Abstract
Classification of enzyme function should be quantitative, computationally accessible, and informed by sequences and structures to enable use of genomic information for functional inference and other applications. Large-scale studies have established that divergently evolved enzymes share conserved elements of structure and common mechanistic steps and that convergently evolved enzymes often converge to similar mechanisms too, suggesting that reaction mechanisms could be used to develop finer-grained functional descriptions than provided by the Enzyme Commission (EC) system currently in use. Here we describe how evolution informs these structure-function mappings and review the databases that store mechanisms of enzyme reactions along with recent developments to measure ligand and mechanistic similarities. Together, these provide a foundation for new classifications of enzyme function.
Introduction
The chemical reactions necessary to support life are catalyzed by enzymes representing a remarkable diversity of substrate and reaction specificities. The classification of their sequences and structures has been facilitated by underlying evolutionary and biophysical models, enabling quantitative assignment of sequence and structural similarity. In contrast, classification of enzyme functions currently relies on the Enzyme Commission (EC) system [1], which is based only on qualitative descriptions of the overall transformation catalyzed, a level of functional granularity too broad to allow direct correlation between enzyme functions and the structural features that are associated with them [2]. We discuss here some features of functionally diverse enzyme superfamilies and of convergently evolved enzymes and explain how the mechanistic steps in their catalytic mechanisms represent a more useful level of functional granularity than overall reactions for linking structure and function. As enzymes are discovered by genome sequencing, creation of resources to manage and enable investigation of their molecular diversity also becomes increasingly important. Thus, we also include here a review of the databases specifically developed to link active site structural features to their mechanistic capabilities in a computationally accessible manner. Finally, models for quantitative comparisons of ligands and mechanisms are described, along with a discussion about how these provide a foundation for a more robust and structurally contextual classification of enzyme functions.
Lessons learned from studying the evolution of enzyme functions
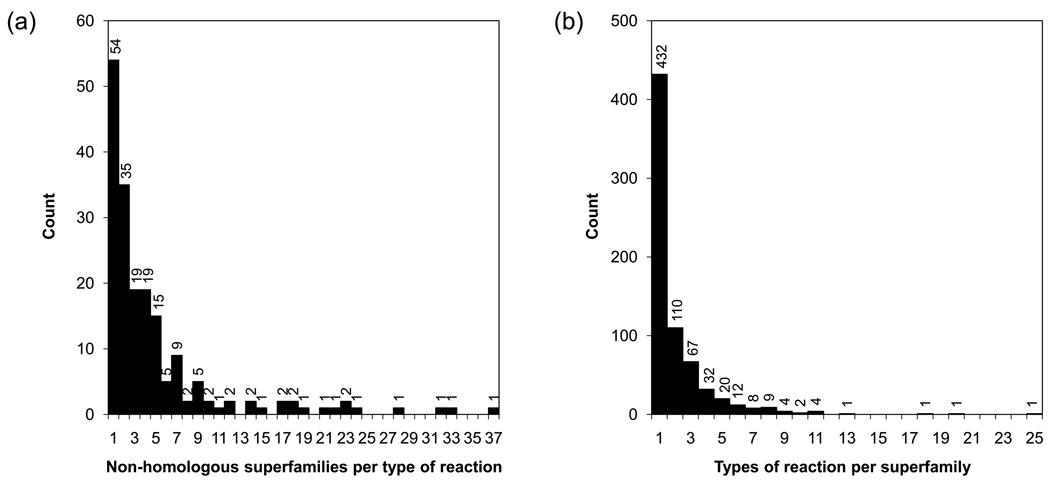
Early views of the enzyme universe assumed that a set of homologous enzymes catalyzed only one type of chemical reaction and that a particular chemical reaction was catalyzed by a single group of homologous enzymes. As more information has accumulated, larger-scale studies of the relationship between enzyme structure and function have shown both that divergent evolution from a common ancestor often generates superfamilies of enzymes catalyzing a diversity of reactions [3,4] and that convergent evolution independently generates unrelated enzymes that catalyze the same type of chemical reaction [5•]. To obtain a current estimate of the incidence of divergent and convergent evolution, we used PDBSprotEC [6] and SCOP [7] to map EC sub-subclasses (reaction types) to structural superfamilies. These counts show that over one third of the structurally characterized enzyme superfamilies are functionally diverse (Figure 1a) and that over two thirds of the currently defined EC sub-subclasses contain analogous enzymes (Figure 1b). Thus, many more enzymes now appear to belong to functionally diverse enzyme superfamilies or evolved via convergent evolution than has been previously appreciated. This underscores the importance of using knowledge of how new enzyme reactions evolve to develop new classifications of enzyme function.
Figure 1.
Estimation of divergent and convergent function evolution in enzymes. In analogy to previous studies within and across genomes [59,60], we have used PDBSprotEC and SCOP to map EC sub-subclasses to structural superfamilies. EC sub-subclasses (first three numbers in the EC system) were used rather than EC serial numbers (all four numbers) to capture reaction specificities irrespective of substrate specificities. (a) The number of different EC sub-subclasses (types of chemical reactions) associated to members of structurally characterized enzyme superfamilies indicates that over one third of superfamilies (272 out of 704) are functionally diverse. (b) The minimum number of non-homologous superfamilies associated to each type of chemical reaction indicates that in over two thirds of the EC sub-subclasses (131 out of 185) Nature has convergently evolved independent enzymes to carry out the same function.
