Abstract
In functionally diverse enzyme superfamilies (SFs), conserved structural and active site features reflect catalytic capabilities “hard-wired” in each SF architecture. Overlaid on this foundation, evolutionary changes in active site machinery, structural topology and other aspects of structural organization and interactions support the emergence of new reactions, mechanisms, and substrate specificity. This review connects topological with functional variation in each of the haloalkanoic acid (HAD) and vicinal oxygen chelate fold (VOC) SFs and a set of redox-active thioredoxin (Trx)-fold SFs to illustrate a few of the varied themes nature has used to evolve new functions from a limited set of structural scaffolds.
Introduction
Although it was originally thought that homologous (related by common ancestry) proteins would share the same fold, the rapidly growing catalogs of protein sequences and structures have revealed an amazing degree of variation among homologs. Clues about similarities in molecular or biological function and the discovery of “missing link” sequences and structures have allowed us to trace distantly related proteins, sometimes with markedly different structures, back to a common ancestor. It has also become clear that as new structural variants of ancestral proteins evolve, their functions can also diverge broadly. Functionally diverse enzyme superfamilies (SFs), the focus of this review, are sets of related enzymes in which some active site features required for fundamental aspects of catalysis are conserved but whose members have diverged to enable catalysis of many different reactions [1]. They represent approximately a third of known enzyme superfamilies 1. While a substantial number of such SFs have now been described in the literature, the structure-function relationships for many more remain to be characterized.
Larger-scale variations in enzyme structure that involve whole domains (see [5] for a review) or significant changes in topology can also play an important role in the evolution of functional variation although these have not been so extensively catalogued. In this article, we briefly review three very different SFs of functionally diverse enzymes to illustrate a few types of topological variation and describe how these are associated with the evolution of new enzymatic functions.
Topological Types and Mechanisms of Structural Change
New proteins are generated by the duplication, divergence, and combination of genes. Topological differences observed between homologous domains include insertions, deletions, and substitutions of secondary structural elements (SSEs), as well as permutations, in which corresponding SSEs occupy approximately the same spatial locations but occur in a different order along the sequence. Grishin has provided clear examples of such changes and further categorized insertions by their effects on beta-sheet topology [6]. Often, these variations are associated with changes in oligomeric state, which also set the stage for domain swapping between monomers and for duplication/fusion events that combine two or more monomers into a single protein.
Another major source of diversification is the combination of domains into different architectures. Recent studies of domain and architecture occurrence across multiple proteomes have shown that whereas most domains were already present in the last universal common ancestor, significant architectural innovation has prevailed throughout the tree of life [7,8*]. A detailed comparison of domain functions in single- versus multi-domain contexts found that domain combination usually preserves function, but makes it more specific or complex [5].
These contrasting themes of significant change and conservation of specific topological features over long evolutionary distances are elaborated below for three diverse groups of enzymes, each illustrating a different type of topological variation associated with the evolution of functional variation.
The Haloalkanoic Acid Dehalogenase (HAD) SF: Cap Domain Variations Enable Divergent Evolution of many Different Reaction and Substrate Specificities
The HAD SF is ubiquitous throughout the biosphere with most of its members thought to have roles in phosphate ester or anhydride hydrolysis in centrally important biological processes. About 20% are ATPases and a huge group of diverse phosphate monoester hydrolases represent almost all of the other known reactions. The SF includes a few known “outlier” reactions as well, such as the SF namesake, haloalkanoic acid dehalogenase, azetidine hydrolase, which cleaves a carbon-nitrogen bond, phosphonatase, which cleaves a phosphorous-carbon bond, and sugar phosphomutases. These reactions are of special interest in illustrating the “outer limits” in reaction and substrate specificity that can be achieved by members of the SF.
HADs are made up of two modules, a core Rossmanoid domain containing conserved active site and catalytic machinery and a “cap” domain (or set of inserts in some variants) represented by several different classes of structures associated with reaction and substrate specificity. Additionally, HAD SF members are also found fused to other proteins, including other enzymes and transporters. A large-scale study by Burroughs et al [9] and another recent review [10**] have detailed the modular nature of the HAD SF architecture and traced in elegant detail the structural, functional and evolutionary relationships across the SF.
