; Box 3). The constraint imposed on sister kinetochores at meiosis I should hinder the ability of these univalents to make stable bipolar attachments to the spindle and, on the basis our understanding of cell cycle control, this should impede cell division. That is, in mitotic cells, all chromosomes must achieve stable bipolar attachments and align at the spindle equator before the cell can initiate anaphase, and the presence of even a single misaligned chromosome is sufficient to activate the spindle assembly checkpoint (SAC) and to delay anaphase onset64. Thus, in meiotic cells, the presence of a univalent chromosome that can form only a monopolar attachment should also activate the SAC. However, the ability of meiocytes to respond to disturbances in chromosome behaviour at metaphase I appears to be sex-specific. In male mice, the response is robust, and the presence of a single univalent chromosome causes metaphase I arrest and death of primary spermatocytes65,66. By contrast, SAC control at the first meiotic division — like pachytene checkpoint mechanisms — appears to be comparatively inefficient in the female. In fact, the presence of one or several univalents at metaphase I is not only compatible with anaphase onset in female mice, but the presence of these aberrant chromosomes induces no detectable cell cycle delay67,68.
At least two factors seem to be key to checkpoint evasion: univalent chromosome behaviour and differences in SAC control. Studies of female mice indicate that at least some univalents can satisfy SAC requirements by making bipolar attachments to the meiosis I spindle68–70. For example, analyses of multiple univalents in females that are deficient for synaptonemal complex protein 3 (Sycp3) indicate that univalents form bipolar attachments before the cell proceeds to anaphase I68. Similar results have recently been reported for females that are homozygous for a null mutation in the crossover-associated gene mutL homologue 1 (Mlh1)70, and it has long been recognized that the single X chromosome in XO female mice can either segregate sister chromatids or segregate intact at the first meiotic division69. Intriguingly, in both Mlh1 mutants and XO females, the ability of univalents to form bipolar attachments to the meiosis I spindle is dependent on genetic background; that is, it is enhanced on some inbred strains69,70. Nevertheless, it seems to be clear that sister centromeres frequently are able to form functionally distinct kinetochores at meiosis I. Importantly, however, although bipolar attachment at the first meiotic division may evade the SAC, premature segregation of sister chromatids during meiosis I predisposes to aneuploidy at the second meiotic division (Box 3).
There is also, however, compelling evidence that the SAC itself is ‘weaker’ in mammalian females than in males, and stable attachment of some, but not all, chromosomes is sufficient to satisfy SAC requirements70–73. Clearly, proteins involved in SAC-mediated control are present in oocytes (for example, Refs 74–78), and both spindle aberrations or an overwhelming number of univalent chromosomes cause metaphase I arrest in the female70,79. Intriguingly, recent studies in the mouse demonstrate that a true metaphase I, with all chromosomes properly aligned at the spindle equator, is not required for anaphase onset in the oocyte70–73. These observations are consistent with other findings from mice (for example, Refs 80–84) and humans85,86, in which the incidence of chromosome misalignment on the first meiotic spindle is correlated with an increased incidence of aneuploidy. Additionally, some cell cycle components appear to be used differently in the oocyte: regulation of the progression from metaphase to anaphase requires an appropriate transition between two distinct forms of the anaphase promoting complex (APC). APC–cadherin 1 (APC–CDH1), which is normally only active during prophase in mitotic cells, regulates chromosome congression during prometaphase I in the oocyte87. The onset of anaphase, however, requires the actions of a separate complex, APC–CDC20 (a version of the APC complexed with the cell division cycle 20 homologue). Successful transition from prometaphase I to anaphase I is achieved through the actions of the SAC protein, BUBR1 (also know as BUB1β), which controls the activity of both forms of the APC88. Importantly, disruptions of either complex result in an increased incidence of aneuploidy83,87.
Environmental effects on the oocyte
The possibility that human aneuploidy may be induced by environmental factors such as smoking, drinking, oral contraceptive use and radiation exposure has been suggested by data from human studies over many decades (for reviews, see Refs 8,42), but confirmatory evidence for these or any other agent has never emerged. During the past decade, however, experimental studies in the mouse and accumulating data from ART have provided compelling evidence of links to endocrine-disrupting chemicals or to exogenous hormones. In this section, we review the available data and outline the types of additional data needed to understand these effects and their ramifications for human reproduction.
Endocrine disruptors and aneuploidy: the BPA story
Perhaps the strongest link between an environmental exposure and aneuploidy comes from studies of a plasticizer to which humans are exposed on an almost continuous basis: bisphenol A (BPA). The first suggestion that this endocrine-disrupting chemical induces aneuploidy was the result of the accidental exposure of mice during the course of meiotic studies in our laboratory82. Although entirely serendipitous, these results supported the hypothesis under investigation: namely, that subtle age-related changes in the hormonal cues that control oocyte maturation contribute to the human maternal age effect81. A number of subsequent studies have confirmed that exposure of female mice to low levels of BPA during the final stages of oocyte growth disrupts meiotic chromosome behaviour89–92, but the endpoints have been disputed. It has been argued that the SAC would cause cell cycle arrest and death of oocytes that exhibit the chromosome alignment failure induced by BPA but would not give rise to aneuploid eggs91. However, an association between chromosome alignment failure — induced by various different mechanisms — and increased aneuploidy has been reported in the mouse (for example Refs 80–82,84). BPA exposure may also alter the likelihood that mature eggs are produced: in rodent studies, BPA has been reported to affect follicle growth93, and studies of women undergoing assisted reproduction suggest that BPA interferes with the stimulation procedures used for oocyte retrieval94 and that levels of BPA in maternal blood and follicular fluid are inversely correlated with oocyte maturity and fertilization rates95. It remains unclear if the observed effects are unique to BPA, and further studies of the effects of other endocrine disrupting chemicals are clearly warranted.