Divergent Evolution of Enzyme Function
The type and extent of functional similarity shared by divergently evolved enzymes depends on which elements of the active site of the common ancestor were more amenable to change and which were more conserved. Building on earlier observations [8,9], Babbitt and Gerlt introduced the model for “chemistry-constrained” evolution of enzyme function [10], where reaction chemistry is conserved among homologs whilst substrate specificity changes. Subsequent large-scale analyses examining pairs of homologous enzymes within and across genomes suggest that this model represents a dominant natural strategy in the evolution of new enzyme functions [11,12]. Several recent reviews continue to enumerate cases where all members of a superfamily conserve at least one common mechanistic aspect linked to conserved features of their active sites [13–16]. Other evidence indicates that most superfamilies also share similarities among their substrates [17], with one article reporting that all but one of 42 such superfamilies indeed share a common substructure in all of their substrates [18]. Diversity of substrate and reaction specificities in divergently evolved superfamilies is thus conscribed by both catalytic and binding capabilities associated with the common active site features in all its members. The result is a specific structure-mechanism paradigm for each superfamily that provides valuable context for classification of their reaction specificities and to guide the prediction of function for new enzymes discovered in the genome projects [19].
Convergent Evolution of Enzyme Function
Several studies have suggested that convergent evolution is also widespread [5,20,21] and that some non-homologous enzymes share structurally and functionally equivalent active site residues and similar mechanisms [20,22]. Recently, a quantitative assessment of convergently evolved enzymes revealed that of those enzymes that have similar overall reactions one third also had similar catalytic mechanisms [23••], concluding that functional analogs, like homologs, often share common mechanistic aspects along their reaction pathways. This finding opens up the possibility of generating structure-mechanism links for functionally analogous enzymes too, along with strategies for function annotation based on analogy rather than homology.
Databases storing mechanisms of enzyme reactions
As the genome projects continue to grow at a rapid pace and more enzymes are cataloged, resources for structure-function mapping in enzymes are needed to aid in leveraging this information for applications that range from functional inference to enzyme design. Four highly curated databases are described below that link information about enzyme sequences and structures to information about their catalytic mechanisms in a computationally accessible form, a necessary prerequisite for the development of functional classifications that are quantitative and can be applied on a large-scale. These include the Structure-Function Linkage Database (SFLD) [24], the Mechanism, Annotation and Classification in Enzymes (MACiE) database [25], the Enzyme Catalysis Database (EzCatDB) [26], and the Hierarchical Classification of Hydrolases Catalytic Sites (HCS) database [27].
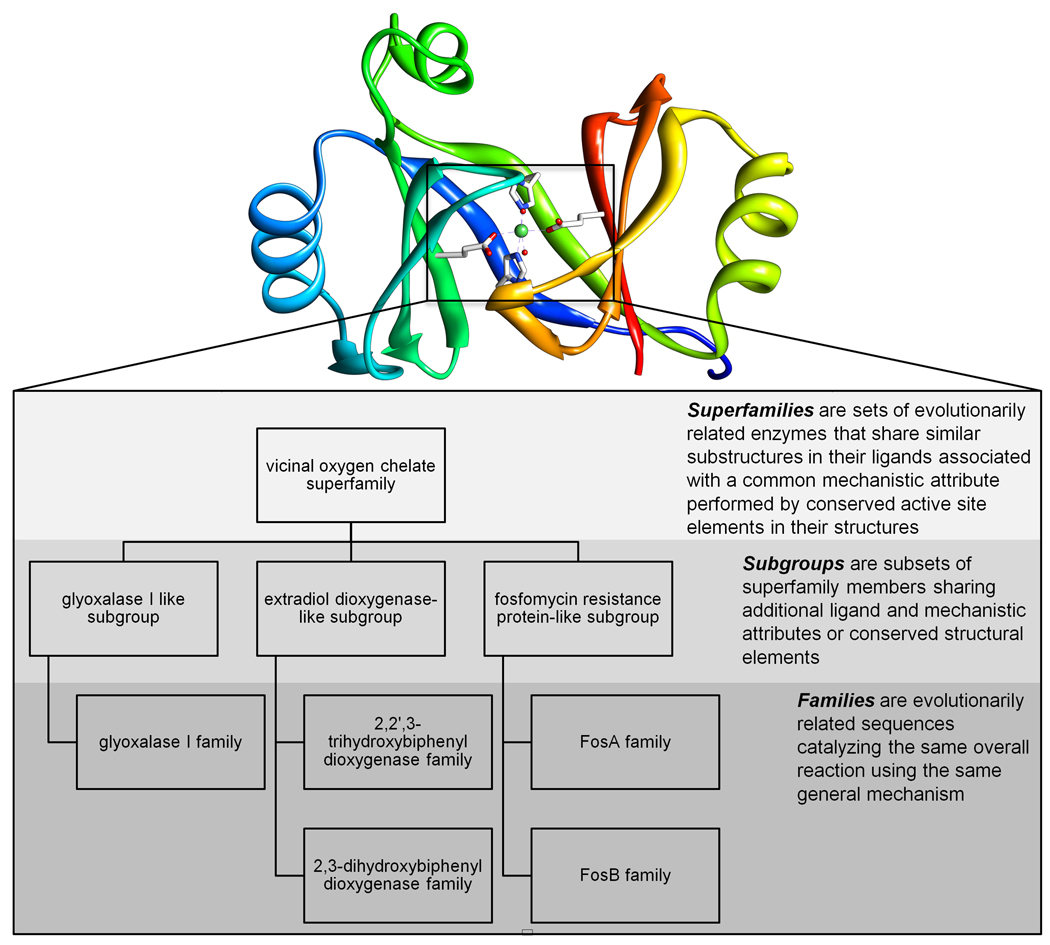
