SUMMARY
It is widely believed that the molecular and cellular features of a tumor reflect its cell of origin and can thus provide clues about treatment targets. The retinoblastoma cell of origin has been debated for over a century. Here, we report that human and mouse retinoblastomas have molecular, cellular, and neurochemical features of multiple cell classes, principally amacrine/horizontal interneurons, retinal progenitor cells, and photoreceptors. Importantly, single-cell gene expression array analysis showed that these multiple cell type-specific developmental programs are coexpressed in individual retinoblastoma cells, which creates a progenitor/neuronal hybrid cell. Furthermore, neurotransmitter receptors, transporters, and biosynthetic enzymes are expressed in human retinoblastoma, and targeted disruption of these pathways reduces retinoblastoma growth in vivo and in vitro.
INTRODUCTION
Mouse models of cancer have become increasingly important in the fields of cancer genetics and translational research. This is particularly true for pediatric cancers, because the patient population is relatively small, and preclinical models are essential for demonstrating the efficacy of new therapeutic approaches before initiating clinical trials in children. In addition, animal models of cancer can provide important insight into the molecular, cellular, and genetic mechanisms underlying tumor initiation and progression.
One challenge of modeling pediatric cancer in mice is that these tumors initiate in the context of developing tissues that change rapidly, and the cells that give rise to the tumors can display an extraordinary degree of plasticity and heterogeneity. This is further complicated by the fact that many tumor suppressor genes and proto-oncogenes play essential roles in regulating cell fate specification and differentiation during development. Specifically, the genetic lesions that contribute to tumor initiation and progression may also alter the intrinsic cell fate specification and differentiation programs in the tumor cells, thereby making it very difficult to infer the cell of origin for that tumor.
As therapeutic agents are developed to target particular molecular pathways in cancer cells, identifying the cell of origin becomes increasingly important. For example, if the cell of origin of a pediatric cancer was a progenitor cell, a very different strategy would be employed to target particular pathways in that cell than if it was a highly specialized differentiated cell from the same tissue. This is particularly true for tumors of the central nervous system, because neurons are incredibly diverse and use a variety of signal transduction pathways that may be co-opted during tumorigenesis.
Retinoblastoma is a rare childhood cancer of the retina that initiates during fetal development and is often diagnosed at birth or during the first few years of life. The first attempt to formulate a link between tumor cell features and a specific retinal cell type (i.e., photoreceptors) as the cell of origin was in 1897 with the description of retinoblastoma rosettes (Wintersteiner, 1897). In subsequent decades, this hypothesis was extended to include morphological analysis using transmission electron microscopy (TEM) (Dickson et al., 1976; Ts'o et al., 1970a; Ts'o et al., 1970b), immunohistochemistry (arrestin and rhodopsin) (Donoso et al., 1985, 1986; Mirshahi et al., 1986; Vrabec et al., 1989), and immunofluorescence (Crx and opsin) (Glubrecht et al., 2009; Xu et al., 2009). However, during this same period, data were published suggesting that retinoblastoma may arise from a progenitor cell (Beemer et al., 1984; Donovan et al., 2006; Glubrecht et al., 2009; Kyritsis et al., 1984a, 1984b; Zhong et al., 2007), a glial cell (Daa Schroder, 1987), or an interneuron such as an amacrine cell (Johnson et al., 2007; Kyritsis et al., 1986; Tsokos et al., 1986). Despite all of these efforts over the past decades, we still do not know which cell type(s) gives rise to retinoblastoma or if analysis of the features of retinoblastoma tumors can shed light on the cell of origin for this developmental tumor of the retina.
In this study, we performed comprehensive, unbiased molecular, cellular, and neurochemical analyses of mouse and human retinoblastoma cells to elucidate their detailed cellular signatures and to determine how these signatures relate to those of normal retinal cell types.
RESULTS
Human Retinoblastomas Have Similar Molecular Profiles
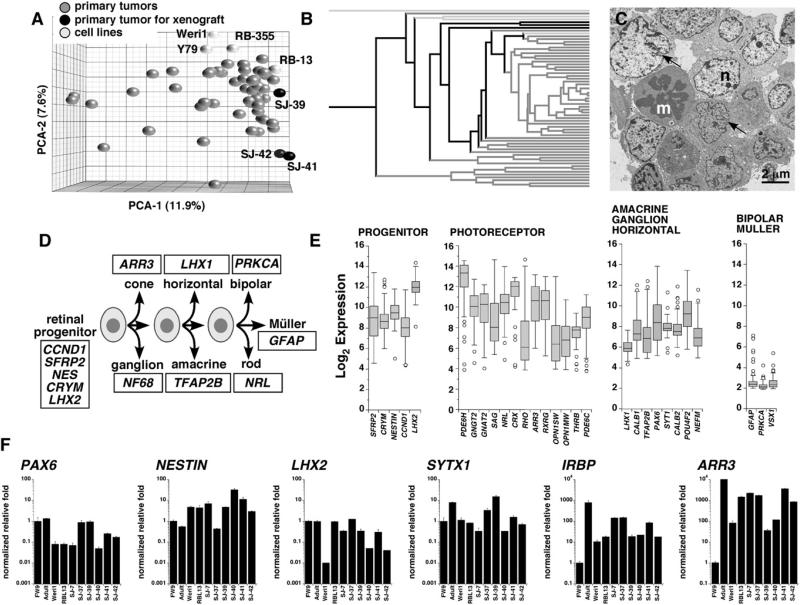
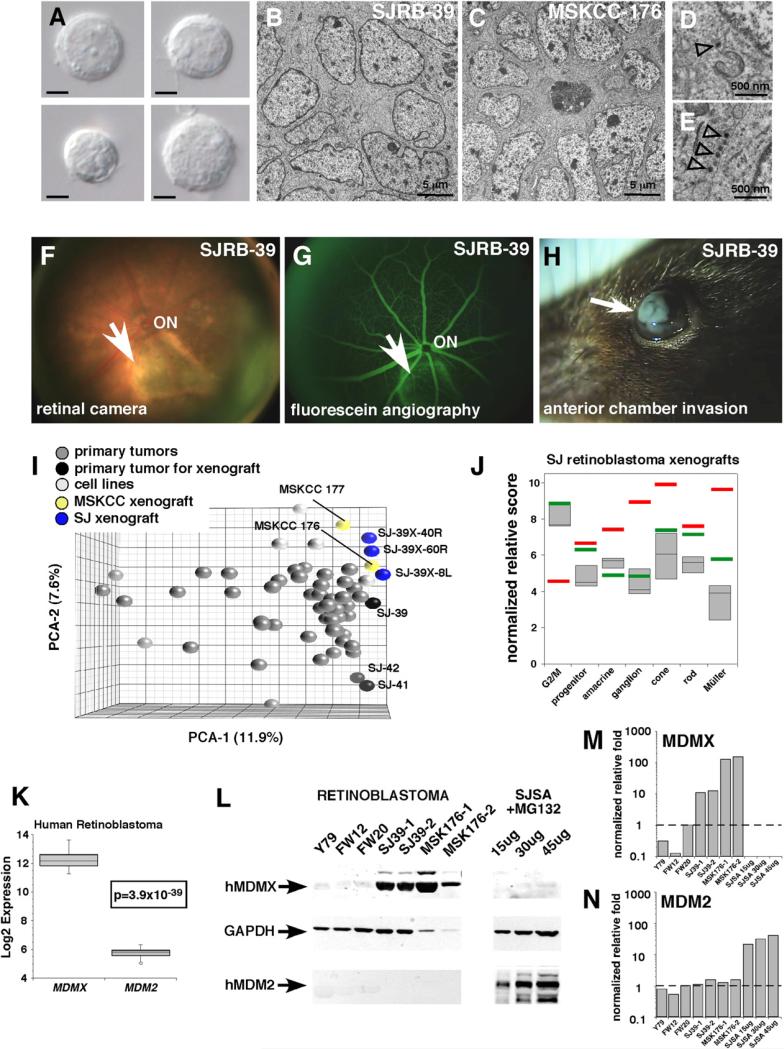
To assemble a molecular profile of human retinoblastoma, we carried out gene expression profiling of 52 fresh retinoblastomas from patients who underwent surgical enucleation prior to anti-cancer therapy (see Table S1 available online). For profile comparisons, we also included four retinoblastoma cell lines (Y79, Weri1, Rb-355, and Rb-13) (Griegel et al., 1990a; Griegel et al., 1990b; McFall et al., 1977; Reid et al., 1974) in this analysis. A principal component analysis (PCA) and hierarchical clustering revealed that the majority of tumor samples (46/52, 88.5%) cluster tightly around a central core molecular signature; no clear subtypes were detected (Figures 1A and 1B). Three tumors that had the same core molecular signature (SJRB-39, SJRB-41, and SJRB-42) were introduced into the vitreous of immunocompromised mice for future orthotopic xenograft studies. Transmission electron microscopy (TEM) analysis of these three primary tumors confirmed that they had many of the cellular features of retinoblastoma, including mitotic figures, abundant mitochondria, cellular processes, and extensive cell-cell junctions (Johnson et al., 2007) (Figure 1C; Figure S1).
Figure 1. Molecular Characterization of Human Retinoblastoma Cells.
(A) Principal component analysis (PCA) of gene expression array data from primary human retinoblastoma cells and four human retinoblastoma cell lines (Y79, Weri1, RB-355, and RB-13). SJ-39, SJ-41, and SJ-42 were subsequently used as a source for orthotopic xenografts.
(B) Hierarchical cluster of human retinoblastoma and cell line gene expression array data. Black lines are those tumors used for orthotopic xenografts, and light gray lines are the cell lines. All other lines (dark gray) are primary tumor samples.
(C) TEM of the primary tumor SJ-39. Arrows indicate invaginations in the nucleus (n) adjacent to a mitotic figure (m).