In addition to effects on the growing oocyte, recent studies in both mice and worms suggest that BPA disrupts the earliest stages of oocyte development, altering synapsis and recombination during meiotic prophase and increasing the incidence of meiotic errors in the adult female96,97. Studies for assessing the mechanisms by which BPA exerts its effects suggest that it interferes with the actions of the classical oestrogen receptor ERβ, implicating oestrogen in the control of the onset of oogenesis in the fetal ovary96. Because disturbances during fetal development may have an impact on the entire cohort of oocytes produced by the female, the implications for human fertility are profound. However, because the disturbances occur in utero, but the effects do not manifest until adulthood, demonstrating cause and effect in humans will be a daunting task. To date, the only suggestion that BPA disturbs prophase events in humans comes from studies of in vitro exposures of cultured fetal ovarian tissues98. However, the striking similarities between the findings in studies of mice and worms96,97 underscore the reasons for concern.
Ovarian stimulation protocols and aneuploidy in ART
It seems likely that at least some of the mechanisms and/or causes of aneuploidy in ART-derived conceptions are unique to this population. Notably, studies of human eggs and embryos almost always involve infertile individuals, and it is conceivable that error rates are intrinsically higher in these couples. More importantly, however, it is also possible that the procedures used in ART increase aneuploidy levels (Box 1). Specifically, during the past decade, evidence that both ovarian stimulation protocols and in vitro culture adversely affect oocyte and embryo quality has accumulated. Most of the available data come from the analysis of epigenetic changes and, as imprints are acquired during the process of oocyte growth, changes in methylation and/or the expression of imprinted genes provide evidence that the late stages of oocyte development and maturation may be affected by ART (for example, Refs 99–104).
Although the data implicating exogenous hormones in the genesis of human aneuploidy are recent, the idea is not a new one. In humans, the introduction of the contraceptive pill raised concerns about an increase in chromosomally abnormal conceptions among women who became pregnant while taking the earliest form of this contraceptive105. Further, early cytogenetic studies suggested that ovarian stimulation protocols used in mice increased the chromosome abnormality rate in eggs106. The effects may even extend to endogenous hormones, as changes in the endocrine environment have been postulated to underlie human age-related aneuploidy in natural pregnancies81.
The introduction of ART made the development of improved stimulation protocols a necessity, and some of the early data from FISH-based analyses suggested that higher aneuploidy rates may be a feature of specific ovarian stimulation regimes107. In recent years, the suggestion that stimulation protocols used for oocyte collection may adversely have an impact on oocyte quality — at least in some women — has raised concern, and some clinics have been experimenting with the use of ‘natural cycles’. Further, the idea that milder stimulation protocols improve oocyte quality is steadily gaining ground, and comparative studies of these new protocols against established protocols have provided the first direct evidence in humans that lower doses of gonadotropins are correlated with lower aneuploidy rates108,109.
These data from ART add to a growing body of evidence81,82, suggesting that subtle changes in the complex interplay of hormonal signals that control oocyte growth and maturation are important in the generation of human aneuploidy. Importantly, the development of array-based approaches for the analysis of human eggs and embryos (Box 2) provides a sensitive means of directly testing at least some of these environmental variables, providing hope for new improvements in ART.
Conclusions and future directions
Recent findings summarized in this Review lend new credence to several old hypotheses: that the human maternal age effect involves different ‘hits’ that conspire to increase the frequency of errors in the egg16; that events occurring in the fetal ovary that influence the prophase interactions between homologous chromosomes have an important role44; that the long prophase arrest in females contributes to aneuploidy because of age-dependent decay of components of the meiotic machinery110; and that environmental effects may act at several different stages of oogenesis to influence the likelihood of mistakes111. Taken together, a unifying theme has emerged: the genesis of human aneuploidy is a multi-step process caused by errors at several distinct stages of oogenesis and exacerbated by a lack of efficient checkpoints. Thus, future attempts to design new clinical strategies to prevent aneuploidy must take into account the fact that no single therapeutic approach will suffice. Although the new findings underscore the complexity of human aneuploidy, they also raise a host of new questions, providing fertile ground for future research. Two of the more intriguing questions are as follows.
First, how do hormonal signals control the onset of meiosis in the fetal ovary, and what types of endocrine disruptors have an impact on these processes? The role of endogenous hormones during the final stages of oocyte development in the adult is well-known, and it is becoming clear that exogenous signals that interfere with the delicate balance of these endocrine signals can cause aneuploidy81,82. However, recent findings suggest that hormonal signals also have a crucial role at the onset of meiosis in the fetal ovary. Given the growing evidence linking environmental factors to aneuploidy, an understanding of the hormonal signals that control both stages of female meiosis — as well as meiosis in males — is imperative.
Second, are the apparent differences between naturally occurring and ART pregnancies a reflection of differences between fertile and subfertile individuals, or could they be induced by ART procedures? The evidence that environmental factors contribute to aneuploidy is growing, and the technology to address this concern is in hand. Array-based procedures for the analysis of human eggs and embryos provide the first means of directly examining the impact of exogenous factors on the genetic quality of the egg (Box 2).
Answers to these questions have direct relevance to the treatment of human infertility and also to the reproductive health of ours and other species. Interest in developing and refining culture systems to support the development of functional gametes from stem cells for the treatment of infertility is intense (for example, Refs 112–119) but, to date, those who are engaged in these endeavours have paid little attention to the meiotic process. Clearly, the successful production of normal gametes in vitro will require great attention to meiotic details and a complete understanding of the differences between the sexes.
Acknowledgements
Research conducted in the Hunt and Hassold laboratories and discussed in this Review was supported by US National Institutes of Health grants HD21341 (to T.J.H.) and ES013527 (to P.A.H.). In addition, the authors would gratefully like to acknowledge the three ‘grand dames’ of human aneuploidy research who sparked our interest and shaped our thinking: D. Warburton, M. Mikkelsen and, most of all, P. Jacobs.