The SFLD [24] focuses explicitly on functionally diverse enzyme superfamilies, linking structure-function relationships at three levels of granularity: superfamilies, subgroups, and families (see Figure 2). At each of these levels, a multiple sequence alignment and a corresponding hidden Markov model are generated, along with interactive protein similarity networks [28•] that provide mapping between sequence similarity clusters and many types of functional properties to facilitate classification and annotation of new sequences (see, for example [29•,30–32]). The SFLD currently contains information on nearly 200 reactions from a number of large, functionally diverse enzyme superfamilies, several of which were reviewed in [4]. An advantage of this approach is that it is structurally contextual, allowing enzymes to be classified according to mechanistic properties directly associated with the structural features it shares with other superfamily, subgroup, or family members. Thus, newly discovered structures and sequences are annotated only at the levels for which strong evidence of membership exists, thereby avoiding misannotation to a family, and thus to an overall reaction specificity, if evidence only exists to link it to the more general mechanistic attributes common to members of a subgroup or superfamily.
Figure 2.
Organization of a subset of subgroups and families of the vicinal oxygen chelate superfamily in the SFLD. The SFLD stores structure-function relationships for functionally diverse enzyme superfamilies at three levels of granularity: superfamilies, subgroups, and families. The structure corresponds to a member of the glyoxalase I family depicting the conserved positioning of the metal binding ligands in many of the members of the superfamily.
The MACiE database [25] focuses on enzymes with known structure and plausible reaction mechanisms published in the literature. It is based on a mostly non-homologous dataset, chosen to represent at least one reaction for each of the EC sub-subclasses. It currently contains nearly 300 fully annotated reactions, each accompanied by a complete stepwise description of catalytic mechanism, including the type of chemistry involved and the functional role of the catalytic amino acid residues participating in each step. New resources focusing exclusively on the role of metal ions [33] and of organic cofactors [34] now complement the mechanistic data presented in MACiE providing a broader view of the chemistry of protein catalysis [35,36,37•,38•]. Because of its design, MACiE does not sample well divergently evolved enzymes, however, it includes several examples of convergently evolved enzymes.
The EzCatDB [26] includes nearly 750 enzyme reactions, each linked with sets of homologous enzyme structures annotated with their catalytic and cofactor binding residues, their cognate ligands, and textual descriptions of their catalytic mechanism. EzCatDB includes examples of both divergently and convergently evolved enzymes, and it has been useful for comparing the catalytic mechanisms of hydrolases and transferases [22] and for comparing active sites and ligand biding residues in functionally diverse superfamilies [39]. Finally, the HCS database [27] classifies active sites based on their catalytic roles and also includes mechanistic information. Currently, the dataset contains over one thousand hydrolases classified into less than one hundred classes, most including a scheme describing their associated catalytic mechanism. The content of the HCS makes it best suited for studying convergently evolved enzymes.
Two additional resources deserve mention in the context of mapping reaction information to structural features in enzymes. The Catalytic Site Atlas (CSA) [40] stores hand-curated information about the catalytic residues of nearly one thousand enzyme structures that by homology can be transferred to nearly 30 thousand structures. The PROCOGNATE database [41], maps cognate ligands to enzyme domains, covering about ten thousand structures from the PDB. Both are suited for the study of divergent and convergent evolution.
Measuring similarity of ligands and mechanisms
Fueled by the increasing availability of enzyme ligand and mechanistic data on a computationally accessible form, several methods have emerged to compare ligands and mechanisms, adding to the toolset required for quantitative analysis of enzyme function.