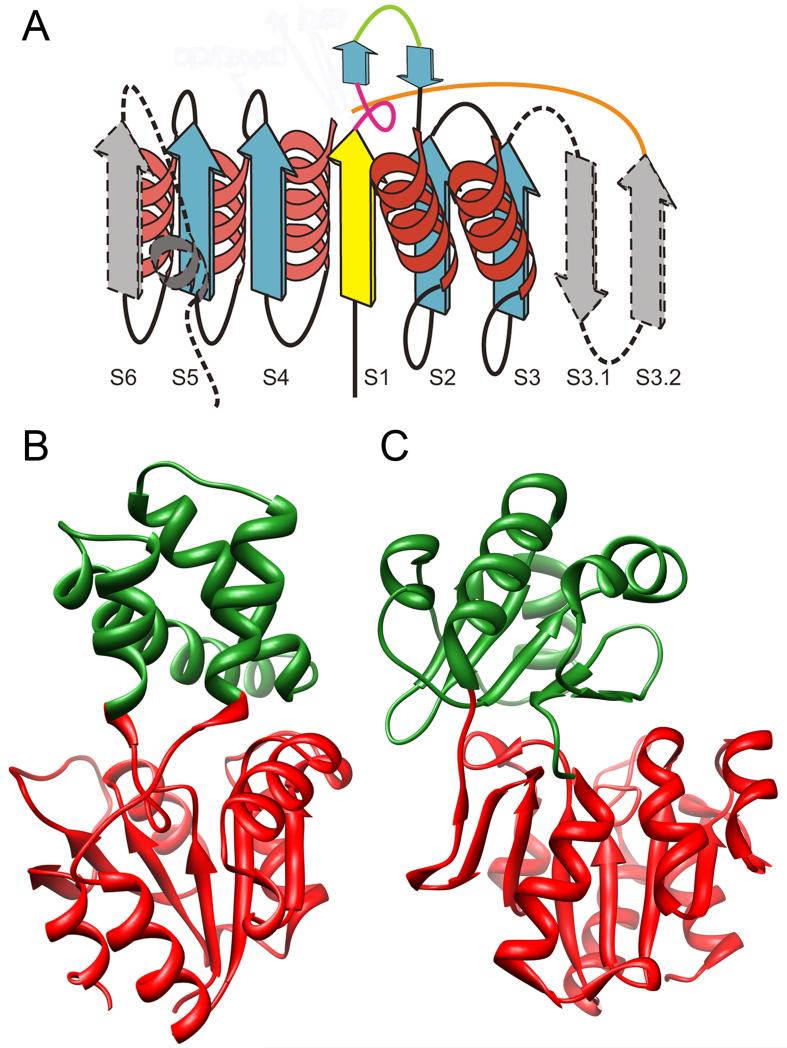
Relative to other Rossmanoid folds, the HAD core domain is distinguished by several features including two structural motifs serving as insertion sites for the caps (Figure 1A), and resulting in different modes by which conformational changes associated with substrate binding deliver specificity. Although the topologies of the HAD domains can vary, the cap domains are more varied with different structural types distinguished by size, topology, associated substrates, and other features (Figures 1B and 1C). The relative topological independence between the catalytic and specificity domains enables an evolutionary paradigm in which the cap domains can evolve to accommodate broad divergence of reaction and substrate specificity while the underlying catalytic machinery presented by the core domain remains relatively conserved. Localization of the active site between these two domains then specifies an architectural organization that makes this modular system work.
Figure 1.
Topologies of HAD and Cap domains. A. Schematic diagram of the classic HAD domain: yellow, strand containing the catalytic asp residue; blue, core strands conserved in all HAD SF members; gray, structural elements that may not have occurred in the ancestral structure; green line shows the insertion point for C1 caps; orange line shows the insertion point for C2 caps. Broken lines indicate secondary structure elements not present in all members containing the HAD domain. Figure adapted from [9]. B. Crystal structure of L-2-haloacid dehalogenase with 2-Chloro-N-Butyrate (ligand not shown) (PDB 1ZRM) showing the HAD domain in red and the C1 cap domain in green. C. Crystal structure of the HAD subclass IIB sugar phosphatases (PDB 1YMQ) showing the HAD domain in red and the C2 cap domain in green.
The so-called “C0 type” HADs are typically considered “capless” as they represent comparatively short loop inserts in the HAD core domain. In contrast, the C1 and C2 caps have evolved as substantial domains, each representing multiple subtypes. The monomeric capless members show a tendency to act on macromolecules (especially protein phosphatases) such that the substrate itself can aid in “capping” the active site to exclude solvent during catalysis [10]. SF members with distinct cap domains, in contrast, mostly appear to catalyze chemistry on small molecules, although exceptions to both trends exist. New catalytic reactions and topological solutions for conferring specificity have been described recently, helping to fill out our understanding of structure-function relationships in this SF [11*,12-14].