Glossary
- Aneuploidy
A chromosome abnormality in which the chromosome number is not a multiple of the haploid number
- Assisted reproductive technology (ART)
Clinical approaches that are used to help infertile couples achieve a normal pregnancy. These include ovarian stimulation protocols using exogenous hormones, in vitro fertilization, intracytoplasmic sperm injection and preimplantation genetic diagnosis
- Nondisjunction
The failure of chromosomes to segregate normally during cell division. Nondisjunction at meiosis I results in products with additional or missing whole chromosomes; nondisjunction at meiosis II results in products with additional or missing sister chromatids
- Pachytene
The stage of meiotic prophase characterized by complete synapsis of all homologues. Importantly, crossover sites can be visualized in pachytene stage cells using appropriate markers
- Bivalent
Paired homologous chromosomes that are tethered by a crossover (or crossovers)
- Synapsis
The intimate pairing of homologous chromosomes that occurs during prophase of meiosis and is essential for meiotic recombination. Synapsis is facilitated by the formation of a meiosis-specific protein scaffold called the synaptonemal complex
- Pseudoautosomal region (PAR)
The small region of homology at the distal ends of the X and Y chromosomes that allows for synapsis and recombination
- Sister chromatid cohesion
Replicated chromosomes, or sister chromatids, are held together by cohesin, which is a protein complex that is loaded onto the chromosomes during S phase. In meiosis, sister chromatid cohesion is sequentially released from the chromosome arms at anaphase I and from sister centromeres at anaphase II, allowing for the orderly segregation of homologues and sister chromatids, respectively
- Univalents
Homologous chromosomes that are not associated with one another (for example, owing to failure to recombine)
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- 1.Lejeune J, Turpin R, Gautier M. Mongolism; a chromosomal disease (trisomy) Bull. Acad. Natl Med. 1959;143:256–265. [PubMed] [Google Scholar]
- 2.Jacobs PA, Baikie AG, Court Brown WM, Strong JA. The somatic chromosomes in mongolism. Lancet. 1959;1:710. doi: 10.1016/s0140-6736(59)91892-6. [DOI] [PubMed] [Google Scholar]
- 3.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nature Rev. Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 4.Gaulden ME. Maternal age effect: the enigma of Down syndrome and other trisomic conditions. Mutat. Res. 1992;296:69–88. doi: 10.1016/0165-1110(92)90033-6. [DOI] [PubMed] [Google Scholar]
- 5.Jacobs PA. The chromosome complement of human gametes. Oxf. Rev. Reprod. Biol. 1992;14:47–72. [PubMed] [Google Scholar]
- 6.Jamieson ME, Coutts JR, Connor JM. The chromosome constitution of human preimplantation embryos fertilized in vitro . Hum. Reprod. 1994;9:709–715. doi: 10.1093/oxfordjournals.humrep.a138575. [DOI] [PubMed] [Google Scholar]
- 7.Pellestor F, Andreo B, Anahory T, Hamamah S. The occurrence of aneuploidy in human: lessons from the cytogenetic studies of human oocytes. Eur. J. Med. Genet. 2006;49:103–116. doi: 10.1016/j.ejmg.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Pacchierotti F, Adler ID, Eichenlaub-Ritter U, Mailhes JB. Gender effects on the incidence of aneuploidy in mammalian germ cells. Environ. Res. 2007;104:46–69. doi: 10.1016/j.envres.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Fragouli E, et al. Comprehensive molecular cytogenetic analysis of the human blastocyst stage. Hum. Reprod. 2008;23:2596–2608. doi: 10.1093/humrep/den287. [DOI] [PubMed] [Google Scholar]
- 10.Fragouli E, et al. The cytogenetics of polar bodies: insights into female meiosis and the diagnosis of aneuploidy. Mol. Hum. Reprod. 2011;17:286–295. doi: 10.1093/molehr/gar024. [DOI] [PubMed] [Google Scholar]
- 11.Gabriel AS, et al. Array comparative genomic hybridisation on first polar bodies suggests that non-disjunction is not the predominant mechanism leading to aneuploidy in humans. J. Med. Genet. 2011;48:433–437. doi: 10.1136/jmg.2010.088070. [DOI] [PubMed] [Google Scholar]
- 12.Gutierrez-Mateo C, et al. Validation of microarray comparative genomic hybridization for comprehensive chromosome analysis of embryos. Fertil. Steril. 2011;95:953–958. doi: 10.1016/j.fertnstert.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 13.Treff NR, et al. Single nucleotide polymorphism microarray-based concurrent screening of 24-chromosome aneuploidy and unbalanced translocations in preimplantation human embryos. Fertil. Steril. 2011;95:1606–1612.e2. doi: 10.1016/j.fertnstert.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 14.Treff NR, et al. SNP microarray-based 24 chromosome aneuploidy screening is significantly more consistent than FISH. Mol. Hum. Reprod. 2010;16:583–589. doi: 10.1093/molehr/gaq039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geraedts J, et al. Polar body array CGH for prediction of the status of the corresponding oocyte. Part I: clinical results. Hum. Reprod. 2011;26:3173–3180. doi: 10.1093/humrep/der294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamb NE, et al. Susceptible chiasmate configurations of chromosome 21 predispose to nondisjunction in both maternal meiosis I and meiosis II. Nature Genet. 1996;14:400–405. doi: 10.1038/ng1296-400. [DOI] [PubMed] [Google Scholar]