Ligand similarity
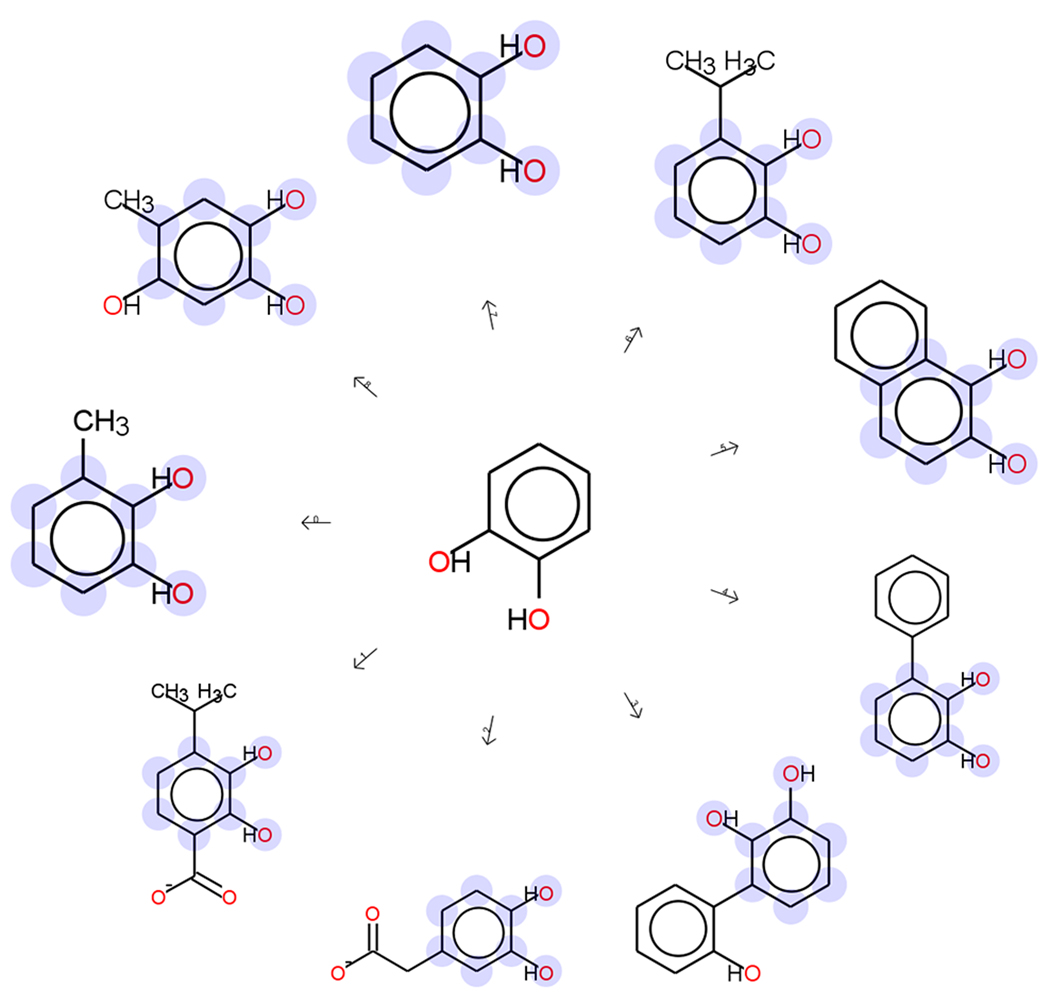
Evaluation of ligand similarity is well developed in cheminformatics and drug design and various methods are available to describe the structures and properties of small molecules and compute their similarities (reviewed in [42]). Of interest here, several reports have clustered cognate ligands of enzymes within and across genomes [17,43–46]. In these works, similarity was usually calculated using Tanimoto coefficients [47] followed by assignment of the molecules to classes using machine learning algorithms, most notably by clustering. For describing the molecules themselves, 2D topological fingerprints and fingerprints containing information from physicochemical properties have been used. Graph-based descriptions have also been used to describe molecules, as these support calculation of the maximum common substructure (MCS) [48] between the ligands compared, providing a visually interpretable output. Among the implementations of the MCS algorithm that are available, we highlight the recent adaptation in the Small Molecule Subgraph Detector (SMDS) toolkit [49••] which incorporates chemical knowledge and was benchmarked on metabolomics data. The SMSD allows MCS searches between sets of molecules, which is valuable for determining common substructures among the ligands of groups of enzymes (Figure 3).
Figure 3.
Common substructure shared among some substrates of enzymes from the extradiol dioxygenase-like subgroup of the vicinal oxygen chelate superfamily curated by the SFLD. The maximum common substructure was computed using the Small Molecule Subgraph Detector (SMDS) toolkit [49]. Highlighting shows the substructures common in all of the substrates.