The Vicinal Oxygen Chelate (VOC) Superfamily: Mixing and Matching Subdomains for Functional Versatility
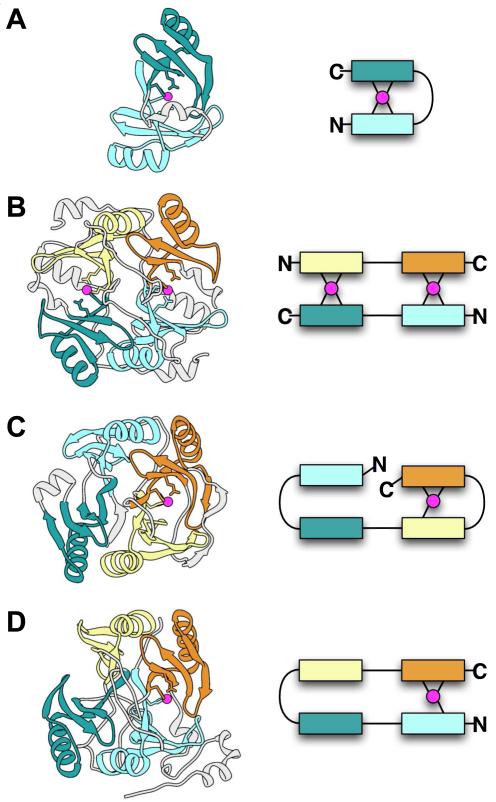
The members of VOC SF are composed of a fundamental structural unit, a short βαβββ subdomain, occurring in pairs and organized in various topological (or domain-swapped) combinations [15]. The known reaction types include isomerizations, epimerizations, oxidative cleavage of C—C bonds as represented by a large and diverse group of extradiol dioxygenases, and nucleophilic substitutions represented by antibiotic resistance proteins such as the fosfomycin resistance proteins [16]. All of these reactions reflect a common mechanistic theme in which a metal plays a direct role in catalysis. The metal coordination sites are supplied by either protein or solvent, with the latter replaced by moieties from the ligand upon binding of substrate or intermediate [17]. The result is an architecture evolved to support catalysis by mixing and matching the paired subdomains in complex patterns (Figure 2). These different topologies can be associated with different reaction types within the SF but also with enzymes catalyzing the same reaction in different species, as exemplified by the two glyoxalase 1 structures from bacteria and human, each of which represents a distinct topological pattern (Figures 2A and 2B).
Figure 2.
Examples of alternate arrangements of paired beta-alpha-beta-beta-beta modules in the VOC SF. Structures are shown on the left and schematic diagrams on the right. In both the structures and schematics, metal ions are magenta and different modules are shown in different colors to highlight the repeating beta-alpha-beta-beta-beta unit. Metal-ligating residues may occur in the first and/or last beta-strands of a module. Parts of the structures not within beta-alpha-beta-beta-beta modules are shown in light gray. (A) Glyoxalase I from Clostridium (PDB 3HDP), in which two modules from a single chain pair to form a metal site. (B) Human glyoxalase I (PDB 1QIN), in which two chains each containing two modules pair in head-to-tail fashion to form two metal sites. (C) 2,3-Dihydroxybiphenyl 1,2-dioxygenase from Burkholderia (PDB 1KMY), in which the four modules in a single chain pair in the order 1-2 and 3-4, and only the latter pair forms a metal site. (D) Protein of unknown function from Bacillus (PDB 1ZSW), in which the four modules in a single chain pair in the order 1-4 and 2-3, and only the former pair forms a metal site.
Another subset of proteins exhibit a dimer topology analogous to that of human glyoxalase I (Figure 2B), but in these, the metal-binding sites have been replaced by antibiotic binding sites. These include the enzymatically inactive bleomycin resistance protein (BRP) that appears to function simply to sequester bleomycin [18]. Recently, an enzymatically inactive thiocoraline (TioX)-binding protein has been structurally characterized [19*] that exhibits a further topological variation. TioX forms a tetramer in which two monomers form a dimer like that of BRP, while the other two monomers do not interact with each other, but are each disulfide-bonded to both monomers of the BRP-like pair.
The extradiol dioxygenases represent the largest reaction class in the VOC SF, two topological variants of which are shown in Figures 2C and 2D. The enzyme 2,3-hydroxybiphenyl 1,2-dioxygenase (Figure 2C) has been structurally and mechanistically characterized [20,21] and serves as a model for understanding relationships in other extradiol dioxygenases acting on aromatic substrates. The ortho OH groups on the aromatic substrate are bound to a required Fe(II) on adjacent coordination sites, accompanied by interactions with two His and a carboxylate active site residue, and with O2, to complete the coordination interactions with iron. In an interesting variation on this theme, a group of sequences that align well with another biochemically characterized homolog of these aromatic extradiol dioxygenases, 2,6-dichlorohydroquinone (DCHQ) dioxygenase, have been identified [22]. Although DCHQ dioxygenase is similar to the 2,3-hydroxybiphenyl 1,2-dioxygenase in its iron- and O2-dependence [23], its substrate cannot chelate the iron in a similar way since its OH groups are para on the aromatic ring rather than ortho. And although DCHQ dioxygenase has not been structurally characterized, a structure of a protein from this sequence alignment group but of unknown reaction specificity has been solved by the Midwest Structure Genomics Consortium (PDB 1ZSW, unpublished). Its topology is illustrated in Figure 2D. The alignment of DCHQ dioxygenase with 1ZSW suggests that both proteins exhibit a similar topological organization. In part because 1ZSW lacks a relevant bound substrate, a structural route for iron-mediated catalysis of substrates such as DCHQ is not obvious. Still, it is intriguing to speculate that the variant topology represented by these two proteins is likely to play a role. Other new extradiol dioxygenases have been recently identified that may represent yet additional topological variants evolved to accommodate functional requirements via mixing and matching of subdomain modules [24,25].