- 17.Lynn A, et al. Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science. 2002;296:2222–2225. doi: 10.1126/science.1071220. [DOI] [PubMed] [Google Scholar]
- 18. Cheng EY, et al. Meiotic recombination in human oocytes. PLoS Genet. 2009;5:e1000661. doi: 10.1371/journal.pgen.1000661. This is a study of human fetal oocytes that provides evidence that recombination errors occurring during fetal development set the stage for nondisjunction in the adult
- 19.Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong A, et al. Sequence variants in the RNF212 gene associate with genome-wide recombination rate. Science. 2008;319:1398–1401. doi: 10.1126/science.1152422. [DOI] [PubMed] [Google Scholar]
- 21.Lynn A, Ashley T, Hassold T. Variation in human meiotic recombination. Annu. Rev. Genom. Hum. Genet. 2004;5:317–349. doi: 10.1146/annurev.genom.4.070802.110217. [DOI] [PubMed] [Google Scholar]
- 22.O’Connell MJ, Walworth NC, Carr AM. The G2-phase DNA-damage checkpoint. Trends Cell Biol. 2000;10:296–303. doi: 10.1016/s0962-8924(00)01773-6. [DOI] [PubMed] [Google Scholar]
- 23.Hochwagen A, Amon A. Checking your breaks: surveillance mechanisms of meiotic recombination. Curr. Biol. 2006;16:R217–R228. doi: 10.1016/j.cub.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 24.Li XC, Schimenti JC. Mouse pachytene checkpoint 2 (trip13) is required for completing meiotic recombination but not synapsis. PLoS Genet. 2007;3:e130. doi: 10.1371/journal.pgen.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunt PA, Hassold TJ. Sex matters in meiosis. Science. 2002;296:2181–2183. doi: 10.1126/science.1071907. [DOI] [PubMed] [Google Scholar]
- 26.Celeste A, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bannister LA, et al. A dominant, recombination-defective allele of Dmc1 causing male-specific sterility. PLoS Biol. 2007;5:e105. doi: 10.1371/journal.pbio.0050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuznetsov S, et al. RAD51C deficiency in mice results in early prophase I arrest in males and sister chromatid separation at metaphase II in females. J. Cell Biol. 2007;176:581–592. doi: 10.1083/jcb.200608130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herran Y, et al. The cohesin subunit RAD21L functions in meiotic synapsis and exhibits sexual dimorphism in fertility. EMBO J. 2011;30:3091–3105. doi: 10.1038/emboj.2011.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKee BD, Handel MA. Sex chromosomes, recombination, and chromatin conformation. Chromosoma. 1993;102:71–80. doi: 10.1007/BF00356023. [DOI] [PubMed] [Google Scholar]
- 31.Turner JM. Meiotic sex chromosome inactivation. Development. 2007;134:1823–1831. doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- 32. Burgoyne PS, Mahadevaiah SK, Turner JM. The consequences of asynapsis for mammalian meiosis. Nature Rev. Genet. 2009;10:207–216. doi: 10.1038/nrg2505. This is an informative Review of the meiotic consequences of synaptic defects that emphasizes the mechanisms and consequences of transcriptional silencing of unsynapsed regions
- 33.Cloutier JM, Turner JM. Meiotic sex chromosome inactivation. Curr. Biol. 2010;20:R962–R963. doi: 10.1016/j.cub.2010.09.041. [DOI] [PubMed] [Google Scholar]
- 34.Baarends WM, et al. Silencing of unpaired chromatin and histone H2A ubiquitination in mammalian meiosis. Mol. Cell. Biol. 2005;25:1041–1053. doi: 10.1128/MCB.25.3.1041-1053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turner JM, et al. Silencing of unsynapsed meiotic chromosomes in the mouse. Nature Genet. 2005;37:41–47. doi: 10.1038/ng1484. [DOI] [PubMed] [Google Scholar]
- 36.Mahadevaiah SK, et al. Extensive meiotic asynapsis in mice antagonizes meiotic silencing of unsynapsed chromatin and consequently disrupts meiotic sex chromosome inactivation. J. Cell Biol. 2008;182:263–276. doi: 10.1083/jcb.200710195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Royo H, et al. Evidence that meiotic sex chromosome inactivation is essential for male fertility. Curr. Biol. 2010;20:2117–2123. doi: 10.1016/j.cub.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 38.Homolka D, Jansa P, Forejt J. Genetically enhanced asynapsis of autosomal chromatin promotes transcriptional dysregulation and meiotic failure. Chromosoma. 2011;121:91–104. doi: 10.1007/s00412-011-0346-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burgoyne PS, Baker TG. Perinatal oocyte loss in XO mice and its implications for the aetiology of gonadal dysgenesis in XO women. J. Reprod. Fertil. 1985;75:633–645. doi: 10.1530/jrf.0.0750633. [DOI] [PubMed] [Google Scholar]
- 40.de Boer P, de Jong JH. In: Fertility and Chromosome Pairing: Recent Studies in Plants and Animals. Gilles CB, editor. CRC Press; 1989. pp. 77–107. [Google Scholar]
- 41.Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell. 2000;6:989–998. doi: 10.1016/s1097-2765(00)00098-8. [DOI] [PubMed] [Google Scholar]
- 42.Hassold TJ, Jacobs PA. Trisomy in man. Annu. Rev. Genet. 1984;18:69–97. doi: 10.1146/annurev.ge.18.120184.000441. [DOI] [PubMed] [Google Scholar]
- 43.Hunt P, Hassold T. Female meiosis: coming unglued with age. Curr. Biol. 2010;20:R699–R702. doi: 10.1016/j.cub.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 44.Henderson SA, Edwards RG. Chiasma frequency and maternal age in mammals. Nature. 1968;218:22–28. doi: 10.1038/218022a0. [DOI] [PubMed] [Google Scholar]