Mechanistic similarity
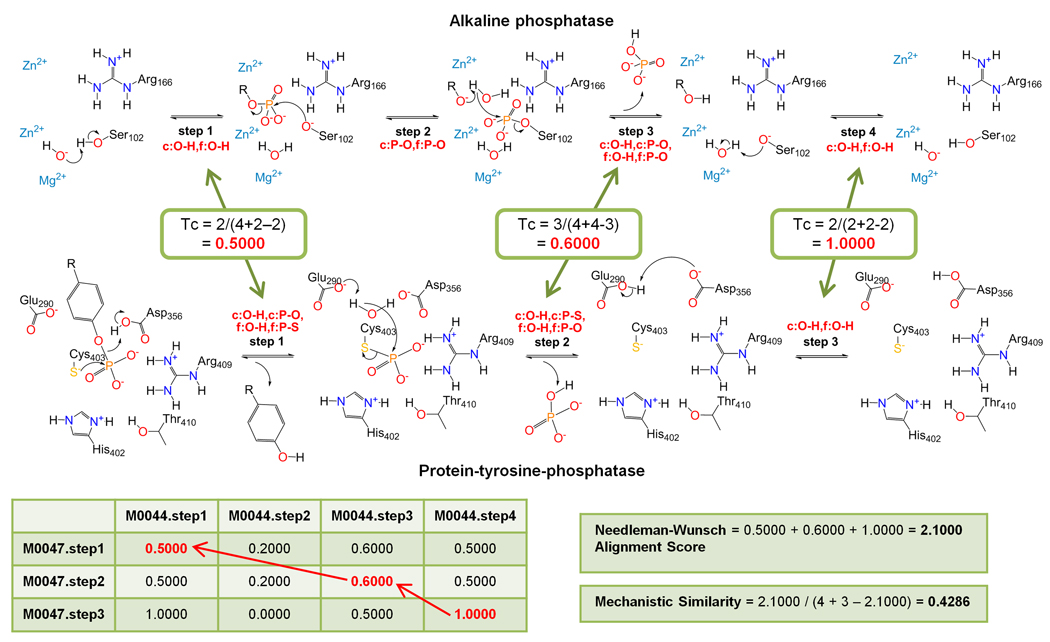
Several methods have been developed to identify similarities among overall reactions of enzymes [50–52,53••], including some that make use of physicochemical descriptors of the ligands involved in the reactions [54,55] as well as of the reacting bonds [56,57], thus indirectly capturing mechanistic information. Interestingly, all these (semi)-automated algorithms, some of which are also quantitative, identified issues with the EC classification and suggested improvements to make it more consistent. Currently, only one algorithm explicitly uses mechanistic information for quantifying similarity. This algorithm [58] encodes the transformation between successive reaction intermediates as either a set of bond changes or as a fingerprint that captures various chemical aspects of a mechanistic step. Similarities among each possible pair of steps between the reactions compared are computed using Tanimoto coefficients (for bond changes) or Euclidean distances (for fingerprints). A global alignment between steps is then generated and a Tanimoto coefficient computed using the similarity data as input. Recently, the algorithm was extended to allow local alignments of mechanistic steps and to handle comparisons of overall reactions [23] (Figure 4). One drawback for these studies has been their dependence on manually annotated bond change information available only from the MACiE database, thus limiting its application. A new algorithm is now available that automatically extracts information about bond changes from the chemical structure of substrates and products of reactions [53] enabling much easier and larger throughput calculations of mechanistic similarities.
Figure 4.
Quantification of mechanistic similarity (adapted from [23]). The convergently evolved reactions catalyzed by alkaline phosphatase (MACiE M0044, EC 3.1.3.1, PDB ID: 1alk), and protein-tyrosine-phosphatase (MACiE M0047, EC 3.1.3.48, PDB ID: 1ytw) are used as examples. Mechanistic step are represented as the set of bond changes occurring in the transformation from substrates to products in that step, with c: bond cleaved, d: bond decreased in order, f: bond formed, and i: bond increased in order. Similarities between the sets of bond changes of the steps of the reactions compared are computed using Tanimoto coefficients (Tc) and stored in a similarity matrix. The maximum-match pathway is then obtained using the Needleman-Wunsch algorithm, and mechanistic similarity is computed as a new Tanimoto coefficient using the number of steps in each reaction and the Needleman-Wunsch alignment score as inputs.
Conclusions
The EC system is the de facto classification scheme for reactions in enzymes. It is based on the overall transformation catalyzed, establishing its worth for linking genes and gene products to reactions. However, because it was created when mechanistic and structural data were sparse and cheminformatics algorithms still in their infancy, classification of reactions using the EC is neither automated nor quantitative. Nor is it linked to sequence and structural information. Recent large-scale studies suggest that both divergently and convergently evolved enzymes share common mechanistic aspects along their reaction pathways, opening an opportunity for new classifications of function based on mechanistic information. In addition, because enzyme reactions are enabled by the structural elements in the enzymes that catalyze them, defining function in terms of catalytic mechanisms and bound ligands seems a more thorough solution. However, we still have a ways to go —while techniques for comparing similarities among ligands are already quite mature, algorithms for computing mechanistic similarity have only recently started to emerge. Still, it seems that the time has come for the widespread utilization of metrics for substrate and mechanistic similarity to complement functional classifications based on overall transformations. We envisage that these methods will help generate structure-function links for all known superfamilies as well as for those convergently evolved enzymes that are mechanistically similar. As information in the enzyme mechanism databases grows and the methods for comparison of mechanistic similarity continue to develop, we can perhaps soon expect to classify the diversity of enzyme functions using these more robust and quantitative measures.
Acknowledgements
This work was supported by NIH GM60595 to PCB. Figure 4 was adapted from reference [23]. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California San Francisco (supported by NIH P41 RR001081).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
• • of outstanding interest
- 1.IUBMB. Enzyme Nomenclature 1992: Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the nomenclature and classification of enzymes. Academic Press; 1992. [Google Scholar]
- 2.Babbitt PC. Definitions of enzyme function for the structural genomics era. Curr Opin Chem Biol. 2003;7:230–237. doi: 10.1016/s1367-5931(03)00028-0. [DOI] [PubMed] [Google Scholar]
- 3.Gerlt JA, Babbitt PC. Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu Rev Biochem. 2001;70:209–246. doi: 10.1146/annurev.biochem.70.1.209. [DOI] [PubMed] [Google Scholar]
- 4.Glasner ME, Gerlt JA, Babbitt PC. Evolution of enzyme superfamilies. Curr Opin Chem Biol. 2006;10:492–497. doi: 10.1016/j.cbpa.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 5. Omelchenko MV, Galperin MY, Wolf YI, Koonin EV. Non-homologous isofunctional enzymes: a systematic analysis of alternative solutions in enzyme evolution. Biol Direct. 2010;5:31. doi: 10.1186/1745-6150-5-31. This is an updated survey of convergently evolved enzymes that identifies 186 groups of non-homologous enzymes that catalyze identical chemical transformations.