The Thioredoxin (Trx)-fold Like SFs: Varied Inserts and Domain Additions Extend the Redox Repertoire of the Canonical Trx Fold
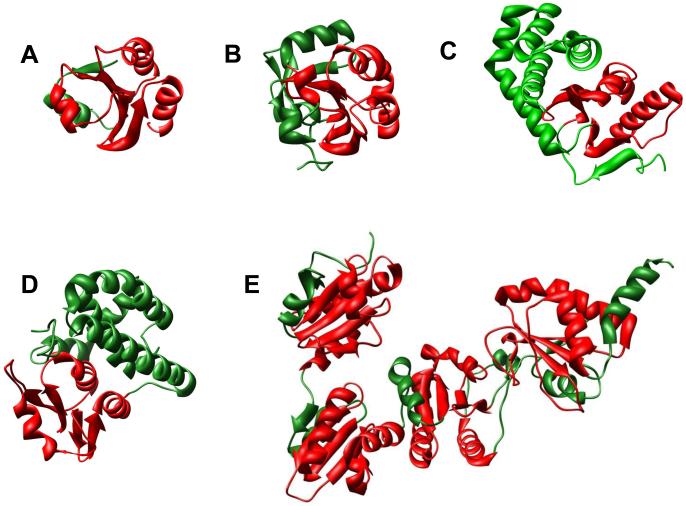
Together, the Trx-fold like SFs represent one of the largest groups of (presumably) homologous proteins known. Figure 3 shows a few of the several topologically distinct SFs that have evolved via decorations and/or duplications of the core Trx domain (found in its most minimal form in glutaredoxin (Grx) [26]). Built around this Trx-like core and variants of an associated CXXC motif, the known functions of these SFs are almost universally involved in redox biology. In almost half of these proteins, however, this core motif is altered, degenerate or missing, as in the glutathione transferases [27]. Nevertheless, some variant of redox machinery is conserved in all of the Trx-fold like SFs, positioned in a similar place in the fold. Large-scale comparisons of structure-function relationships across these SFs reveal functional variations associated with the catalytic redox machinery in particular [28**,29]. Multidomain proteins that combine different Trx domains have also been characterized [30] and some of these have been recently reviewed with a focus on how the combination of domains modulates catalysis [31*]. Using structure comparisons, more remotely similar proteins containing circular permutations of Trx-like domains have also been identified [32].
Figure 3.
Examples of topological variations for Trx-fold SFs. Red: Trx-fold, Green: inserts or domain pairing additions to the Trx fold. A. Human Trx (PDB 1AUC). B. Human Prx V (PDB 1HD2). C. E. coli disulfide binding protein A (PDB 1FVK). D. Human glutathione transferase M2-2 (PDB 1XW5). E. Yeast protein disulfide isomerase (PDB 2B53).
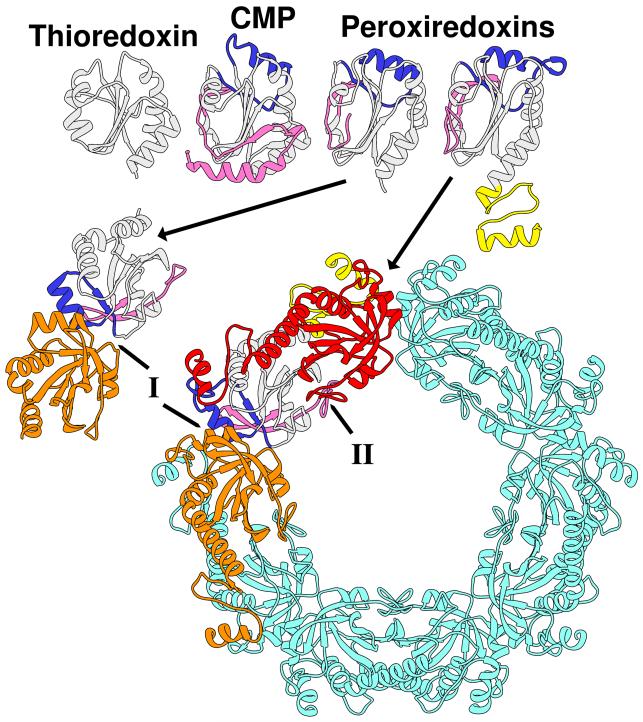
It has been suggested that many SFs containing the Trx-like fold and associated redox catalytic machinery may have evolved from a Trx-like ancestor [28,33,34]. One of these SFs, the peroxiredoxins (Prx), can be most closely linked to Trxs through a group of “bridging” proteins with similarities to both [28,33]. As shown in Figure 4 (top), this bridging group, the cytochrome maturation proteins (CMPs), exhibit two insertions relative to the canonical Trx fold, one at the N-terminus and a second after the β2 strand, [35,36]. Like Trxs, CMPs catalyze thiol oxidoreductase reactions using a CXXC catalytic center. Both inserts are also found in Prxs but in these proteins they are accompanied by changes in the active sites consistent with evolution of Prx chemistry (using a conserved TXXC active site, for example) from a Trx-like ancestor. The bottom left of Figure 4 shows the involvement of the CMP- and Prx-specifying inserts in the formation of the most prevalent dimerization interface seen across Prx subfamilies. Figure 4 (bottom right) shows a decameric structure associated with 2-Cys Prxs, which is built from this interface plus an additional oligomerization interface that brings together the Trx-like beta sheets edge to edge. Formation of the decamer has been linked to redox state [37,38].