(D) Overview of retinal development from multipotent progenitor cells; representative cell type-restricted genes appear in the boxes.
(E) Box plots of the Log2 expression from the microarrays for a series of genes found in retinal progenitor cells, rod and cone photoreceptors, amacrine/ganglion/ horizontal cells, or bipolar/Müller cells. Each box includes the data for all 52 human gene expression arrays. The bottom and top of the boxes are the 25th and 75th percentile and the line in the middle is the 50th percentile. The lines represent one standard deviation above and below the mean and data points outside of those lines represent outliers.
(F) Real-time RT-PCR data using TaqMan probes of genes selected from (E). Each bar is the mean and standard deviation of triplicate samples. Abbreviations: Adult, adult human retina; FW9, human fetal week 9; m, mitotic figure; n, nucleus.
To gain insight into the differentiation features of human retinoblastomas, we analyzed the expression of 28 genes from the microarrays that are restricted to different retinal cell classes (Figure 1D). A mixture of genes that are normally expressed in retinal progenitor cells and differentiated retinal neurons (i.e., photoreceptors and amacrine cells) were expressed in human retinoblastomas (Figure 1E). Real-time RT-PCR was used to confirm these data (Figure 1F; data not shown). These results together suggested that human retinoblastomas are relatively homogenous and express a hybrid differentiation phenotype.
Mouse Retinoblastomas from Six Genotypes Show Striking Similarities
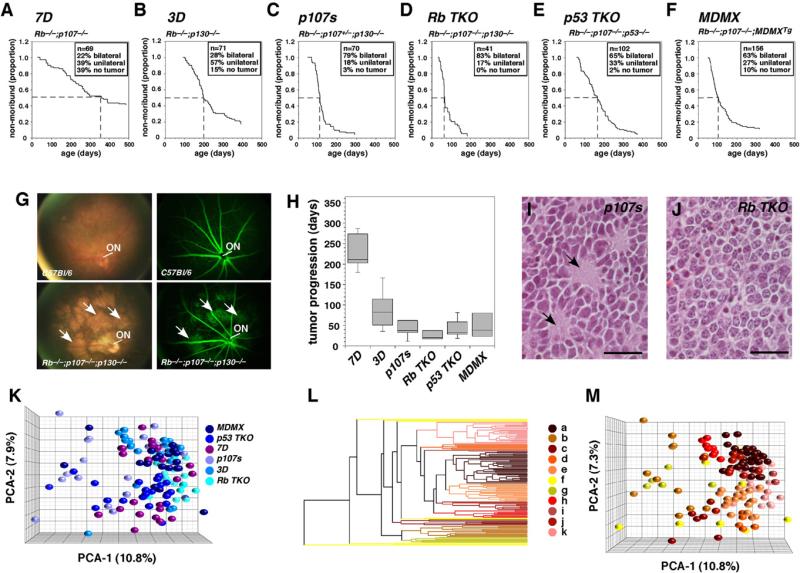
We have generated six mouse lines that develop retinoblastoma by using the Chx10-Cre to conditionally inactivated tumor suppressor pathways in their developing retinae (Rowan and Cepko, 2004) (Figures 2A–2F). Chx10-Cre;RbLox/Lox;p107–/– (7D), Chx10-Cre;RbLox/Lox;p130–/– (3D), Chx10-Cre;RbLox/Lox; p107+/–;p130–/– (p107s), and Chx10-Cre;RbLox/Lox;p107–/–; p130Lox/Lox(RbTKO) develop retinoblastoma when different combinations of Rb family members are inactivated in retinal progenitor cells (Figures 2A–2D). The other two strains developed retinoblastoma when Rb-pathway inactivation was combined with p53 pathway inactivation. The Chx10-Cre;RbLox/Lox; p107–/–;p53Lox/Lox (p53TKO) mice lack p53, Rb, and p107, and the Chx10-Cre;RbLox/Lox;p107–/–;MDMXTg (MDMX) mice conditionally overexpress the MDMX gene (Xiong et al., 2010) in retinal progenitor cells that lack Rb and p107 (Figures 2E and 2F).
Figure 2. Characterization of Mouse Retinoblastoma Cells from Six Different Strains.
(A–F) Kaplan-Meyer survival curves for 509 mice from six genotypes. The endpoint is moribund status, as defined by imminent ocular rupture. Abbreviations for each genotype are indicated above each curve. The period required for 50% of mice from each strain to reach moribund status is indicated with a dashed line.
(G) Representative images of mouse retinoblastomas and normal retinae tissue made by using a digital retinal camera and fluorescein angiography. Arrows indicate tumor foci.
(H) Box plot of time from diagnosis to moribund status for a small cohort of 10–12 animals per genotype. The bottom and top of the boxes are the 25th and 75th percentile and the line in the middle is the 50th percentile. The lines represent one standard deviation above and below the mean and data points outside of those lines represent outliers.
(I and J) Hematoxylin and eosin staining of a representative p107s (Rb–/–;p107+/–;p130–/–) tumor featuring rosettes with central plexi (arrows) and an RbTKO (Rb–/–;p107–/–;p130–/–) tumor lacking a plexus. Scale bars: 25 μm.
(K) PCA of gene expression arrays for 120 mouse retinoblastomas (20 from each of the 6 indicated strains).
(L) Hierarchical cluster of mouse retinoblastomas with individual clusters designated by letters (a–k).
(M) PCA of mouse clusters from (L).
To characterize tumor incidence and progression in these mouse models of retinoblastoma, we monitored 509 mice weekly from 3 weeks of age until advanced tumor progression necessitated euthanasia (Figures 2A–2F). Retinal camera images and fluorescein angiography of early-stage tumors (Figure 2G) showed that in mice, retinoblastoma initiates in a manner similar to that seen in humans. Tumors originate in multiple foci across the retina and efficiently reorganize the retinal vasculature. The time from tumor diagnosis to moribund status reflected the ages at which the six strains developed advanced-stage retinoblastoma (Figure 2H). This finding suggests that the difference between strains was due to differences in the rate of tumor progression rather than the rate of initiation. Histopathologic analysis of five mice from each strain (60 eyes total) confirmed that optic nerve, anterior chamber, subretinal, and choroidal invasion were present in each strain (Figures S2A–S2F). Anterior chamber and subretinal invasion was the most common (80%–100% of eyes examined), and optic nerve and choroidal invasion was the least common (30%–60% of eyes examined). Necropsy on 22 of the p53TKO mice revealed that 23% (5/22) of the mice had metastases. Four of the metastatic lesions were in the brain, and one was in a lymph node, which is consistent with the pattern seen in patients with metastatic retinoblastoma. All of the mouse retinoblastoma strains displayed abundant rosettes, except for the RbTKO strain, which had only a few areas with rosettes (Figures 2I and 2J and data not shown).
We performed immunostaining for markers of proliferation (Ki67), apoptosis (caspase 3), neuronal differentiation (Tuj1, synaptophysin), astrocytes and activated Müller glia (GFAP), vascular endothelial cells (CD34), and macrophages (Mac2, F4/80) on paraffin-embedded sections from five mice per geno-type and scored the proportion of immunopositive cells (Figures S2G–S2I). At the stage when these tumors were isolated (moribund status), there were no dramatic differences across strains in the expression of these markers by genotype. In addition, TEM analysis of another 30 tumors across the six genotypes indicated that there were no significant differences in cellular morphology, including differentiation features such as processes and junctions (Figure S2J).
To develop a molecular profile of our retinoblastoma mice, we performed gene expression array analysis on 120 tumors (20 per strain). When combined on a PCA plot, the six retinoblastoma mouse strains exhibited extensive overlap and intermixing of their gene expression profiles (Figure 2K). The gene expression profiles were highly correlated (95%–99%) across all strains but the most distinct were p53 TKO and Rb TKO (Figures S2K and S2L and Table S2). The orthologs of the 28 genes used to characterize the retinal differentiation properties of human retinoblastomas (Figure 1) were also coexpressed in the mouse retinoblastomas (Figure S2M). Cell type scoring and real time RT-PCR confirmed a hybrid molecular signature (Figures S2N and S2O). Hierarchical cluster analysis (Figures 2L and 2M) identified two clusters (j and k) that exhibited gene expression differences in photoreceptor and amacrine/horizontal neuron genes that were validated by real-time RT-PCR (Figures S2P–S2S and Table S3). However, even in the clusters with enriched gene expression of particular retinal differentiation pathways, multiple developmental programs were coexpressed (Figures S2P–S2S).
Mouse and Human Retinoblastomas Share Molecular Profiles
To directly compare the molecular signatures of human and mouse retinoblastomas, we focused our analysis on the 15,646 unique ortholog-matched unigene pairs present on the human and mouse gene expression arrays. There are three potential sources of variance in comparing human and mouse array data: (1) differences in individual probe sets of the ortholog-matched unigenes; (2) general biological differences across species; and (3) biological differences between human and mouse tumors, which provide insight into the biology of retinoblastoma and how well the mouse models recapitulate the human disease.
To minimize the contribution of the first two sources of variance and highlight the major differences between the human and mouse retinoblastomas, we developed an algorithm for cross-species comparisons of array data (see Supplemental Experimental Procedures). Using this method, we found that 12,267 of 15,646 (78%) ortholog-matched unigene pairs exhibited similar levels of expression (<1 quartile difference) across species. To establish the significance of this correlation, we randomized the probe sets in a series of 50,000 simulations per chip and performed the same analysis. These simulations demonstrated that any correlation greater than 3.5% is significant with a p value of at least 0.00002. The majority of the differences between human and mouse retinoblastoma unigene pairs (3094/3379; 92%) was a 1-quartile difference in expression; 280 of 3379 (8.3%) were a 2-quartile difference; and 5 of 3379 (0.1%) were a 3-quartile difference in expression. Among those unigene pairs that were most distinct across species (p = 2.3 × 10–17), 269 differed across all mouse genotypes (Table S4), and an additional 525 differed in subsets of the six genotypes (Table S4).