- 45.Koehler KE, et al. Spontaneous X chromosome MI and MII nondisjunction events in Drosophila melanogaster oocytes have different recombinational histories. Nature Genet. 1996;14:406–414. doi: 10.1038/ng1296-406. [DOI] [PubMed] [Google Scholar]
- 46.Ross LO, Maxfield R, Dawson D. Exchanges are not equally able to enhance meiotic chromosome segregation in yeast. Proc. Natl Acad. Sci. USA. 1996;93:4979–4983. doi: 10.1073/pnas.93.10.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Angell RR. Predivision in human oocytes at meiosis I: a mechanism for trisomy formation in man. Hum. Genet. 1991;86:383–387. doi: 10.1007/BF00201839. [DOI] [PubMed] [Google Scholar]
- 48.Jeffreys CA, Burrage PS, Bickel SE. A model system for increased meiotic nondisjunction in older oocytes. Curr. Biol. 2003;13:498–503. doi: 10.1016/s0960-9822(03)00134-9. [DOI] [PubMed] [Google Scholar]
- 49.Hodges CA, Revenkova E, Jessberger R, Hassold TJ, Hunt PA. SMC1β-deficient female mice provide evidence that cohesins are a missing link in age-related nondisjunction. Nature Genet. 2005;37:1351–1355. doi: 10.1038/ng1672. [DOI] [PubMed] [Google Scholar]
- 50.Liu L, Keefe DL. Defective cohesin is associated with age-dependent misaligned chromosomes in oocytes. Reprod. Biomed. Online. 2008;16:103–112. doi: 10.1016/s1472-6483(10)60562-7. [DOI] [PubMed] [Google Scholar]
- 51.Chiang T, Duncan FE, Schindler K, Schultz RM, Lampson MA. Evidence that weakened centromere cohesion is a leading cause of age-related aneuploidy in oocytes. Curr. Biol. 2010;20:1522–1528. doi: 10.1016/j.cub.2010.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lister LM, et al. Age-related meiotic segregation errors in Mammalian oocytes are preceded by depletion of cohesin and Sgo2. Curr. Biol. 2010;20:1511–1521. doi: 10.1016/j.cub.2010.08.023. [DOI] [PubMed] [Google Scholar]
- 53.Chiang T, Schultz RM, Lampson M. Meiotic origins of maternal age-related aneuploidy. Biol. Reprod. 2011;86:1–7. doi: 10.1095/biolreprod.111.094367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Revenkova E, Herrmann K, Adelfalk C, Jessberger R. Oocyte cohesin expression restricted to predictyate stages provides full fertility and prevents aneuploidy. Curr. Biol. 2010;20:1529–1533. doi: 10.1016/j.cub.2010.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tachibana-Konwalski K, et al. Rec8-containing cohesin maintains bivalents without turnover during the growing phase of mouse oocytes. Genes Dev. 2010;24:2505–2516. doi: 10.1101/gad.605910. This series of recent papers focuses on meiotic cohesins during oogenesis in the mouse. References 50–53 link loss of cohesin proteins with maternal age-dependent aneuploidy. References 54 and 55 provide evidence that cohesin proteins loaded during fetal development are necessary and sufficient to orchestrate meiotic chromosome segregation in the adult.
- 56.Angell RR, Xian J, Keith J, Ledger W, Baird DT. First meiotic division abnormalities in human oocytes: mechanism of trisomy formation. Cytogenet. Cell Genet. 1994;65:194–202. doi: 10.1159/000133631. [DOI] [PubMed] [Google Scholar]
- 57.Pellestor F, Andreo B, Arnal F, Humeau C, Demaille J. Maternal aging and chromosomal abnormalities: new data drawn from in vitro unfertilized human oocytes. Hum. Genet. 2003;112:195–203. doi: 10.1007/s00439-002-0852-x. [DOI] [PubMed] [Google Scholar]
- 58.Fisher JM, Harvey JF, Morton NE, Jacobs PA. Trisomy 18: studies of the parent and cell division of origin and the effect of aberrant recombination on nondisjunction. Am. J. Hum. Genet. 1995;56:669–675. [PMC free article] [PubMed] [Google Scholar]
- 59.Bugge M, et al. Non-disjunction of chromosome 18. Hum. Mol. Genet. 1998;7:661–669. doi: 10.1093/hmg/7.4.661. [DOI] [PubMed] [Google Scholar]
- 60.Hassold T, Merrill M, Adkins K, Freeman S, Sherman S. Recombination and maternal age-dependent nondisjunction: molecular studies of trisomy 16. Am. J. Hum. Genet. 1995;57:867–874. [PMC free article] [PubMed] [Google Scholar]
- 61.Oliver TR, et al. New insights into human nondisjunction of chromosome 21 in oocytes. PLoS Genet. 2008;4:e1000033. doi: 10.1371/journal.pgen.1000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bond DJ, Chandley AC. Aneuploidy. Oxford Univ. Press; 1983. pp. 83–90. [Google Scholar]
- 63.Garcia-Cruz R, et al. Dynamics of cohesin proteins REC8, STAG3, SMC1 β and SMC3 are consistent with a role in sister chromatid cohesion during meiosis in human oocytes. Hum. Reprod. 2010;25:2316–2327. doi: 10.1093/humrep/deq180. [DOI] [PubMed] [Google Scholar]
- 64.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nature Rev. Mol. Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 65.Kot MC, Handel MA. Spermatogenesis in XO, Sxr mice: role of the Y chromosome. J. Exp. Zool. 1990;256:92–105. doi: 10.1002/jez.1402560112. [DOI] [PubMed] [Google Scholar]
- 66.Sutcliffe MJ, Darling SM, Burgoyne PS. Spermatogenesis in XY, XYSxra and XOSxra mice: a quantitative analysis of spermatogenesis throughout puberty. Mol. Reprod. Dev. 1991;30:81–89. doi: 10.1002/mrd.1080300202. [DOI] [PubMed] [Google Scholar]