Figure 4.
Structures of Trx, CMP, and Prxs are consistent with their connections in sequence space, in which CMPs link the other two SFs (adapted from [23]). Top row, from left to right: Trx (PDB 1XOA), CMP (PDB 1JFU, chain A), and Prxs (left, PDB 1XXU chain A; right, PDB 1QMV chain A). The canonical Trx fold is shown in light gray, an N-terminal extension in dark pink, an insert that includes a helix in blue, and a C-terminal extension in yellow. The bottom row shows the same two Prxs as in the top row, but in homomultimeric assemblies. Bottom left, Prx dimer (PDB 1XXU chains A and B) with one subunit colored as in the top row, the other subunit in orange. This dimer uses one type of interface (marked I) that primarily involves the insert helix (blue). Right, Prx decamer (PDB 1QMV) with one monomer colored as in the top row, a second monomer in orange interacting with the first through the type I interface, a third monomer in red interacting with the first through the type II interface, and the remaining seven monomers in aqua. The type II interface is primarily a concatenation of the central beta-sheet of the Trx fold, but in this decamer, the C-terminal extension (yellow) is also involved.
As with other SFs exhibiting a Trx-fold, Prxs can be further subdivided into several subgroups distinguished by structural and functional features. Nelson et al. have recently classified in careful detail the PRX SF into six subfamilies [39**]. This classification shows that each can be distinguished by variations in the locations of the Cys outside of the peroxidatic active site that is involved in recycling the enzymes back to the active form following catalysis (the “resolving” Cys), along with other active site and topological features. The latter include variations in oligomeric interactions in some Prxs that appear to be important for achieving a correct active site geometry. A recent review summarizes catalytic transitions in the context of available structures for these Prx subfamilies [40**].
Conclusions
In this review, we describe variations in the topologies of three functionally diverse enzyme superfamilies to illustrate some themes by which these features contribute to the diversification of function. Each involves a different primary way in which topology has been varied by evolution and the functional consequences that have resulted. Thus, the HAD superfamily typifies the use of two different domains to separate the catalytic machinery from the more varied domains that provide binding of specific substrates and positioning of labile bonds appropriately for chemistry to occur. This separation of catalytic and substrate “presentation” domains is a basic theme for topological variation across large SFs representing widely different folds, for example, the glutathione transferases [27] and the two-dinucleotide binding domains flavoproteins (tDBDF) SF [41]. Further, this SF also typifies additional levels of topological variation associated with duplications of subdomains that result in different topologies within each of the C1 and C2 capping domain types as well as the many different multidomain architectures (fusion proteins) in which they are included [9]. Topological variations in the VOC SF are more profoundly different, involving major permutations and/or re-organization of duplicated subdomains in the protein core of each subgroup. This enables evolution of complex arrangements of a SF-common catalytic machinery often located according to the binding position in which the labile bonds in the substrates are presented. Finally, the Trx-like fold SFs represent a large-scale and diverse set of topological variations across many different SFs, while maintaining in each the core Trx-like domain and some variant of its associated redox machinery.
As we look toward the future, it is becoming clear that large-scale studies such as some of those described in this work will be increasingly important in the investigation of topological variation and its role in enabling and supporting functional diversification. We note that many functionally diverse enzyme SFs now represent many thousands of sequences and sometimes scores of structures. Moreover, these numbers continue to increase rapidly. Currently, we count >40,000 members of the HAD SF in Genbank [42], up from ~20,000 noted in [10]. Likewise, the latest sequence count from Pfam (release 24.0) for the VOC SF totals >13,000 sequences and we estimate that the >30,000 sequences of the Trx-fold class SFs described in [28] have now increased by at least 15-20%.