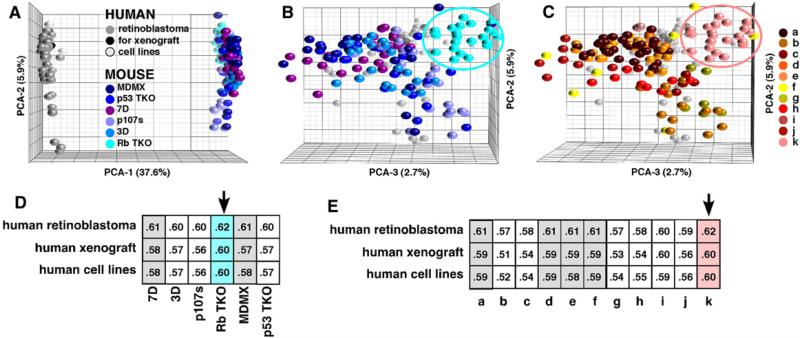
PCA analysis demonstrated that most of the difference between the profiles of human and mouse retinoblastoma cells was in a single dimension (Figure 3). A correlation analysis of all mouse genotypes and clusters to human tumors indicated that the RbTKO strain was the best match to human retinoblastomas by a small margin (Figure 3).
Figure 3. Comparison of Gene Expression in Human and Mouse Retinoblastoma.
(A) PCA of mouse and human gene expression array data for the ortholog-matched unigene pairs.
(B) The view is rotated 90° around the y axis to show the overlay of the human and mouse data. The Rb TKO strain is encircled.
(C) The same view of the mouse-human PCA that is shown in (B), but the colors now reflect the hierarchical clusters. Cluster k represents the best match for human retinoblastoma and is primarily made up of Rb TKO.
(D and E) Correlation table of human retinoblastoma, human retinoblastoma xenografts (SJ-39, SJ-41, SJ-42, and MSKCC 176), and human retinoblastoma cell lines compared to each genotype (D) or cluster (E). The arrows indicate the best match, and the gray-shaded columns indicate the second-best match.
See also Table S4.
Mouse and Human Retinoblastomas Express Multiple Differentiation Pathways
To perform an unbiased analysis of retinal cell type molecular signatures, we took advantage of a recent series of publications using gene expression array analysis of individually isolated retinal progenitor cells and differentiated retinal neurons and glia (Cherry et al., 2009; Roesch et al., 2008; Trimarchi et al., 2008). Cone photoreceptors have not been analyzed using this single-cell technique, so we used in situ-validated microarray data from purified pools of cone photoreceptors (Akimoto et al., 2006).
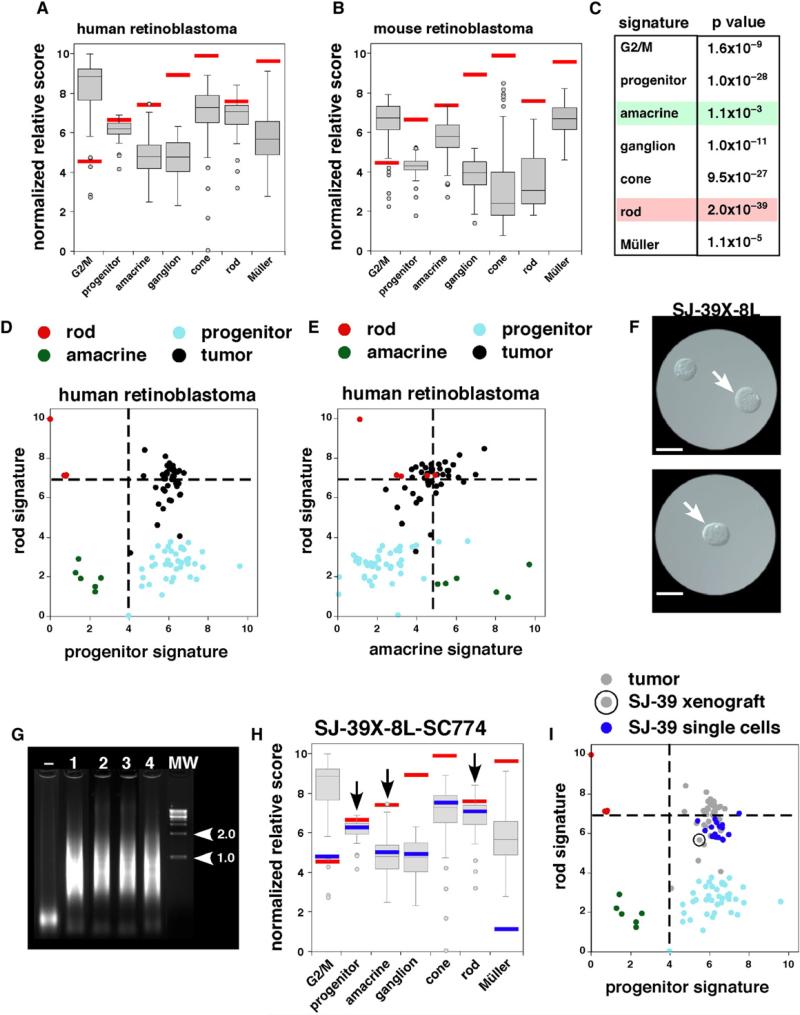
Cell type signatures of retinal progenitor cells, rod photoreceptors, cone photoreceptors, amacrine cells, ganglion cells, and Müller glia (Table S5) were used to characterize the differentiation state of all 172 mouse and human retinoblastomas in our microarray study, as described previously (Cherry et al., 2009; Roesch et al., 2008; Trimarchi et al., 2008). We also analyzed a group of previously characterized genes that regulate the G2/M phase of the cell cycle to compare the proliferation signature in the retinoblastomas to that of normal retinal progenitor cells. The most robust cell type-specific signatures for human retinoblastomas were those of progenitor cells, G2/M genes, and rod photoreceptors (Figure 4A; Figures S3A and S3B). The rod signature was indistinguishable from normal differentiated rods, and the progenitor signature was indistinguishable from that of normal progenitor cells. More importantly, the human retinoblastomas also had robust signatures for amacrine cells and cone photoreceptors (Figure 4A). The strongest cell type-specific signature for mouse retinoblastomas was the amacrine cell score; these tumors also had a pronounced expression of genes that regulated the G2/M phase in progenitor cells (Figure 4B; Figures S3A and S3B). Statistical analysis of the human and mouse retinal cell type signatures identified the amacrine cell signature and the rod cell signature as the most similar and the most distinct, respectively, across species (Figure 4C; Table S6 and Supplemental Experimental Procedures).
Figure 4. Retinal Cell Type-Specific Gene Signatures for Human and Mouse Retinoblastomas.
(A and B) Box plots of normalized relative scores for cell type-specific gene signatures for individual retinal cell types and the G2/M signature. Red bars indicate the median of the actual cell type score to serve as a benchmark. Ortholog-matched unigenes were used for these comparisons. All 52 human arrays were used in (A), and all 120 mouse arrays were used in (B). The bottom and top of the boxes are the 25th and 75th percentile and the line in the middle is the 50th percentile. The lines represent one standard deviation above and below the mean and data points outside of those lines represent outliers.
(C) Statistical analysis of cell type-specific score differences for mouse and human retinoblastomas based on a t test.
(D and E) Two-dimensional plots of human retinoblastoma gene expression array scores for retinal cell type signatures. Vertical dashed lines represent the lower limit for progenitor signature for retinal progenitor cells or amacrine signature for retinal amacrine cells. Horizontal dashed line represents the lower limit for rod signature for rod photoreceptors. Rods are red, amacrine cells are green, progenitor cells are light blue and retinoblastomas are black.
(F) Differential interference contrast (DIC) image of dissociated SJ-39 xenograft used for single-cell microarrays. In the upper panel, a tumor cell is picked (arrow) and transferred to a new dish (lower panel) to verify that a single cell was isolated (arrow). Scale bars: 5 μm.
(G) Ethidium bromide-stained agarose gel with the amplification product from four isolated SJ-39 xenograft cells prior to hybridization to gene expression arrays. Molecular weight markers (1.0 and 2.0 kb) are indicated with arrowheads.
(H) Cell type-specific scores for 1 of the 20 individual SJ-39 xenograft tumor cells that was profiled in this study. The blue bar represents the scores for this single cell; the gray box plot is the data from all 52 human retinoblastomas; and the red bar is the median score for normal cell populations used in this study. The bottom and top of the boxes are the 25th and 75th percentile and the line in the middle is the 50th percentile. The lines represent one standard deviation above and below the mean and data points outside of those lines represent outliers.
(I) Two-dimensional plot of SJ-39 xenograft single-cell gene expression array scores for rod photoreceptor and progenitor signatures. Rods are red, amacrine cells are green, progenitor cells are light blue and retinoblastomas are gray. The primary tumor used to generate the xenograft is circled and the individual cells derived from this xenograft are shown in blue.
When signatures were compared, correlations became evident between rod and cone scores, G2/M and progenitor scores, and between amacrine and ganglion cell scores along a continuum for both mouse and human retinoblastomas (Figure S3B). The cell type signatures that are normally incompatible (i.e., progenitor versus rod or amacrine versus rod) coexisted in the same tumors (Figures 4D and 4E; Figure S3B). Immunostaining for cell type markers and scoring of dissociated cells provided additional evidence that individual cells express multiple retinal differentiation pathways (Figures S3C–S3F; data not shown). These analyses suggest that human and mouse retinoblastomas represent a true hybrid of progenitor cells, photoreceptors, and amacrine interneurons.