- 6.Martin ACR. PDBSprotEC: a Web-accessible database linking PDB chains to EC numbers via SwissProt. Bioinformatics. 2004;20:986–988. doi: 10.1093/bioinformatics/bth048. [DOI] [PubMed] [Google Scholar]
- 7.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 8.Jensen RA. Enzyme recruitment in evolution of new function. Annu Rev Microbiol. 1976;30:409–425. doi: 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- 9.Petsko GA, Kenyon GL, Gerlt JA, Ringe D, Kozarich JW. On the origin of enzymatic species. Trends Biochem Sci. 1993;18:372–376. doi: 10.1016/0968-0004(93)90091-z. [DOI] [PubMed] [Google Scholar]
- 10.Babbitt PC, Gerlt JA. Understanding enzyme superfamilies. Chemistry As the fundamental determinant in the evolution of new catalytic activities. J Biol Chem. 1997;272:30591–30594. doi: 10.1074/jbc.272.49.30591. [DOI] [PubMed] [Google Scholar]
- 11.Teichmann SA, Rison SC, Thornton JM, Riley M, Gough J, Chothia C. The evolution and structural anatomy of the small molecule metabolic pathways in Escherichia coli. J Mol Biol. 2001;311:693–708. doi: 10.1006/jmbi.2001.4912. [DOI] [PubMed] [Google Scholar]
- 12.Bartlett GJ, Borkakoti N, Thornton JM. Catalysing new reactions during evolution: economy of residues and mechanism. J Mol Biol. 2003;331:829–860. doi: 10.1016/s0022-2836(03)00734-4. [DOI] [PubMed] [Google Scholar]
- 13.Nowotny M. Retroviral integrase superfamily: the structural perspective. EMBO Rep. 2009;10:144–151. doi: 10.1038/embor.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mindnich RD, Penning TM. Aldo-keto reductase (AKR) superfamily: genomics and annotation. Hum Genomics. 2009;3:362–370. doi: 10.1186/1479-7364-3-4-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allen KN, Dunaway-Mariano D. Markers of fitness in a successful enzyme superfamily. Curr Opin Struct Biol. 2009;19:658–665. doi: 10.1016/j.sbi.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linsky T, Fast W. Mechanistic similarity and diversity among the guanidine-modifying members of the pentein superfamily. Biochim Biophys Acta. 2010;1804:1943–1953. doi: 10.1016/j.bbapap.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nobeli I, Spriggs RV, George RA, Thornton JM. A ligand-centric analysis of the diversity and evolution of protein-ligand relationships in E.coli. J Mol Biol. 2005;347:415–436. doi: 10.1016/j.jmb.2005.01.061. [DOI] [PubMed] [Google Scholar]
- 18.Chiang RA, Sali A, Babbitt PC. Evolutionarily conserved substrate substructures for automated annotation of enzyme superfamilies. PLoS Comput Biol. 2008;4:e1000142. doi: 10.1371/journal.pcbi.1000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerlt JA. A Protein Structure (or Function ?) Initiative. Structure. 2007;15:1353–1356. doi: 10.1016/j.str.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Gherardini PF, Wass MN, Helmer-Citterich M, Sternberg MJE. Convergent evolution of enzyme active sites is not a rare phenomenon. J Mol Biol. 2007;372:817–845. doi: 10.1016/j.jmb.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 21.Otto TD, Guimarães ACR, Degrave WM, de Miranda AB. AnEnPi: identification and annotation of analogous enzymes. BMC Bioinformatics. 2008;9:544. doi: 10.1186/1471-2105-9-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagano N, Noguchi T, Akiyama Y. Systematic comparison of catalytic mechanisms of hydrolysis and transfer reactions classified in the EzCatDB database. Proteins. 2007;66:147–159. doi: 10.1002/prot.21193. [DOI] [PubMed] [Google Scholar]
- 23. Almonacid DE, Yera ER, Mitchell JBO, Babbitt PC. Quantitative comparison of catalytic mechanisms and overall reactions in convergently evolved enzymes: implications for classification of enzyme function. PLoS Comput Biol. 2010;6:e1000700. doi: 10.1371/journal.pcbi.1000700. This article concludes that functionally analogous enzymes often use similar catalytic mechanisms along their reaction pathway, even in the absence of active site similarities. The article also implements new functionality to an existing algorithm to quantify similarities among overall reactions and mechanisms.