While these data provide an enormously rich resource, structural characterization of many more sequences is going to be required to achieve a truly useful and interpretable census of their topological variations. Thus, it is only as many more structures, including liganded structures, are solved (or adequately modeled) and their reaction and substrate specificities discovered, that we can expect to fill in the picture. This is going to be a major challenge since even high-throughput experimental methods for characterizing 3D structures and reaction specificities are orders of magnitude slower than the generation of sequence data. Still, by using effective strategies for choosing targets for characterization (see [43] for example), it may be possible to leverage new experimental information to inform us broadly about how both topology and active site structural features track with functional divergence.
Highlights.
> We examine how topological variations contribute to natural evolution of new enzymes.
> These variations often allow changes in specificity, conserving primary mechanism.
> Variations span permutations of a primordial subdomain to inserts in a core domain.
> Large-scale studies across a superfamily can be needed to reveal these variations.
Acknowledgements
This work was supported by NIH GM60595 to PCB. Figure 1A was adapted from reference [9]; we thank L. Aravind for supplying the original for our use. Figure 3 was adapted from a figure generated by Dr. Holly J. Atkinson while a PhD student in the Babbitt lab; we thank her for its use. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR001081).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Of the 704 enzyme SFs classified in SCOP vs. 1.75 [2], we count 272 as functionally diverse (D. Almonacid and P. Babbitt, unpublished), defined as SFs containing two or more enzymes that differ in their Enzyme Commission (EC) nomenclature [3] sub-subclass (third digit), which specifies the type of chemical reaction represented. This is certainly an undercount since additional reactions in enzyme SFs can be expected to be discovered among the many enzymes of unknown reaction specificity still awaiting annotation [4].
References and Recommended Reading
- 1.Gerlt JA, Babbitt PC. Divergent evolution of enzymatic function: Mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu Rev Biochem. 2001;70:209–246. doi: 10.1146/annurev.biochem.70.1.209. [DOI] [PubMed] [Google Scholar]
- 2.Andreeva A, Murzin AG. Evolution of protein fold in the presence of functional constraints. Curr Opin Struct Biol. 2006;16:399–408. doi: 10.1016/j.sbi.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Tipton KF. Enzyme Nomenclature: Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (IUBMB) NC-IUBMB; New York: 1992. [DOI] [PubMed] [Google Scholar]
- 4.Gerlt JA. A Protein Structure (or Function ?) Initiative. Structure. 2007;15:1353–1356. doi: 10.1016/j.str.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Bashton M, Chothia C. The generation of new protein functions by the combination of domains. Structure. 2007;15:85–99. doi: 10.1016/j.str.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 6.Grishin NV. Fold change in evolution of protein structures. J Struct Biol. 2001;134:167–185. doi: 10.1006/jsbi.2001.4335. [DOI] [PubMed] [Google Scholar]
- 7.Caetano-Anolles G, Wang M, Caetano-Anolles D, Mittenthal JE. The origin, evolution and structure of the protein world. Biochem J. 2009;417:621–637. doi: 10.1042/BJ20082063. [DOI] [PubMed] [Google Scholar]
- *8.Chothia C, Gough J. Genomic and structural aspects of protein evolution. Biochem J. 2009;419:15–28. doi: 10.1042/BJ20090122. An excellent and up-to-date introduction to the duplication and divergence of protein domains to form new proteins. This paper provides a good foundation for understanding topological variation in proteins and includes a section on enzymes, the focus of this review.
- 9.Burroughs AM, Allen KN, Dunaway-Mariano D, Aravind L. Evolutionary genomics of the HAD superfamily: understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J Mol Biol. 2006;361:1003–1034. doi: 10.1016/j.jmb.2006.06.049. [DOI] [PubMed] [Google Scholar]
- **10.Allen KN, Dunaway-Mariano D. Markers of fitness in a successful enzyme superfamily. Curr Opin Struct Biol. 2009;19:658–665. doi: 10.1016/j.sbi.2009.09.008. This is a recent update of the HAD SF since it was initially described in [9]. This review extends our understanding to new members of the SF and provides a concise reprise on the structural modularity and relationship between the catalytic and specificity domains in enabling diversification of reaction specificity throughout the biosphere.
- *11.Nguyen HH, Wang L, Huang H, Peisach E, Dunaway-Mariano D, Allen KN. Structural determinants of substrate recognition in the HAD superfamily member D-glycero-D-manno-heptose-1,7-bisphosphate phosphatase (GmhB) Biochemistry. 2010;49:1082–1092. doi: 10.1021/bi902019q. This description of a capless HAD extends our understanding about how substrate recognition can be achieved in SF members that lack a defined cap domain and provides a new level of detail for understanding structure-function relationships in this less-well studied subset of HAD SF members.