To extend these observations and perform a broader, unbiased analysis of individual tumor cells, we isolated retinoblastoma cells from one of our orthotopic xenografts (SJ-RB39) and performed single-cell gene expression array analysis, as done previously (Cherry et al., 2009; Roesch et al., 2008; Trimarchi et al., 2008). These data confirmed that individual retinoblastoma cells in humans or mice express a mixture of differentiation programs for photoreceptors, amacrine cells, and progenitor cells (Figures 4F–4I; Figures S3G–S3I).
Multiple Differentiation Pathways in Human Orthotopic Retinoblastoma Xenografts
The expression of multiple differentiation pathways in primary retinoblastomas may reflect contamination of normal retinal neurons in the tumors. To test this possibility, we established orthotopic xenografts of three human retinoblastomas (SJ-RB39, SJ-RB41, and SJ-RB42; see Figure 1) by injecting primary tumor cells into the vitreous of SCID mice. For comparison, we also obtained a previously described orthotopic xenograft (MSKCC176) (Xu et al., 2009). If normal retinal cells contaminate primary tumors, then they would be rapidly overgrown in the orthotopic xenografts, and any contribution from the recipient mouse retina would not be detected because our probes and primers are specific for the human sequences. We carefully documented the morphology of the live cells and tissue specimens by TEM and intraocular histopathologic analyses (Figures 5A–5E; Figure S4). Tumor initiation, expansion, and progression were monitored using a digital retinal camera (Figure 5F), fluorescein angiography (Figure 5G), and weekly inspection for anterior chamber invasion (Figure 5H). The gene expression profile for the primary tumors used for the SJ-RB xenografts were representative of the overall population of human retinoblastomas in our study; minimal change occurred over the three passages in the vitreous of the eyes of the SCID mice for the SJ-RB xeno-grafts or the MSKCC176 xenograft (Figure 5I; Table S7). The cell type signatures for these orthotopic xenografts were very similar to that of the primary human tumors (Figure 5J).
Figure 5. Orthotopic Xenografts of Human Retinoblastomas.
(A) Differential interference contrast microscopy images of single dissociated cells from a primary human retinoblastoma (SJ-39) that was injected into the vitreous of immunocompromised mice. Scale bars: 5 μm.
(B–E) Representative TEM images of the SJ-39 xenograft and MSKCC176 xenograft showing tightly packed clusters of cells and rosettes. Dense core vesicles were present in both xenografts (open arrowheads in D and E).
(F and G) Retinal camera image of the SJ-39 xenograft with a large focal tumor (arrow) from an early-stage tumor with corresponding fluorescein angiography.
(H) The SJ-39 xenograft is highly invasive and aggressive, as shown by anterior chamber invasion (arrow).
(I) PCA of human retinoblastomas showing the addition of third-passage xenografted samples (blue) and the MSKCC176 and 177 samples (yellow). SJ39X-40R, -60R, and -8L are sublines of SJ-39.
(J) Box plot of the three SJ-RB retinoblastoma xenografts (SJ-RB39, SJ-RB41, SJ-RB42), with respect to their cell type scores. The bottom and top of the boxes are the 25th and 75th percentile and the line in the middle is the 50th percentile. The lines represent one standard deviation above and below the mean and data points outside of those lines represent outliers. The red bars represent the median for the cell types, and the green bars represent the median of the human retinoblastomas.
(K) Log2 expression of MDMX and MDM2 across all 52 human retinoblastomas.
(L) Immunoblot for MDMX and MDM2 in human retinoblastoma xenografts (SJ-RB and MSKCC), a retinoblastoma cell line (Y79), and human fetal retinae (fetal week [FW] 12 and FW20). SJSA cells treated with MG132 were used as a positive control for MDM2 immunoblotting.
(M and N) Quantification of the protein levels of the blots in (L). Each bar was the normalized value relative to GAPDH expression. The data were plotted relative to the level of expression in FW20 retina, which was set to 1.0 (dashed line). Abbreviation: ON, optic nerve.
Human retinoblastomas express high levels of the MDMX gene and the MDMX protein (Laurie et al., 2006). We confirmed that primary human retinoblastomas and the SJ-RB and MSKCC176 orthotopic xenografts express high levels of MDMX mRNA and protein (Figures 5K–5M and data not shown). In contrast, MDM2 levels were below the limit of detection in any of the human tumor xenografts or cell lines in our study, including the MSKCC176 xenograft (Figures 5K–5N). These data suggest that the orthotopic xenografts provide an ideal model in which to study inhibitors of the MDMX-p53 interaction.
Neuroanatomical and Neurochemical Features of Amacrine Cells in Retinoblastoma
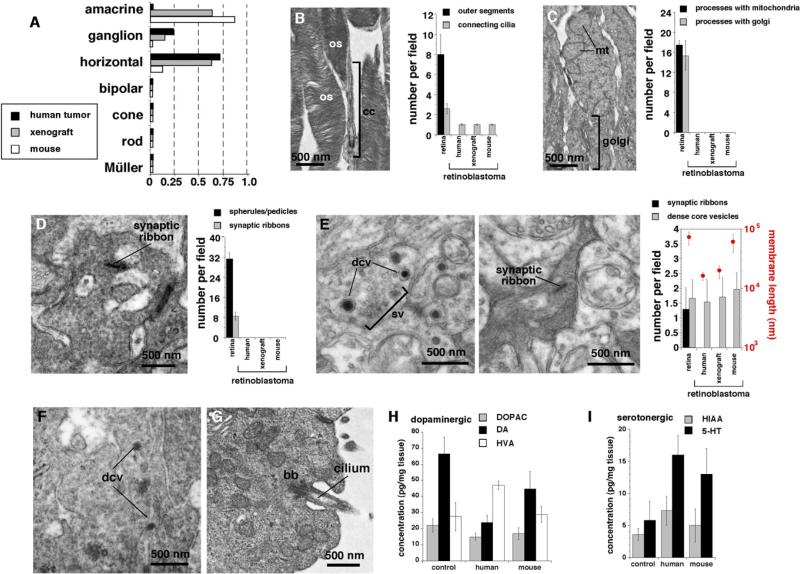
In addition to their molecular signatures, each retinal cell type exhibits distinct morphological and anatomical features that can be quantified on TEM images. We established a series of six morphometric features of cell bodies and ten neuroanatomical features of cellular processes that allowed us to unambiguously distinguish the different classes of retinal cell types (Figures 6A–6E; Table S8 and Table S9). On the basis of these morphometric and neuroanatomical measurements, the human and mouse retinoblastomas most closely resembled amacrine interneurons than photoreceptors, bipolar cells, or Müller glia (Figures 6A–6G; Table S8 and Table S9). One of the most striking features of the neuroanatomical analysis of plexus regions was the presence of dense core vesicles in human and mouse retinoblastomas (Figures 6E and 6F). These structures were present in the same tumors with connecting cilia and basal bodies (Figure 6G). Dense core vesicles are a hallmark of monoamine neurotransmitter secretion, and the only cell type in the retina that has dense core vesicles and produces monoamines are amacrine cells (Macneil et al., 1999; MacNeil and Masland, 1998). We used HPLC-ED to measure the levels of dopamine, serotonin, and their metabolites in normal retinae, mouse retinoblastoma, and human retinoblastoma xenografts (Figures 6H and 6I). The levels of these molecules in human and mouse retinoblastoma were similar to those found in normal retinae.
Figure 6. Morphometric, Neuroanatomical, and Neurochemical Analyses of Retinoblastoma Cells.
(A) Morphometric scoring of six measurements in more than 1000 human or mouse retinoblastoma cells on transmission electron micrographs. Each bar is the sum of the score for the tumors, as compared with those of the normal cell populations. At least five independent fields from five independent biological replicates were used for each sample.
(B–E) Neuroanatomical characterization of human and mouse retinoblastomas compared to those of normal retinae by using TEM. Representative images and scoring of outer segments (B), inner segments (C), synaptic elements at the outer plexiform layer (OPL) (D), and those of the inner plexiform layer (IPL) (E) are shown. Scoring for nine neuroanatomical features representative of different retinal cell types is shown. Each bar represents at least five independent fields from five independent samples.
(F) Representative dense core vesicles from a human retinoblastoma.
(G) Representative basal body and connecting cilia from a human retinoblastoma.
(H and I) HPLC-ED measurements of dopamine and its metabolites (H) and serotonin and its metabolites (I). All histograms are mean ± SD. Abbreviations: bb, basal body; cc, connecting cilium; dcv, dense core vesicle; os, outer segments; sv, synaptic vesicles.
Functional Significance of Neuronal Differentiation in Retinoblastoma
To begin to explore the functional significance of the features of neuronal differentiation in retinoblastoma, we searched our database of pediatric solid tumor cell line sensitivity to FDA-approved drugs and well-characterized cytotoxic agents (A.A. Shelat and R.K. Guy, unpublished data). This database includes dose-response data in triplicate for more than 600 compounds on 21 different solid tumor cell lines including retinoblastoma, osteosarcoma, rhabdomyosarcoma, and neuroblastoma. The Y79 and Weri1 retinoblastoma cell lines were found to be uniquely sensitive to a subset of antipsychotic agents. Cheminformatic analysis showed that the agents were all derivatives of phenothiazines, which are broad-acting monoamine receptor inhibitors, including dopamine, serotonin, histamine, and epinephrine receptors. These intriguing results prompted us to analyze neurotransmitter receptors, transporters, and regulators in detail and to study their functional significance for retinoblastoma growth and survival.