- 67.LeMaire-Adkins R, Radke K, Hunt PA. Lack of checkpoint control at the metaphase/anaphase transition: a mechanism of meiotic nondisjunction in mammalian females. J. Cell Biol. 1997;139:1611–1619. doi: 10.1083/jcb.139.7.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kouznetsova A, Lister L, Nordenskjold M, Herbert M, Hoog C. Bi-orientation of achiasmatic chromosomes in meiosis I oocytes contributes to aneuploidy in mice. Nature Genet. 2007;39:966–968. doi: 10.1038/ng2065. [DOI] [PubMed] [Google Scholar]
- 69.LeMaire-Adkins R, Hunt PA. Nonrandom segregation of the mouse univalent X chromosome: evidence of spindle-mediated meiotic drive. Genetics. 2000;156:775–783. doi: 10.1093/genetics/156.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nagaoka SI, Hodges CA, Albertini DF, Hunt PA. Oocyte-specific differences in cell-cycle control create an innate susceptibility to meiotic errors. Curr. Biol. 2011;21:651–657. doi: 10.1016/j.cub.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gui L, Homer H. Spindle assembly checkpoint signalling is uncoupled from chromosomal position in mouse oocytes. Development. 2012;139:1941–1946. doi: 10.1242/dev.078352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kolano A, Brunet S, Silk AD, Cleveland DW, Verlhac MH. Error prone mammalian female meiosis from silencing the spindle assembly checkpoint without normal interkinetochore tension. Proc. Natl Acad. Sci. USA. 2012 May 2; doi: 10.1073/pnas.1204686109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lane SIR, Yun Y, Jones KT. Timing of anaphase promoting complex activation in mouse oocytes is predicted by microtubule–kinetochore attachment, but not by bivalent alignment or tension. Development. 2012;139:1947–1955. doi: 10.1242/dev.077040. The studies described in references 70–73 provide evidence that the spindle assembly checkpoint mechanism differs in the oocyte and that metaphase alignment of all chromosomes is not a prerequisite for anaphase onset. This difference provides a mechanism whereby various different factors can all lead to aneuploidy
- 74.Steuerwald N, Cohen J, Herrera RJ, Sandalinas M, Brenner CA. Association between spindle assembly checkpoint expression and maternal age in human oocytes. Mol. Hum. Reprod. 2001;7:49–55. doi: 10.1093/molehr/7.1.49. [DOI] [PubMed] [Google Scholar]
- 75.Brunet S, Pahlavan G, Taylor S, Maro B. Functionality of the spindle checkpoint during the first meiotic division of mammalian oocytes. Reproduction. 2003;126:443–450. doi: 10.1530/rep.0.1260443. [DOI] [PubMed] [Google Scholar]
- 76.Wassmann K, Niault T, Maro B. Metaphase I arrest upon activation of the Mad2-dependent spindle checkpoint in mouse oocytes. Curr. Biol. 2003;13:1596–1608. doi: 10.1016/j.cub.2003.08.052. [DOI] [PubMed] [Google Scholar]
- 77.Homer HA, McDougall A, Levasseur M, Murdoch AP, Herbert M. Mad2 is required for inhibiting securin and cyclin B degradation following spindle depolymerisation in meiosis I mouse oocytes. Reproduction. 2005;130:829–843. doi: 10.1530/rep.1.00856. [DOI] [PubMed] [Google Scholar]
- 78.McGuinness BE, et al. Regulation of APC/C activity in oocytes by a Bub1-dependent spindle assembly checkpoint. Curr. Biol. 2009;19:369–380. doi: 10.1016/j.cub.2009.01.064. [DOI] [PubMed] [Google Scholar]
- 79.Woods LM, et al. Chromosomal influence on meiotic spindle assembly: abnormal meiosis I in female Mlh1 mutant mice. J. Cell Biol. 1999;145:1395–1406. doi: 10.1083/jcb.145.7.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yin H, Cukurcam S, Betzendahl I, Adler ID, Eichenlaub-Ritter U. Trichlorfon exposure, spindle aberrations and nondisjunction in mammalian oocytes. Chromosoma. 1998;107:514–522. doi: 10.1007/s004120050337. [DOI] [PubMed] [Google Scholar]
- 81.Hodges CA, et al. Experimental evidence that changes in oocyte growth influence meiotic chromosome segregation. Hum. Reprod. 2002;17:1171–1180. doi: 10.1093/humrep/17.5.1171. [DOI] [PubMed] [Google Scholar]
- 82.Hunt PA, et al. Bisphenol a exposure causes meiotic aneuploidy in the female mouse. Curr. Biol. 2003;13:546–553. doi: 10.1016/s0960-9822(03)00189-1. [DOI] [PubMed] [Google Scholar]
- 83.Jin F, et al. Cdc20 is critical for meiosis I and fertility of female mice. PLoS Genet. 2010;6:e1001147. doi: 10.1371/journal.pgen.1001147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Selesniemi K, Lee HJ, Muhlhauser A, Tilly JL. Prevention of maternal aging-associated oocyte aneuploidy and meiotic spindle defects in mice by dietary and genetic strategies. Proc. Natl Acad. Sci. USA. 2011;108:12319–12324. doi: 10.1073/pnas.1018793108. This provocative report links caloric restriction with decreased levels of aneuploidy in the ageing female mouse
- 85.Battaglia DE, Goodwin P, Klein NA, Soules MR. Influence of maternal age on meiotic spindle assembly in oocytes from naturally cycling women. Hum. Reprod. 1996;11:2217–2222. doi: 10.1093/oxfordjournals.humrep.a019080. [DOI] [PubMed] [Google Scholar]
- 86.Volarcik K, et al. The meiotic competence of in-vitro matured human oocytes is influenced by donor age: evidence that folliculogenesis is compromised in the reproductively aged ovary. Hum. Reprod. 1998;13:154–160. doi: 10.1093/humrep/13.1.154. [DOI] [PubMed] [Google Scholar]
- 87.Reis A, et al. Prometaphase APCcdh1 activity prevents non-disjunction in mammalian oocytes. Nature Cell Biol. 2007;9:1192–1198. doi: 10.1038/ncb1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Homer H, Gui L, Carroll J. A spindle assembly checkpoint protein functions in prophase I arrest and prometaphase progression. Science. 2009;326:991–994. doi: 10.1126/science.1175326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Can A, Semiz O, Cinar O. Bisphenol-A induces cell cycle delay and alters centrosome and spindle microtubular organization in oocytes during meiosis. Mol. Hum. Reprod. 2005;11:389–396. doi: 10.1093/molehr/gah179. [DOI] [PubMed] [Google Scholar]