- 12.Rangarajan ES, Proteau A, Wagner J, Hung MN, Matte A, Cygler M. Structural snapshots of Escherichia coli histidinol phosphate phosphatase along the reaction pathway. J Biol Chem. 2006;281:37930–37941. doi: 10.1074/jbc.M604916200. [DOI] [PubMed] [Google Scholar]
- 13.Toyoda M, Jitsumori K, Mikami B, Wackett LP, Kurihara T, Esaki N. Crystallization and preliminary X-ray analysis of L-azetidine-2-carboxylate hydrolase from Pseudomonas sp. strain A2C. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:801–804. doi: 10.1107/S1744309110017045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Huang H, Nguyen HH, Allen KN, Mariano PS, Dunaway-Mariano D. Divergence of biochemical function in the HAD superfamily: D-glycero-D-manno-heptose-1,7-bisphosphate phosphatase (GmhB) Biochemistry. 2010;49:1072–1081. doi: 10.1021/bi902018y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergdoll M, Eltis LD, Cameron AD, Dumas P, Bolin JT. All in the family: structural and evolutionary relationships among three modular proteins with diverse functions and variable assembly. Protein Sci. 1998;7:1661–1670. doi: 10.1002/pro.5560070801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernat BA, Laughlin LT, Armstrong RN. Fosfomycin resistance protein (FosA) is a manganese metalloglutathione transferase related to glyoxalase I and the extradiol dioxygenases. Biochemistry. 1997;36:3050–3055. doi: 10.1021/bi963172a. [DOI] [PubMed] [Google Scholar]
- 17.Armstrong RN. Mechanistic diversity in a metalloenzyme superfamily. Biochemistry. 2000;39:13625–13632. doi: 10.1021/bi001814v. [DOI] [PubMed] [Google Scholar]
- 18.Dumas P, Bergdoll M, Cagnon C, Masson JM. Crystal structure and site-directed mutagenesis of a bleomycin resistance protein and their significance for drug sequestering. EMBO J. 1994;13:2483–2492. doi: 10.1002/j.1460-2075.1994.tb06535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *19.Biswas T, Zolova OE, Lombo F, de la Calle F, Salas JA, Tsodikov OV, Garneau-Tsodikova S. A new scaffold of an old protein fold ensures binding to the bisintercalator thiocoraline. J Mol Biol. 2010;397:495–507. doi: 10.1016/j.jmb.2010.01.053. Recent structural and biochemical characterization of this antibiotic resistance protein and potent antitumor agent establishes its membership in the VOC SF. The paper describes a new topological arrangement of the core subdomain elements that nature has evolved to generate new SF functions.
- 20.Han S, Eltis LD, Timmis KN, Muchmore SW, Bolin JT. Crystal structure of the biphenyl-cleaving extradiol dioxygenase from a PCB-degrading pseudomonad. Science. 1995;270:976–980. doi: 10.1126/science.270.5238.976. [DOI] [PubMed] [Google Scholar]
- 21.Lipscomb JD. Mechanism of extradiol aromatic ring-cleaving dioxygenases. Curr Opin Struct Biol. 2008;18:644–649. doi: 10.1016/j.sbi.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pegg SC, Brown SD, Ojha S, Seffernick J, Meng EC, Morris JH, Chang PJ, Huang CC, Ferrin TE, Babbitt PC. Leveraging enzyme structure-function relationships for functional inference and experimental design: the structure-function linkage database. Biochemistry. 2006;45:2545–2555. doi: 10.1021/bi052101l. [DOI] [PubMed] [Google Scholar]
- 23.Xu L, Resing K, Lawson SL, Babbitt PC, Copley SD. Evidence that pcpA encodes 2,6-dichlorohydroquinone dioxygenase, the ring cleavage enzyme required for pentachlorophenol degradation in Sphingomonas chlorophenolica strain ATCC 39723. Biochemistry. 1999;38:7659–7669. doi: 10.1021/bi990103y. [DOI] [PubMed] [Google Scholar]
- 24.Mulako I, Farrant JM, Collett H, Illing N. Expression of Xhdsi-1VOC, a novel member of the vicinal oxygen chelate (VOC) metalloenzyme superfamily, is up-regulated in leaves and roots during desiccation in the resurrection plant Xerophyta humilis (Bak) Dur and Schinz. J Exp Bot. 2008;59:3885–3901. doi: 10.1093/jxb/ern226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu H, Zhang Y, Yang J, Mahmud T, Bai L, Deng Z. Alternative epimerization in C(7)N-aminocyclitol biosynthesis is catalyzed by ValD, a large protein of the vicinal oxygen chelate superfamily. Chem Biol. 2009;16:567–576. doi: 10.1016/j.chembiol.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin JL. Thioredoxin--a fold for all reasons. Structure. 1995;3:245–250. doi: 10.1016/s0969-2126(01)00154-x. [DOI] [PubMed] [Google Scholar]
- 27.Atkinson HJ, Babbitt PC. Glutathione transferases are structural and functional outliers in the thioredoxin fold. Biochemistry. 2009;48:11108–11116. doi: 10.1021/bi901180v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **28.Atkinson HJ, Babbitt PC. An atlas of the thioredoxin fold class reveals the complexity of function-enabling adaptations. PLoS Comput Biol. 2009;5:e1000541. doi: 10.1371/journal.pcbi.1000541. This large-scale study describes and compares the many Trx-fold class superfamilies distinguished by their variations in topology and functional roles in redox biology. Structure and similarity networks are used to establish structural connections among the SFs and subgroups within them and provide a census across this set of species representation, active site motifs, and other features.