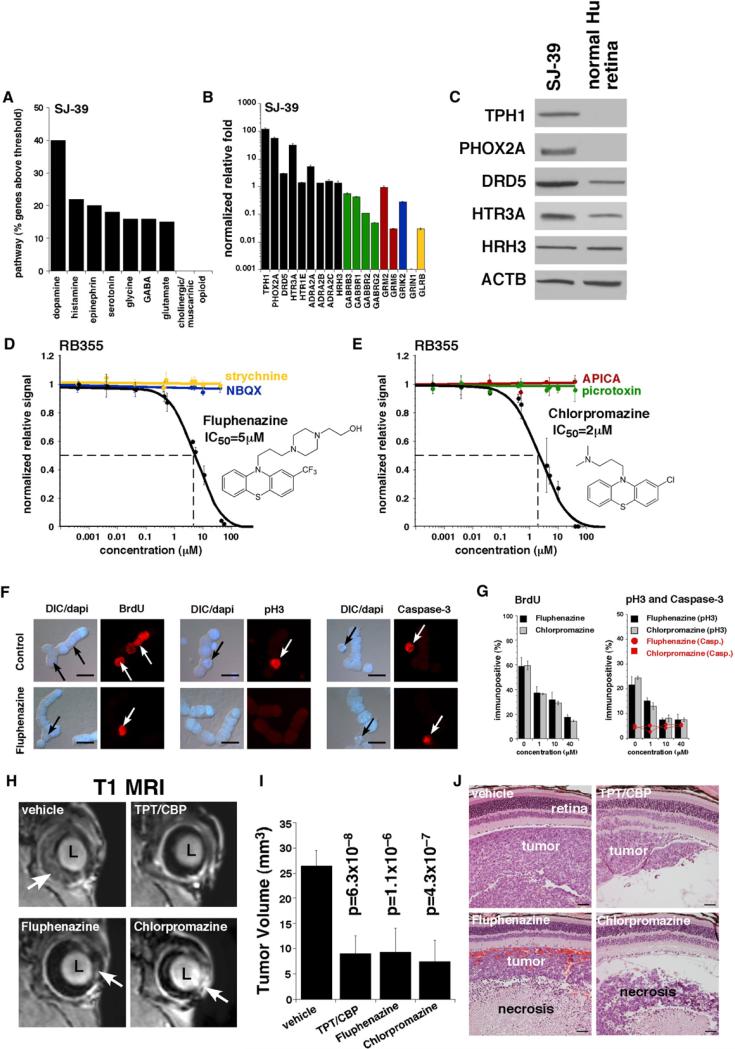
We first identified genes encoding all major neurotransmitter receptors and transporters in the retina that were represented on the gene expression microarrays (175 genes, 212 probe sets, Table S10). For example, we reviewed the data for 29 genes (43 probe sets) involved in GABA neurotransmission. Among those 29 genes, only 6 had expression levels for either tumor or normal retina greater than Log2 = 8, and only 3 genes (GABBR1, GABBR2, and GABRB3) were expressed in primary human retinoblastomas or cell lines at levels similar to or greater than that in normal retina (Figure 7A). Those three genes were selected for further analysis (see below). Identical criteria were used to select genes involved in glutamate, glycine, melatonin, histamine, epinephrine, dopamine, serotonin, cholinergic/ muscarinic, and opioid signaling in neurons (Figure 7A). In the gene expression array analysis, the monoamine receptors and pathways (dopamine, histamine, epinephrine, and serotonin) were most abundant in retinoblastoma, but other pathways (GABA, glycine, and glutamate) were also expressed (Figures 7A and 7B).
Figure 7. Inhibition of Neurotransmitter Receptors Reduces Retinoblastoma Growth In Vitro and In Vivo.
(A) Analysis of neurotransmitter pathway gene expression in human retinoblastoma tumors and orthotopic xenograft. In this analysis, genes that were expressed at levels equal to or greater than that in human retina and had a Log2 value of ≥8.0 were included.
(B) Real-time RT-PCR for representative genes from the pathways shown in (A). Each bar represents the mean and standard deviation of duplicate experiments. The data were normalized to β-actin levels for each sample and plotted relative to normal human retina. Green bars correspond to the GABA neurotransmitter pathway; red bars correspond to the metabotropic glutamate receptors; the blue bar corresponds to the kainite glutamate receptor subclass; and the yellow bars correspond to the glycine receptor-signaling pathway.
(C) Immunoblot for a subset of the proteins encoded by the genes in (B). Some of these proteins (TPH1 and PHOX2A) are not normally expressed in the mammalian retina at detectable levels.
(D and E) Representative dose response for RB355 cells to fluphenazine and chlorpromazine. Each data point represents the mean and standard deviation of triplicate samples. Viability was measured with Cell Titer-Glo and normalized to the vehicle-treated cells. The cells were maintained in culture for 72 hr in the presence of the compound.
(F) Representative immunostaining results for vehicle-treated (control) and fluphenazine-treated retinoblastoma cells. Red fluorescence indicates BrdU, phosphohistone H3 (pH3), and activated caspase-3 as indicated. The DAPI nuclear stain (blue) was overlaid with DIC to show cell morphology. Arrows indicate typical immunopositive cells. In this study, the cells were treated for 24 hr with each agent. (G) Scoring of the proportions of BrdU+, pH3+, and activated caspase-3+ following treatment with fluphenazine or chlorpromazine. For each sample and drug concentration, 250 cells were scored in duplicate. The bars represent the mean and standard deviation of triplicate experiments.
(H) Representative MRIs of the eyes of retinoblastoma mice with SJ-39 orthotopic xenograft after three courses of indicated treatment. Arrows indicate tumor (light area) in the normally dark vitreous or anterior chamber.
(I) The tumor volume was measured on MRIs of ten eyes for each treatment group, and the bars represent the mean and standard deviation for each treatment group.
(J) Representative H&E staining of each treatment group showing retina, tumor, and vitreous for each cohort. All histograms are mean ± SD. Abbreviations: CBP, carboplatin; DIC, differential interference contrast microscopy; TPT, topotecan.
Scale bars: (F), 10 μm; (J), 50 μm. See also Table S10.
To validate the gene expression microarray data, we generated TaqMan real-time RT-PCR probes for each gene that was expressed in the human retinoblastomas and analyzed expression in the SJ39 xenograft, human retinoblastoma cell lines, and normal retinae. In total, 96 genes were analyzed in duplicate for each sample (complete data set available upon request). Several of the monoamine/catecholamine receptors including serotonin receptors (HTR3A, HTR1E), a dopamine receptor (DRD5), and a histamine receptor (HRH3) were expressed in retinoblastoma at levels equal to or greater than normal human retina (Figure 7B). In addition, several of the genes involved in the biosynthesis of catecholamines such as TPH1, which encodes the rate-limiting enzyme for serotonin synthesis, and PHOX2A, which regulates genes that are important for dopamine neurotransmission, were expressed at high levels in the human retinoblastomas (Figure 7B). All three alpha-2 adrenergic receptors (ADRA2A, ADRA2B, and ADRA2C) were expressed in the tumors (Figure 7B). These proteins regulate the catecholamine-mediated inhibition of adenylate cyclase through the action of G-proteins in neurons. Immunoblotting confirmed the expression of these proteins (Figure 7C and data not shown).
Although most genes involved in neurotransmission that are upregulated in retinoblastomas are those found in amacrine cells and involved in catecholamine/monoamine signaling, there were some that are from other pathways in the retina including GABA, glycine, and glutamate. It is possible that proteins important in other neurotransmitter pathways are expressed in retinoblastoma and are functionally important but were expressed at levels below the limit of detection in our molecular studies. Therefore, to determine which of the major neurotransmitter pathways are functionally relevant, we performed a series of pharmacological experiments with retinoblastoma cell lines in culture. We tested 13 well-characterized pharmacological agents targeting each pathway in all 3 retinoblastoma cell lines (Weri1, Y79, and RB355) and a nonneuronal cell line (BJ) in triplicate dose-response experiments (Table 1 and data not shown). Only the broad-acting monoamine transporter inhibitors showed any activity in these experiments, and fluphenazine and chlorpromazine were the most active among those agents (Figures 7D and 7E and Table 1). We analyzed the receptor-binding profile of these two agents by using data from the PDSP Ki database (http://pdsp.med.unc.edu/pdsp.php). Using this approach, we identified another pharmacological agent with a similar profile (thioridazine) and tested it in dose response studies against all three retinoblastoma cell lines in triplicate. This compound gave results similar to fluphenazine and chlorpromazine (Table 1). None of the agents were active against the BJ cells (data not shown). In addition, more specific catecholamine pathway inhibitors such as the selective serotonin reuptake inhibitors (SSRIs) had no effect on retinoblastoma cell lines (data not shown). These data suggest that broad inhibition of multiple classes of monoamine receptors is required to affect retinoblastoma cells in culture.
Table 1.
Inhibition of Neurotransmitter Receptors in Retinoblastoma
| Compound | Target | Y79 EC50 (μM) | Weri1 EC50 (μM) | RB355 EC50 (μM) |
|---|---|---|---|---|
| Fluphenazine | Monoamine receptors | 7 | 7 | 5 |
| Chlorpromazine | Monoamine receptors | 8 | 6 | 2 |
| Thioridazine | Monoamine receptors | 10.5 | 8 | 7 |
| Promazine | Monoamine receptors | 40 | 20 | 20 |
| Triflupromazine | Monoamine receptors | 27 | 25 | 11 |
| Promethazine | Monoamine receptors | 40 | 20 | 20 |
| Levomepromazine | Monoamine receptors | 17 | 18 | 7 |
| NBQX | AMPA receptors | No activity | No activity | No activity |
| Picrotoxin | GABAA receptors | No activity | No activity | No activity |
| Strychnine | Glycine receptors | No activity | No activity | No activity |
| APICA | Metabotrpic receptors | No activity | No activity | No activity |
| SKF97541 | GABAB receptors | No activity | No activity | No activity |
To test whether the inhibition of monoamine receptors affected retinoblastoma cell proliferation and/or cell viability, we exposed the three retinoblastoma cell lines to different concentrations of fluphenazine and chlorpromazine for 24 hr and then labeled the cells in S-phase with a 1 hr BrdU pulse. We performed immunostaining for BrdU and phospho histone H3 (pH3) to monitor proliferation and activated caspase-3 to monitor cell death (Figure 7F). Overall, the retinoblastoma cells exhibited reduced BrdU and pH3 labeling with increasing concentration of fluphenazine or chlorpromazine (Figure 7G). There was no significant effect on the proportion of activated caspase-3+ cells (Figure 7G).