- 90.Pacchierotti F, Ranaldi R, Eichenlaub-Ritter U, Attia S, Adler ID. Evaluation of aneugenic effects of bisphenol A in somatic and germ cells of the mouse. Mutat. Res. 2008;651:64–70. doi: 10.1016/j.mrgentox.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 91.Eichenlaub-Ritter U, et al. Exposure of mouse oocytes to bisphenol A causes meiotic arrest but not aneuploidy. Mutat. Res. 2008;651:82–92. doi: 10.1016/j.mrgentox.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 92.Lenie S, Cortvrindt R, Eichenlaub-Ritter U, Smitz J. Continuous exposure to bisphenol A during in vitro follicular development induces meiotic abnormalities. Mutat. Res. 2008;651:71–81. doi: 10.1016/j.mrgentox.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 93.Peretz J, Gupta RK, Singh J, Hernandez-Ochoa I, Flaws JA. Bisphenol A impairs follicle growth, inhibits steroidogenesis, and downregulates rate-limiting enzymes in the estradiol biosynthesis pathway. Toxicol. Sci. 2011;119:209–217. doi: 10.1093/toxsci/kfq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bloom MS, et al. Bisphenol A exposure reduces the estradiol response to gonadotropin stimulation during in vitro fertilization. Fertil. Steril. 2011;96:672–677.e2. doi: 10.1016/j.fertnstert.2011.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fujimoto VY, et al. Serum unconjugated bisphenol A concentrations in women may adversely influence oocyte quality during in vitro fertilization. Fertil. Steril. 2011;95:1816–1819. doi: 10.1016/j.fertnstert.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 96.Susiarjo M, Hassold TJ, Freeman E, Hunt PA. Bisphenol A exposure in utero disrupts early oogenesis in the mouse. PLoS Genet. 2007;3 doi: 10.1371/journal.pgen.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Allard P, Colaiacovo MP. Bisphenol A impairs the double-strand break repair machinery in the germline and causes chromosome abnormalities. Proc. Natl Acad. Sci. USA. 2010;107:20405–20410. doi: 10.1073/pnas.1010386107. These studies provide evidence that exposure to the bisphenol A (BPA) disrupts the prophase events of meiosis in the ovaries of mice and worms, setting the stage for nondisjunctional events during the meiotic divisions
- 98.Brieno-Enriquez MA, et al. Human meiotic progression and recombination are affected by bisphenol A exposure during in vitro human oocyte development. Hum. Reprod. 2011;26:2807–2818. doi: 10.1093/humrep/der249. [DOI] [PubMed] [Google Scholar]
- 99.Doherty AS, Mann MR, Tremblay KD, Bartolomei MS, Schultz RM. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol. Reprod. 2000;62:1526–1535. doi: 10.1095/biolreprod62.6.1526. [DOI] [PubMed] [Google Scholar]
- 100.Khosla S, Dean W, Brown D, Reik W, Feil R. Culture of preimplantation mouse embryos affects fetal development and the expression of imprinted genes. Biol. Reprod. 2001;64:918–926. doi: 10.1095/biolreprod64.3.918. [DOI] [PubMed] [Google Scholar]
- 101.Mann MR, et al. Selective loss of imprinting in the placenta following preimplantation development in culture. Development. 2004;131:3727–3735. doi: 10.1242/dev.01241. [DOI] [PubMed] [Google Scholar]
- 102.Rivera RM, et al. Manipulations of mouse embryos prior to implantation result in aberrant expression of imprinted genes on day 9.5 of development. Hum. Mol. Genet. 2008;17:1–14. doi: 10.1093/hmg/ddm280. [DOI] [PubMed] [Google Scholar]
- 103.Market-Velker BA, Zhang L, Magri LS, Bonvissuto AC, Mann MR. Dual effects of superovulation: loss of maternal and paternal imprinted methylation in a dose-dependent manner. Hum. Mol. Genet. 2010;19:36–51. doi: 10.1093/hmg/ddp465. [DOI] [PubMed] [Google Scholar]
- 104.Denomme MM, Zhang L, Mann MR. Embryonic imprinting perturbations do not originate from superovulation-induced defects in DNA methylation acquisition. Fertil. Steril. 2011;96:734–738.e2. doi: 10.1016/j.fertnstert.2011.06.055. [DOI] [PubMed] [Google Scholar]
- 105.Harlap S, et al. Chromosome abnormalities in oral contraceptive breakthrough pregnancies. Lancet. 1979;1:1342–1343. doi: 10.1016/s0140-6736(79)91969-x. [DOI] [PubMed] [Google Scholar]
- 106.Maudlin I, Fraser LR. The effect of PMSG dose on the incidence of chromosomal anomalies in mouse embryos fertilized in vitro. J Reprod. Fertil. 1977;50:275–280. doi: 10.1530/jrf.0.0500275. [DOI] [PubMed] [Google Scholar]
- 107.Munne S, et al. Treatment-related chromosome abnormalities in human embryos. Hum. Reprod. 1997;12:780–784. doi: 10.1093/humrep/12.4.780. [DOI] [PubMed] [Google Scholar]
- 108.Baart EB, et al. Milder ovarian stimulation for in-vitro fertilization reduces aneuploidy in the human preimplantation embryo: a randomized controlled trial. Hum. Reprod. 2007;22:980–988. doi: 10.1093/humrep/del484. [DOI] [PubMed] [Google Scholar]
- 109.Rubio C, et al. Prospective cohort study in high responder oocyte donors using two hormonal stimulation protocols: impact on embryo aneuploidy and development. Hum. Reprod. 2010;25:2290–2297. doi: 10.1093/humrep/deq174. [DOI] [PubMed] [Google Scholar]