- 24.Pegg SC, Brown SD, Ojha S, Seffernick J, Meng EC, Morris JH, Chang P, Huang CC, Ferrin TE, Babbitt PC. Leveraging enzyme structure-function relationships for functional inference and experimental design: the structure-function linkage database. Biochemistry. 2006;45:2545–2555. doi: 10.1021/bi052101l. [DOI] [PubMed] [Google Scholar]
- 25.Holliday GL, Almonacid DE, Bartlett GJ, O'Boyle NM, Torrance JW, Murray-Rust P, Mitchell JBO, Thornton JM. MACiE (Mechanism, Annotation and Classification in Enzymes): novel tools for searching catalytic mechanisms. Nucleic Acids Res. 2007;35:D515–D520. doi: 10.1093/nar/gkl774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagano N. EzCatDB: the Enzyme Catalytic-mechanism Database. Nucleic Acids Res. 2005;33:D407–D412. doi: 10.1093/nar/gki080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gariev IA, Varfolomeev SD. Hierarchical classification of hydrolases catalytic sites. Bioinformatics. 2006;22:2574–2576. doi: 10.1093/bioinformatics/btl413. [DOI] [PubMed] [Google Scholar]
- 28. Atkinson HJ, Morris JH, Ferrin TE, Babbitt PC. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS One. 2009;4:e4345. doi: 10.1371/journal.pone.0004345. This study overlays sequence similarity networks (SSNs) with orthogonal sources of information for analyzing functional trends in protein superfamilies. It demonstrates that SSNs quantitatively correlate with phylogenetic trees but allow exploration of much larger datasets.
- 29. Seffernick JL, Dodge AG, Sadowsky MJ, Bumpus JA, Wackett LP. Bacterial ammeline metabolism via guanine deaminase. J Bacteriol. 2010;192:1106–1112. doi: 10.1128/JB.01243-09. This study uses the known structure-mechanism relationship for the amidohydrolase superfamily of enzymes to hypothesize that a member of this superfamily is responsible for ammeline deamination. It turned out that the only amidohydrolase member common to all the studied organisms known to have ammeline deaminase activity was the widespread enzyme guanine deaminase. Genomic and biochemical data confirmed that ammeline deaminase is indeed a promiscuous activity of guanine deaminase.
- 30.Cummings JA, Fedorov AA, Xu C, Brown S, Fedorov E, Babbitt PC, Almo SC, Raushel FM. Annotating enzymes of uncertain function: the deacylation of D-amino acids by members of the amidohydrolase superfamily. Biochemistry. 2009;48:6469–6481. doi: 10.1021/bi900661b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiang DF, Xu C, Kumaran D, Brown AC, Sauder JM, Burley SK, Swaminathan S, Raushel FM. Functional annotation of two new carboxypeptidases from the amidohydrolase superfamily of enzymes. Biochemistry. 2009;48:4567–4576. doi: 10.1021/bi900453u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiang DF, Kolb P, Fedorov AA, Meier MM, Fedorov LV, Nguyen TT, Sterner R, Almo SC, Shoichet BK, Raushel FM. Functional annotation and three-dimensional structure of Dr0930 from Deinococcus radiodurans, a close relative of phosphotriesterase in the amidohydrolase superfamily. Biochemistry. 2009;48:2237–2247. doi: 10.1021/bi802274f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal-MACiE: a database of metals involved in biological catalysis. Bioinformatics. 2009;25:2088–2089. doi: 10.1093/bioinformatics/btp256. [DOI] [PubMed] [Google Scholar]
- 34.Fischer JD, Holliday GL, Thornton JM. The CoFactor database: organic cofactors in enzyme catalysis. Bioinformatics. 2010;26:2496–2497. doi: 10.1093/bioinformatics/btq442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holliday GL, Almonacid DE, Mitchell JBO, Thornton JM. The chemistry of protein catalysis. J Mol Biol. 2007;372:1261–1267. doi: 10.1016/j.jmb.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 36.Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal ions in biological catalysis: from enzyme databases to general principles. J Biol Inorg Chem. 2008;13:1205–1218. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- 37. Holliday GL, Mitchell JBO, Thornton JM. Understanding the functional roles of amino acid residues in enzyme catalysis. J Mol Biol. 2009;390:560–577. doi: 10.1016/j.jmb.2009.05.015. This article identifies the dominant residues involved in protein catalysis together with their functional roles. It also correlates classes of EC reactions with catalytic residues (and their functional roles) and with types of reaction mechanisms.
- 38. Fischer JD, Holliday GL, Rahman SA, Thornton JM. The Structures and Physicochemical Properties of Organic Cofactors in Biocatalysis. J Mol Biol. 2010;403:803–824. doi: 10.1016/j.jmb.2010.09.018. This work clusters organic cofactors based on their structures and properties, analyzes the distribution of organic cofactors among EC classes, and compares their functional roles to those of inorganic cofactors and amino acid residues.