To extend our observations to a more relevant in vivo model, we tested the effect of these monoamine transporter inhibitors on the orthotopic xenograft SJ-39. Twenty immunocompromised mice were injected intravitreally with the SJ-39 retinoblastoma xenograft, and after 3 weeks, engraftment was confirmed with retinal camera and fluorescein angiography. The 20 animals were divided into four groups, and each group received a subconjunctival injection of vehicle, topotecan/carboplatin, fluphenazine, or chropromazine once weekly for 3 consecutive weeks in a double-blind study design. The tumor volume was then measured in each of the 40 eyes by using MRI (Figures 7H and 7I), and histological analysis was performed (Figure 7J). Fluphenazine and chlorpromazine injections appeared to significantly reduce tumor volume in vivo (Figure 7I), and this effect was confirmed by histopathologic analysis (Figure 7J).
DISCUSSION
Since 1897, when Wintersteiner proposed that rosettes in retinoblastoma reflect photoreceptor differentiation (Wintersteiner, 1897), the cell of origin and the cellular features of retinoblastoma have been debated. Initially, the focus was on those features that could be identified by histologic analysis such as the presence of rosettes. With advances in microscopy, histology, and molecular biology, the debate about the retinoblastoma cell of origin has been revisited with a strong predilection toward photoreceptors being the cell of origin (Dickson et al., 1976; Donoso et al., 1985, 1986; Glubrecht et al., 2009; Mirshahi et al., 1986; Ts'o et al., 1970a, 1970b; Vrabec et al., 1989; Xu et al., 2009). However, a number of independent studies suggested that retinoblastomas may also have features of progenitor cells or interneurons such as amacrine cells (Beemer et al., 1984; Donovan et al., 2006; Glubrecht et al., 2009; Johnson et al., 2007; Kyritsis et al., 1984a; Kyritsis et al., 1984b; Kyritsis et al., 1986; Tsokos et al., 1986; Zhong et al., 2007). The possibility of a hybrid cell with features of multiple cell types was not explored in these previous studies. Our ability to perform single-cell gene expression array analysis provided us with an opportunity to distinguish between these different possibilities.
Our analyses of human and mouse retinoblastomas showed that individual retinoblastoma tumor cells express multiple developmental programs for photoreceptors, interneurons, and progenitor cells. This was surprising and unexpected, because these developmental programs are never coexpressed in differentiated cells or progenitor cells during retinogenesis. This discovery underscores the challenges of assigning cell-of-origin status without first establishing the full molecular and cellular signature of a tumor. It is important to emphasize that these studies do not necessarily indicate that a photoreceptor, interneuron, or progenitor cell is the cell of origin for retinoblastoma. Detailed cell-lineage studies will be required to definitively identify the cell of origin for retinoblastoma.
Molecular Characterization of Human and Mouse Retinoblastomas
Unlike other human cancers that are readily divided into distinct subtypes based on their molecular profiles, human retinoblastomas are remarkably homogenous, and our data are in good agreement with a small study of six tumors (Ganguly and Shields, 2010). In addition, retinoblastomas from six different mouse strains were very similar to each other and to the human tumors. We were surprised to learn that no single mouse strain emerged as statistically more similar to the human tumors than the others. However, subtle differences in the expression of photoreceptor genes was found among some of the mouse tumors using unsupervised hierarchical cluster analysis. There are several possible explanations for these differences in photoreceptor gene expression. The cluster with the higher photoreceptor gene expression may have had some contamination from the normal retina. Alternatively, the different clusters may have had different cells of origin, and further lineage studies on the different genetic backgrounds will be required to distinguish between these possibilities.
Photoreceptor Signature in Retinoblastoma
Many of the studies on retinoblastoma differentiation conducted over the past century have focused on photoreceptor features, including a recent study linking retinoblastomas origin to cone photoreceptors (Xu et al., 2009). Our data show that retinoblastomas express molecular features of photoreceptors and that the rod signature is more robust than the cone signature. More importantly, our morphometric and neuroanatomical studies of these cells showed few cellular features of photoreceptors. This raises the possibility that the gene regulatory network that controls photoreceptors is deregulated in human retinoblastomas, and to a lesser degree, in mouse retinoblastomas, but it does not necessarily mean that the tumors originated from these cells. Interestingly, a large number of photoreceptor genes are also expressed in medulloblastomas (Kool et al., 2008; Vincent et al., 2003); yet, those tumors clearly do not originate from a photoreceptor. This may indicate that the transcriptional pathways involved in photoreceptor differentiation are nonspecifically deregulated in a variety of tumors of the CNS leading to false conclusions about the cell of origin.
Amacrine and Progenitor Cell Features in Retinoblastoma
In a previous study of mouse retinoblastomas, we suggested that these tumors have features of amacrine, horizontal, and progenitor cells (Macpherson and Dyer, 2007). Very few genes distinguish amacrine cells from horizontal cells, thereby making these cells difficult to distinguish in our analysis. The unexpected result from this study was the degree of amacrine cell differentiation in human retinoblastomas and xenografts derived from primary tumors. Indeed, the cell type signature, morphometric, neuroanatomical, and neurochemical features that are most common across human and mouse retinoblastomas are those related to amacrine cell differentiation. To our knowledge, no other tumors in the CNS exhibit this unique combination of neuronal differentiation, which suggests that it is specific to retinoblastoma. However, this does not mean that retinoblastomas in humans or mice arise from amacrine cells. The tumor's cell of origin may be completely independent of amacrine interneurons, and because tumor suppressor or oncogenic pathways are deregulated during tumorigenesis, the amacrine differentiation program may be activated.
It is widely believed that the initiating genetic lesion in the RB1 gene occurs in proliferating retinal progenitor cells during fetal development. Retinal progenitor cells may be the cell of origin for retinoblastoma, and the multiple differentiation programs that are found in human and mouse retinoblastomas may reflect the multipotent competence of retinal progenitor cells. Indeed, in the single-cell gene expression array analysis of mouse retinal progenitor cells, photoreceptor and interneuron genes were often expressed in proliferating retinal progenitor cells (Trimarchi et al., 2008).
Single-Cell Gene Expression Profiling for Retinoblastoma
The single-cell gene expression profiling for the human retinoblastoma xenograft provided important validation for the coexpression of multiple differentiation programs in individual tumor cells. However, the PCR-amplification protocol that is required for this method (35 cycles of amplification) could affect the final representation of transcript abundance. A previous study specifically designed to investigate this question found a significant level (~25%) of false positives associated with exponential amplification of small amounts of starting material (Iscove et al., 2002). In that same study, however, the authors found a significant level of false negatives associated with a linear-based amplification approach (Iscove et al., 2002). Using a PCR-based single-cell profiling method in tissue culture cells and the olfactory system, Tietjen et al. did not encounter a high false-positive rate (Tietjen et al., 2003). This same single-cell profiling method has been used extensively to examine the transcriptomes of many different cells from retinal progenitor cells (Trimarchi et al., 2008), retinal amacrine cells (Cherry et al., 2009), neuronal progenitor cells (Kawaguchi et al., 2008), hematopoietic stem cells (Ramos et al., 2006), and olfactory neuron progenitors (Tietjen et al., 2003). Expression analyses of a large number of retinal progenitor and amacrine genes (>200) revealed a very high degree of correlation (~90%) between the microarray results and in situ hybridization (ISH) experiments using both retinal cryosections and dissociated retinal cells (Cherry et al., 2009; Trimarchi et al., 2007, 2008). For approximately 10% of the genes examined, the observed lack of correlation between the predicted gene expression pattern from the single-cell microarray data and the subsequent ISH staining pattern involved contamination in the single-cell preparation (Trimarchi et al., 2007). Such contamination arises from two main sources in retinal preparations. First, rod photoreceptor cell outer segments are quite fragile and can break during the dissociation, leading to the presence of rod mRNAs in the media. Second, a tight connection between two cell types, as there is between the RPE and PR cells, can also lead to contamination (Trimarchi et al., 2007). Neither of these is relevant to our study because the human orthotopic xenograft was injected into the vitreous of the eye, and when the tumor was removed for single-cell isolation, the retina and RPE were left intact and connected to the sclera. Moreover, gene expression arrays were performed using human gene chips, because the tumor was a human orthotopic xenograft; thus, any contaminating mouse cells or transcripts would not have been detected in this analysis.
The single biggest caveat to interpreting the single-cell data is the difficulty in understanding whether the absence of a signal arises due to a cell not expressing that particular gene or the failure to sample that mRNA in the single-cell reverse-transcription reaction. Thus, while we can say that retinoblastoma cells clearly express multiple differentiation programs, we cannot absolutely rule-out the possibility that we failed to detect low levels of transcripts for some of the other cell types.
MDM2 and MDMX in Retinoblastoma
A recent hypothesis related to the retinoblastoma cell of origin came from the observation that cell type-specific expression of MDM2 occurs in cone photoreceptors in human retina and in amacrine cells in mouse retina (Xu et al., 2009). This finding provided a plausible mechanism in which cones are the cell of origin for retinoblastoma in humans, and amacrine cells are the cell of origin in mice. It was hypothesized that high levels of MDM2 in these cells predisposed them to tumorigenesis, because MDM2 silences the p53 pathway and prevents cell death. According to this hypothesis, the tumors maintain MDM2 expression to suppress the p53 pathway, and the tumor grows and spreads. In our study, we found that MDMX was the predominant family member expressed in human retinoblastomas. We propose that instead of MDM2, MDMX silences p53 in human retinoblastomas, and future therapeutic efforts focused on inhibiting MDMX may prove beneficial for these patients.