- 110.Penrose L. The Early Conceptus, Normal and Abnormal. Univ. St Andrews; 1964. pp. 94–97. [Google Scholar]
- 111.Bond DJ, Chandley AC. Aneuploidy. Oxford Univ. Press; 1983. pp. 67–75. [Google Scholar]
- 112.Hubner K, et al. Derivation of oocytes from mouse embryonic stem cells. Science. 2003;300:1251–1256. doi: 10.1126/science.1083452. [DOI] [PubMed] [Google Scholar]
- 113.Toyooka Y, Tsunekawa N, Akasu R, Noce T. Embryonic stem cells can form germ cells in vitro . Proc. Natl Acad. Sci. USA. 2003;100:11457–11462. doi: 10.1073/pnas.1932826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Geijsen N, et al. Derivation of embryonic germ cells and male gametes from embryonic stem cells. Nature. 2004;427:148–154. doi: 10.1038/nature02247. [DOI] [PubMed] [Google Scholar]
- 115.Nayernia K, et al. In vitro-differentiated embryonic stem cells give rise to male gametes that can generate offspring in mice. Dev. Cell. 2006;11:125–132. doi: 10.1016/j.devcel.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 116.Aflatoonian B, et al. In vitro post-meiotic germ cell development from human embryonic stem cells. Hum. Reprod. 2009;24:3150–3159. doi: 10.1093/humrep/dep334. [DOI] [PubMed] [Google Scholar]
- 117.Kee K, Angeles VT, Flores M, Nguyen HN, Reijo Pera RA. Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature. 2009;462:222–225. doi: 10.1038/nature08562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nicholas CR, Haston KM, Grewall AK, Longacre TA, Reijo Pera RA. Transplantation directs oocyte maturation from embryonic stem cells and provides a therapeutic strategy for female infertility. Hum. Mol. Genet. 2009;18:4376–4389. doi: 10.1093/hmg/ddp393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.White YA, et al. Oocyte formation by mitotically active germ cells purified from ovaries of reproductive-age women. Nature Med. 2012;18:413–421. doi: 10.1038/nm.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.The Practice Committee of the Society for Assisted Reproductive Technology, Practice Committee of the American Society for Reproductive Medicine. Preimplantation genetic testing: a Practice Committee opinion. Fertil. Steril. 2008;90:S136–S143. doi: 10.1016/j.fertnstert.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 121.Committee on Genetics. ACOG Committee Opinion No. 430: preimplantation genetic screening for aneuploidy. Obstet. Gynecol. 2009;113:766–767. doi: 10.1097/AOG.0b013e31819e9f05. [DOI] [PubMed] [Google Scholar]
- 122.Munne S, Wells D, Cohen J. Technology requirements for preimplantation genetic diagnosis to improve assisted reproduction outcomes. Fertil. Steril. 2010;94:408–430. doi: 10.1016/j.fertnstert.2009.02.091. [DOI] [PubMed] [Google Scholar]
- 123.Handyside AH. PGD and aneuploidy screening for 24 chromosomes by genome-wide SNP analysis: seeing the wood and the trees. Reprod. Biomed. Online. 2011;23:686–691. doi: 10.1016/j.rbmo.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 124.Lamb NE, et al. Characterization of susceptible chiasma configurations that increase the risk for maternal nondisjunction of chromosome 21. Hum. Mol. Genet. 1997;6:1391–1399. doi: 10.1093/hmg/6.9.1391. [DOI] [PubMed] [Google Scholar]
- 125.Hassold T, et al. Human aneuploidy: incidence, origin, and etiology. Environ. Mol. Mutagen. 1996;28:167–175. doi: 10.1002/(SICI)1098-2280(1996)28:3<167::AID-EM2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 126.Zenzes MT, Casper RF. Cytogenetics of human oocytes, zygotes, and embryos after in vitro fertilization. Hum. Genet. 1992;88:367–375. doi: 10.1007/BF00215667. [DOI] [PubMed] [Google Scholar]
- 127.Magli MC, Gianaroli L, Ferraretti AP. Chromosomal abnormalities in embryos. Mol. Cell Endocrinol. 2001;183:S29–S34. doi: 10.1016/s0303-7207(01)00574-3. [DOI] [PubMed] [Google Scholar]
- 128.Staessen C, et al. Comparison of blastocyst transfer with or without preimplantation genetic diagnosis for aneuploidy screening in couples with advanced maternal age: a prospective randomized controlled trial. Hum. Reprod. 2004;19:2849–2858. doi: 10.1093/humrep/deh536. [DOI] [PubMed] [Google Scholar]
- 129.Munne S, et al. Maternal age, morphology, development and chromosome abnormalities in over 6000 cleavage-stage embryos. Reprod. Biomed. Online. 2007;14:628–634. doi: 10.1016/s1472-6483(10)61057-7. [DOI] [PubMed] [Google Scholar]
- 130.Ercelen N, et al. Successful preimplantation genetic aneuploidy screening in Turkish patients. Genet. Mol. Res. 2011;10:4093–4103. doi: 10.4238/2011.November.17.6. [DOI] [PubMed] [Google Scholar]
- 131.Obradors A, et al. Whole-chromosome aneuploidy analysis in human oocytes: focus on comparative genomic hybridization. Cytogenet. Genome Res. 2011;133:119–126. doi: 10.1159/000324233. [DOI] [PubMed] [Google Scholar]
- 132.Martin RH, Rademaker A. The frequency of aneuploidy among individual chromosomes in 6,821 human sperm chromosome complements. Cytogenet. Cell Genet. 1990;53:103–107. doi: 10.1159/000132905. [DOI] [PubMed] [Google Scholar]
- 133.Martin RH, Ko E, Rademaker A. Distribution of aneuploidy in human gametes: comparison between human sperm and oocytes. Am. J. Med. Genet. 1991;39:321–331. doi: 10.1002/ajmg.1320390315. [DOI] [PubMed] [Google Scholar]
- 134.Templado C, Vidal F, Estop A. Aneuploidy in human spermatozoa. Cytogenet. Genome Res. 2011;133:91–99. doi: 10.1159/000323795. [DOI] [PubMed] [Google Scholar]