- 29.Pan JL, Bardwell JC. The origami of thioredoxin-like folds. Protein Sci. 2006;15:2217–2227. doi: 10.1110/ps.062268106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim SJ, Woo JR, Hwang YS, Jeong DG, Shin DH, Kim K, Ryu SE. The tetrameric structure of Haemophilus influenza hybrid Prx5 reveals interactions between electron donor and acceptor proteins. J Biol Chem. 2003;278:10790–10798. doi: 10.1074/jbc.M209553200. [DOI] [PubMed] [Google Scholar]
- *31.Pedone E, Limauro D, D’Ambrosio K, De Simone G, Bartolucci S. Multiple catalytically active thioredoxin folds: a winning strategy for many functions. Cell Mol Life Sci. 2010;67:3797–3814. doi: 10.1007/s00018-010-0449-9. This work describes some examples of protein families comprised of multiple Trx domains, their modes of interaction, and ways in which their combination affects their functions.
- 32.Qi Y, Grishin NV. Structural classification of thioredoxin-like fold proteins. Proteins. 2005;58:376–388. doi: 10.1002/prot.20329. [DOI] [PubMed] [Google Scholar]
- 33.Copley SD, Novak WR, Babbitt PC. Divergence of function in the thioredoxin fold suprafamily: evidence for evolution of peroxiredoxins from a thioredoxin-like ancestor. Biochemistry. 2004;43:13981–13995. doi: 10.1021/bi048947r. [DOI] [PubMed] [Google Scholar]
- 34.Schroder E, Ponting CP. Evidence that peroxiredoxins are novel members of the thioredoxin fold superfamily. Protein Sci. 1998;7:2465–2468. doi: 10.1002/pro.5560071125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Capitani G, Rossmann R, Sargent DF, Grutter MG, Richmond TJ, Hennecke H. Structure of the soluble domain of a membrane-anchored thioredoxin-like protein from Bradyrhizobium japonicum reveals unusual properties. J Mol Biol. 2001;311:1037–1048. doi: 10.1006/jmbi.2001.4913. [DOI] [PubMed] [Google Scholar]
- 36.Edeling MA, Guddat LW, Fabianek RA, Thony-Meyer L, Martin JL. Structure of CcmG/DsbE at 1.14 A resolution: high-fidelity reducing activity in an indiscriminately oxidizing environment. Structure. 2002;10:973–979. doi: 10.1016/s0969-2126(02)00794-3. [DOI] [PubMed] [Google Scholar]
- 37.Hall A, Karplus PA, Poole LB. Typical 2-Cys peroxiredoxins--structures, mechanisms and functions. FEBS J. 2009;276:2469–2477. doi: 10.1111/j.1742-4658.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wood ZA, Poole LB, Hantgan RR, Karplus PA. Dimers to doughnuts: redox-sensitive oligomerization of 2-cysteine peroxiredoxins. Biochemistry. 2002;41:5493–5504. doi: 10.1021/bi012173m. [DOI] [PubMed] [Google Scholar]
- **39.Nelson KJ, Knutson ST, Soito L, Klomsiri C, Poole LB, Fetrow JS. Analysis of the peroxiredoxin family: Using active-site structure and sequence information for global classification and residue analysis. Proteins. 2010;79:947–964. doi: 10.1002/prot.22936. This work uses active site profiles to classify >3500 putative Prxs into subfamilies, allowing these groupings to be more effectively associated with functional properties and used in other structure-function studies.
- **40.Hall A, Nelson K, Poole L, Karplus PA. Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid Redox Signal. 2011 doi: 10.1089/ars.2010.3624. This paper reviews classification of Prxs into subfamilies, providing a lucid and useful big picture view of their structure-function mappings with an emphasis on relating how their functional properties track with structural features, including quaternary structure.
- 41.Ojha S, Meng EC, Babbitt PC. Evolution of Function in the “Two Dinucleotide Binding Domains” Flavoproteins. PLoS Comput Biol. 2007;3:e121. doi: 10.1371/journal.pcbi.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2011;39:D32–37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pieper U, Chiang R, Seffernick JJ, Brown SD, Glasner ME, Kelly L, Eswar N, Sauder JM, Bonanno JB, Swaminathan S, et al. Target selection and annotation for the structural genomics of the amidohydrolase and enolase superfamilies. J Struct Funct Genomics. 2009 doi: 10.1007/s10969-008-9056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]