Role of Neurotransmitter Signaling in Retinoblastoma
The amacrine differentiation of human and mouse retinoblastoma may be functionally significant for tumor survival and progression. The most striking evidence for amacrine differentiation at the cellular level came from the identification of dense core vesicles in human and mouse retinoblastomas. However, an important distinction is that the retinoblastoma cells appear to express a mixture of receptors, transporters, and metabolic genes for catecholamines, whereas individual amacrine neurons are more restricted and can be subdivided based on catecholamine accumulation (i.e., serotonergic versus dopaminergic). This hybrid-amacrine phenotype is consistent with the broader theme of deregulation of multiple differentiation pathways in the retinoblastoma tumors.
As reported previously, retinoblastomas produce catecholamines and other monoamines (Klaidman et al., 1993; Madtes et al., 1985; Yorek et al., 1987). We found that several of the receptors for monoamines are expressed, as are the genes involved in the biosynthesis of monoamines and the regulation of signaling through these receptors. However, out data show that only the blockade of monoamine receptors, but not other major neurotranmitter signaling pathways, reduces retinoblastoma growth and survival. Our interpretation of these data is that retinoblastoma cells are engaged in an autocrine induction of this pathway that is selective for catecholamines. All of the major catecholamine/monoamine receptors stimulate the MAPK pathway and provide an important mitogenic signal for retinoblastoma. It may also explain why retinoblastomas form in the retina because the amacrine-differentiation pathway can be co-opted for mitogenic signaling following RB1 gene inactivation. Additional studies will be required to determine whether catecholamine signaling influences survival and growth through the MAPK pathway or if it is required for some other aspect of tumorigenesis.
One major caveat of these studies is that we were unable to definitively identify an individual receptor that is important for retinoblastoma proliferation. This suggests that the retinoblastoma tumors have co-opted several monoamine neurotransmitter signaling pathways from amacrine neurons. The monoamine neurotransmitter receptors themselves may not be the critical autocrine receptor, but some other receptor family may be expressed on the tumor cells that share downstream effectors. By perturbing the monoamine neurotransmitter receptors, we may be interfering with other autocrine signaling cascades by competing for the same downstream effectors. In either case, these data provide important insight into potential therapeutic targets for retinoblastoma.
EXPERIMENTAL PROCEDURES
Human Retinoblastoma Samples
Immediately following enucleation, a small incision was made in the sclera, and forceps were used to extract a sample of tumor (3–5 mm3). Tumor specimens from patients treated at St. Jude Children's Research Hospital (St. Jude) were placed in RPMI medium on ice and immediately processed for RNA isolation, DNA isolation, TEM, and/or orthotopic xenografts. Samples from other institutions were immediately placed in RNAlater (QIAGEN) and shipped to St. Jude on ice. The tumors were removed from a diverse patient population, with respect to clinical presentation, sex, and race. All experiments involving human subjects were approved by the St. Jude Children's Research Hospital Institutional Review Board and informed consent was obtained.
Orthotopic Xenografts
Primary human retinoblastoma specimens were dissociated in a solution of trypsin and phosphate-buffered saline (PBS) at 37°C until a single-cell suspension was obtained. Total cell number was determined using a hemacytometer, and the viability of the cells was estimated using a Guava cell analyzer. The cells were collected by centrifugation and resuspended in warm RPMI with 10% fetal calf serum (FCS) at approximately 10,000 cells/μl. Approximately 50,000 cells (5 μl) were injected into each eye of anesthetized SCID mice (B6.CB17-Prkdc < scid > SzJ; Jackson Labs). Virtually all tumor cells engrafted; tumor progression was monitored with a digital retinal camera, and intraocular pressure was regularly measured. No tumor cells were out of the intraocular environment for more than 60 min, and none of the tumors used for orthotopic xenografts were maintained in culture. The MSKCC176 cells were kindly provided by Dr. David Cobrinik (Memorial Sloan-Kettering Cancer Center) (Xu et al., 2009).
Mouse Retinoblastoma
The p107- and p130-knockout mice were obtained from Dr. Tyler Jacks (Massachusetts Institute of Technology); Chx10-Cre mice were obtained from Dr. Connie Cepko (Harvard Medical School); p53Lox/Lox and RbLox/Lox mice were obtained from the Mouse Models of Human Cancer Consortium at the National Cancer Institute; MDMXTg mice were obtained from Dr. Gigi Lozano (MD Anderson Cancer Center); and p130Lox/Lox mice were obtained from Dr. Julien Sage (Stanford Medical School). Mice were monitored weekly for signs of retinoblastoma and anterior chamber invasion. Moribund status was defined as the point when tumor cells invaded the anterior chamber, and intraocular pressure increased to the point of imminent ocular rupture. The St. Jude Institutional Animal Care and Use Committee approved all animal procedures.
Gene Expression Arrays
RNA was extracted using TRIzol reagent (Invitrogen) per the manufacturer's instructions. RNA concentration was quantified using a NanoDrop 100 Spectrophotomer (Thermo Scientific) or a SpectroMax Plus384 spectrophotometer (Molecular Devices). RNA integrity was measured using Agilent Technologies 2100 Bioanalyzer Lab-on-a-chip system. Total RNA (100 ng) was amplified using the Affymetrix Two-Cycle Eukaryotic Target Labeling Assay per the manufacturer's instructions (GeneChip Expression Analysis Technical Manual). The detailed hybridization procedure for the Affymetrix GeneChip for mouse 430_2.0 and human U133_2 are described in the Supplemental Experimental Procedures. The Hartwell Center for Bioinformatics and Biotechnology at St. Jude performed all Affymetrix chip procedures and data analysis.
Supplementary Material
Significance.
Our finding that retinoblastoma tumor cells express multiple neuronal differentiation programs that are normally incompatible in development suggests that the pathways that control retinal development and establish distinct cell types are perturbed during tumorigenesis. Therefore, the cell of origin for retinoblastoma cannot be inferred from the features of the tumor cells themselves. However, we now have a detailed understanding of the neuronal pathways that are deregulated in retinoblastoma and targeting the catecholamine and indolamine receptors or downstream components could provide useful therapeutic approaches in future studies. This example highlights the importance of comprehensive molecular, cellular, and physiological characterization of human cancers with single cell resolution as we incorporate molecular targeted therapy into treatment regimens.
ACKNOWLEDGMENTS
We thank Kip Guy and Anang Shelat for assistance with high-throughput screening and cheminformatics; Sharon Frase and Marina Kedrov for assistance with electron microscopy and morphometric scoring; Geoff Neale for assistance with bioinformatics, Connie Cepko for sharing data, methods, and reagents and for many helpful discussions; David Cobrinik for providing the MSKCC176 cells and for helpful discussions; Justin Thurman for assistance with the animal colony and sectioning; Angela McArthur for editing the manuscript; and Wei Zhao for assistance with biostatistics. This work was supported in part by Cancer Center Support (CA21765) from the NCI; grants to M.A.D from the NIH (EY014867 and EY018599), the American Cancer Society, and Research to Prevent Blindness Foundation; a grant to J.S. (CA114102); and the American Lebanese Syrian Associated Charities (ALSAC). M.A.D. is a Howard Hughes Medical Institute Early Career Scientist.
Footnotes
ACCESSION NUMBERS
The array data were deposited in the GEO public database (accession number: GSE29686).
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, four figures, and ten tables and can be found with this article online at doi:10.1016/j.ccr.2011.07.005.
REFERENCES
- Akimoto M, Cheng H, Zhu D, Brzezinski JA, Khanna R, Filippova E, Oh EC, Jing Y, Linares JL, Brooks M, et al. Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. Proc. Natl. Acad. Sci. USA. 2006;103:3890–3895. doi: 10.1073/pnas.0508214103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemer FA, Van Veelen CW, Staal GE. How did retinoblastoma arise? Nature. 1984;310:731. doi: 10.1038/310731a0. [DOI] [PubMed] [Google Scholar]
- Cherry TJ, Trimarchi JM, Stadler MB, Cepko CL. Development and diversification of retinal amacrine interneurons at single cell resolution. Proc. Natl. Acad. Sci. USA. 2009;106:9495–9500. doi: 10.1073/pnas.0903264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daa Schroder H. Immunohistochemical demonstration of glial markers in retinoblastomas. Vichows Arch A. 1987;411:67–72. doi: 10.1007/BF00734516. [DOI] [PubMed] [Google Scholar]
- Dickson DH, Ramsey MS, Tonus JG. Synapse formation in retinoblastoma tumours. Br. J. Ophthalmol. 1976;60:371–375. doi: 10.1136/bjo.60.5.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoso LA, Folberg R, Arbizo V. Retinal S antigen and retinoblastoma. A monoclonal antibody histopathologic study. Arch. Ophthalmol. 1985;103:855–857. doi: 10.1001/archopht.1985.01050060115039. [DOI] [PubMed] [Google Scholar]
- Donoso LA, Hamm H, Dietzschold B, Augsburger JJ, Shields JA, Arbizo V. Rhodopsin and retinoblastoma. A monoclonal antibody histopathologic study. Arch. Ophthalmol. 1986;104:111–113. doi: 10.1001/archopht.1986.01050130125035. [DOI] [PubMed] [Google Scholar]
- Donovan SL, Schweers B, Martins R, Johnson D, Dyer MA. Compensation by tumor suppressor genes during retinal development in mice and humans. BMC Biol. 2006;4:14. doi: 10.1186/1741-7007-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A, Shields CL. Differential gene expression profile of retinoblastoma compared to normal retina. Mol. Vis. 2010;16:1292–1303. [PMC free article] [PubMed] [Google Scholar]
- Glubrecht DD, Kim JH, Russell L, Bamforth JS, Godbout R. Differential CRX and OTX2 expression in human retina and retinoblastoma. J. Neurochem. 2009;111:250–263. doi: 10.1111/j.1471-4159.2009.06322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griegel S, Heise K, Kindler-Röhrborn A, Rajewsky MF. In vitro differentiation of human retinoblastoma cells into neuronal phenotypes. Differentiation. 1990a;45:250–257. doi: 10.1111/j.1432-0436.1990.tb00479.x. [DOI] [PubMed] [Google Scholar]
- Griegel S, Hong C, Frötschl R, Hülser DF, Greger V, Horsthemke B, Rajewsky MF. Newly established human retinoblastoma cell lines exhibit an “immortalized” but not an invasive phenotype in vitro. Int. J. Cancer. 1990b;46:125–132. doi: 10.1002/ijc.2910460123. [DOI] [PubMed] [Google Scholar]
- Iscove NN, Barbara M, Gu M, Gibson M, Modi C, Winegarden N. Representation is faithfully preserved in global cDNA amplified exponentially from sub-picogram quantities of mRNA. Nat. Biotechnol. 2002;20:940–943. doi: 10.1038/nbt729. [DOI] [PubMed] [Google Scholar]
- Johnson DA, Zhang J, Frase S, Wilson M, Rodriguez-Galindo C, Dyer MA. Neuronal differentiation and synaptogenesis in retinoblastoma. Cancer Res. 2007;67:2701–2711. doi: 10.1158/0008-5472.CAN-06-3754. [DOI] [PubMed] [Google Scholar]
- Kawaguchi A, Ikawa T, Kasukawa T, Ueda HR, Kurimoto K, Saitou M, Matsuzaki F. Single-cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development. 2008;135:3113–3124. doi: 10.1242/dev.022616. [DOI] [PubMed] [Google Scholar]
- Klaidman LK, Tombran-Tink J, Adams JD, Jr., Johnson LV. Effects of medium conditioned by retinal pigmented epithelial cells on neurotransmitter phenotype in retinoblastoma cells. Cancer Lett. 1993;68:207–213. doi: 10.1016/0304-3835(93)90148-3. [DOI] [PubMed] [Google Scholar]
- Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, van Sluis P, Troost D, Meeteren NS, Caron HN, Cloos J, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS ONE. 2008;3:e3088. doi: 10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyritsis A, Tsokos M, Chader G. Attachment culture of human retinoblastoma cells: long-term culture conditions and effects of dibutyryl cyclic AMP. Exp. Eye Res. 1984a;38:411–421. doi: 10.1016/0014-4835(84)90196-9. [DOI] [PubMed] [Google Scholar]
- Kyritsis AP, Tsokos M, Triche TJ, Chader GJ. Retinoblastoma—origin from a primitive neuroectodermal cell? Nature. 1984b;307:471–473. doi: 10.1038/307471a0. [DOI] [PubMed] [Google Scholar]
- Kyritsis AP, Tsokos M, Triche TJ, Chader GJ. Retinoblastoma: a primitive tumor with multipotential characteristics. Invest. Ophthalmol. Vis. Sci. 1986;27:1760–1764. [PubMed] [Google Scholar]
- Laurie NA, Donovan SL, Shih CS, Zhang J, Mills N, Fuller C, Teunisse A, Lam S, Ramos Y, Mohan A, et al. Inactivation of the p53 pathway in retinoblastoma. Nature. 2006;444:61–66. doi: 10.1038/nature05194. [DOI] [PubMed] [Google Scholar]
- MacNeil MA, Masland RH. Extreme diversity among amacrine cells: implications for function. Neuron. 1998;20:971–982. doi: 10.1016/s0896-6273(00)80478-x. [DOI] [PubMed] [Google Scholar]
- MacNeil MA, Heussy JK, Dacheux RF, Raviola E, Masland RH. The shapes and numbers of amacrine cells: matching of photofilled with Golgi-stained cells in the rabbit retina and comparison with other mammalian species. J. Comp. Neurol. 1999;413:305–326. [PubMed] [Google Scholar]
- MacPherson D, Dyer MA. Retinoblastoma: from the two-hit hypothesis to targeted chemotherapy. Cancer Res. 2007;67:7547–7550. doi: 10.1158/0008-5472.CAN-07-0276. [DOI] [PubMed] [Google Scholar]
- Madtes P, Jr., Kyritsis A, Chader GJ. Neurotransmitter systems in morphologically undifferentiated human Y-79 retinoblastoma cells: studies of GABAergic, glycinergic, and beta-adrenergic systems. J. Neurochem. 1985;45:1836–1841. doi: 10.1111/j.1471-4159.1985.tb10541.x. [DOI] [PubMed] [Google Scholar]
- McFall RC, Sery TW, Makadon M. Characterization of a new continuous cell line derived from a human retinoblastoma. Cancer Res. 1977;37:1003–1010. [PubMed] [Google Scholar]
- Mirshahi M, Boucheix C, Dhermy P, Haye C, Faure JP. Expression of the photoreceptor-specific S-antigen in human retinoblastoma. Cancer. 1986;57:1497–1500. doi: 10.1002/1097-0142(19860415)57:8<1497::aid-cncr2820570810>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Ramos CA, Bowman TA, Boles NC, Merchant AA, Zheng Y, Parra I, Fuqua SA, Shaw CA, Goodell MA. Evidence for diversity in transcriptional profiles of single hematopoietic stem cells. PLoS Genet. 2006;2:e159. doi: 10.1371/journal.pgen.0020159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid TW, Albert DM, Rabson AS, Russell P, Craft J, Chu EW, Tralka TS, Wilcox JL. Characteristics of an established cell line of retinoblastoma. J. Natl. Cancer Inst. 1974;53:347–360. doi: 10.1093/jnci/53.2.347. [DOI] [PubMed] [Google Scholar]
- Roesch K, Jadhav AP, Trimarchi JM, Stadler MB, Roska B, Sun BB, Cepko CL. The transcriptome of retinal Müller glial cells. J. Comp. Neurol. 2008;509:225–238. doi: 10.1002/cne.21730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S, Cepko CL. Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev. Biol. 2004;271:388–402. doi: 10.1016/j.ydbio.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Tietjen I, Rihel JM, Cao Y, Koentges G, Zakhary L, Dulac C. Single-cell transcriptional analysis of neuronal progenitors. Neuron. 2003;38:161–175. doi: 10.1016/s0896-6273(03)00229-0. [DOI] [PubMed] [Google Scholar]
- Trimarchi JM, Stadler MB, Roska B, Billings N, Sun B, Bartch B, Cepko CL. Molecular heterogeneity of developing retinal ganglion and amacrine cells revealed through single cell gene expression profiling. J. Comp. Neurol. 2007;502:1047–1065. doi: 10.1002/cne.21368. [DOI] [PubMed] [Google Scholar]
- Trimarchi JM, Stadler MB, Cepko CL. Individual retinal progenitor cells display extensive heterogeneity of gene expression. PLoS One. 2008;3:e1588. doi: 10.1371/journal.pone.0001588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ts'o MO, Fine BS, Zimmerman LE. The nature of retinoblastoma. II. Photoreceptor differentiation: an electron microscopic study. Am. J. Ophthalmol. 1970a;69:350–359. doi: 10.1016/0002-9394(70)92264-6. [DOI] [PubMed] [Google Scholar]
- Ts'o MO, Zimmerman LE, Fine BS. The nature of retinoblastoma. I. Photoreceptor differentiation: a clinical and histopathologic study. Am. J. Ophthalmol. 1970b;69:339–349. doi: 10.1016/0002-9394(70)92263-4. [DOI] [PubMed] [Google Scholar]
- Tsokos M, Kyritsis AP, Chader GJ, Triche TJ. Differentiation of human retinoblastoma in vitro into cell types with characteristics observed in embryonal or mature retina. Am. J. Pathol. 1986;123:542–552. [PMC free article] [PubMed] [Google Scholar]
- Vincent S, Mirshari M, Nicolas C, Adenis C, Dhellemmes P, Soto Ares G, Maurage CA, Baranzelli MC, Giangaspero F, Ruchoux MM. Large-cell medulloblastoma with arrestin-like protein expression. Clin. Neuropathol. 2003;22:1–9. [PubMed] [Google Scholar]
- Vrabec T, Arbizo V, Adamus G, McDowell JH, Hargrave PA, Donoso LA. Rod cell-specific antigens in retinoblastoma. Arch. Ophthalmol. 1989;107:1061–1063. doi: 10.1001/archopht.1989.01070020123044. [DOI] [PubMed] [Google Scholar]
- Wintersteiner H. Das Neuroepithelioma Retinae. Franz Deutioke; Vienna: 1897. [Google Scholar]
- Xiong S, Pant V, Suh YA, Van Pelt CS, Wang Y, Valentin-Vega YA, Post SM, Lozano G. Spontaneous tumorigenesis in mice over-expressing the p53-negative regulator Mdm4. Cancer Res. 2010;70:7148–7154. doi: 10.1158/0008-5472.CAN-10-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XL, Fang Y, Lee TC, Forrest D, Gregory-Evans C, Almeida D, Liu A, Jhanwar SC, Abramson DH, Cobrinik D. Retinoblastoma has properties of a cone precursor tumor and depends upon cone-specific MDM2 signaling. Cell. 2009;137:1018–1031. doi: 10.1016/j.cell.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorek MA, Strom DK, Spector AA. Synthesis and high affinity uptake of serotonin and dopamine by human Y79 retinoblastoma cells. J. Neurochem. 1987;49:1316–1323. doi: 10.1111/j.1471-4159.1987.tb10026.x. [DOI] [PubMed] [Google Scholar]
- Zhong X, Li Y, Peng F, Huang B, Lin J, Zhang W, Zheng J, Jiang R, Song G, Ge J. Identification of tumorigenic retinal stem-like cells in human solid retinoblastomas. Int. J. Cancer. 2007;121:2125–2131. doi: 10.1002/ijc.22880. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.