- 39.Nagao C, Nagano N, Mizuguchi K. Relationships between functional subclasses and information contained in active-site and ligand-binding residues in diverse superfamilies. Proteins. 2010;78:2369–2384. doi: 10.1002/prot.22750. [DOI] [PubMed] [Google Scholar]
- 40.Porter CT, Bartlett GJ, Thornton JM. The Catalytic Site Atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res. 2004;32:D129–D133. doi: 10.1093/nar/gkh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bashton M, Nobeli I, Thornton JM. PROCOGNATE: a cognate ligand domain mapping for enzymes. Nucleic Acids Res. 2008;36:D618–D622. doi: 10.1093/nar/gkm611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willett P. Similarity methods in chemoinformatics. Annu Rev Inform Sci. 2009;43:1–117. [Google Scholar]
- 43.Nobeli I, Ponstingl H, Krissinel EB, Thornton JM. A structure-based anatomy of the E.coli metabolome. J Mol Biol. 2003;334:697–719. doi: 10.1016/j.jmb.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 44.Koch MA, Schuffenhauer A, Scheck M, Wetzel S, Casaulta M, Odermatt A, Ertl P, Waldmann H. Charting biologically relevant chemical space: a structural classification of natural products (SCONP) Proc Natl Acad Sci U S A. 2005;102:17272–17277. doi: 10.1073/pnas.0503647102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macchiarulo A, Thornton JM, Nobeli I. Mapping human metabolic pathways in the small molecule chemical space. J Chem Inf Model. 2009;49:2272–2289. doi: 10.1021/ci900196u. [DOI] [PubMed] [Google Scholar]
- 46.Adams JC, Keiser MJ, Basuino L, Chambers HF, Lee D, Wiest OG, Babbitt PC. A mapping of drug space from the viewpoint of small molecule metabolism. PLoS Comput Biol. 2009;5:e1000474. doi: 10.1371/journal.pcbi.1000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jaccard P. La distribution de la flore dans la zone alpine. Rev Gen Sci Pures Appl. 1907;18:961–967. [Google Scholar]
- 48.Raymond JW, Willett P. Maximum common subgraph isomorphism algorithms for the matching of chemical structures. J Comput Aided Mol Des. 2002;16:521–533. doi: 10.1023/a:1021271615909. [DOI] [PubMed] [Google Scholar]
- 49. Rahman SA, Bashton M, Holliday GL, Schrader R, Thornton JM. Small Molecule Subgraph Detector (SMSD) toolkit. J Cheminform. 2009;1:12. doi: 10.1186/1758-2946-1-12. This toolkit implements a chemically-aware version of the maximum common substructure (MCS) algorithm. It also allows identification of the MCS between sets of molecules, as opposed to just pairs of molecules.
- 50.Yamanishi Y, Hattori M, Kotera M, Goto S, Kanehisa M. E-zyme: predicting potential EC numbers from the chemical transformation pattern of substrate-product pairs. Bioinformatics. 2009;25:i179–i186. doi: 10.1093/bioinformatics/btp223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saigo H, Hattori M, Kashima H, Tsuda K. Reaction graph kernels predict EC numbers of unknown enzymatic reactions in plant secondary metabolism. BMC Bioinformatics. 2010;11(Suppl 1):S31. doi: 10.1186/1471-2105-11-S1-S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Egelhofer V, Schomburg I, Schomburg D. Automatic assignment of EC numbers. PLoS Comput Biol. 2010;6:e1000661. doi: 10.1371/journal.pcbi.1000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Leber M, Egelhofer V, Schomburg I, Schomburg D. Automatic assignment of reaction operators to enzymatic reactions. Bioinformatics. 2009;25:3135–3142. doi: 10.1093/bioinformatics/btp549. The authors developed an algorithm that given the chemical structure of substrates and products in a reaction automatically extracts information about changes in bonds and valence electrons. The algorithm was applied to overall reactions classified in the EC system identifying several groups of EC sub-subclasses with identical sets of changes.
- 54.Latino DARS, Zhang Q, Aires-de-Sousa J. Genome-scale classification of metabolic reactions and assignment of EC numbers with self-organizing maps. Bioinformatics. 2008;24:2236–2244. doi: 10.1093/bioinformatics/btn405. [DOI] [PubMed] [Google Scholar]
- 55.Latino DARS, Aires-de-Sousa J. Assignment of EC Numbers to Enzymatic Reactions with MOLMAP Reaction Descriptors and Random Forests. J Chem Inf Model. 2009;49:1839–1846. doi: 10.1021/ci900104b. [DOI] [PubMed] [Google Scholar]
- 56.Sacher O, Reitz M, Gasteiger J. Investigations of enzyme-catalyzed reactions based on physicochemical descriptors applied to hydrolases. J Chem Inf Model. 2009;49:1525–1534. doi: 10.1021/ci800277f. [DOI] [PubMed] [Google Scholar]
- 57.Hu X, Yan A, Tan T, Sacher O, Gasteiger J. Similarity perception of reactions catalyzed by oxidoreductases and hydrolases using different classification methods. J Chem Inf Model. 2010;50:1089–1100. doi: 10.1021/ci9004833. [DOI] [PubMed] [Google Scholar]
- 58.O'Boyle NM, Holliday GL, Almonacid DE, Mitchell JBO. Using reaction mechanism to measure enzyme similarity. J Mol Biol. 2007;368:1484–1499. doi: 10.1016/j.jmb.2007.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.George RA, Spriggs RV, Thornton JM, Al-Lazikani B, Swindells MB. SCOPEC: a database of protein catalytic domains. Bioinformatics. 2004;20(Suppl 1):I130–I136. doi: 10.1093/bioinformatics/bth948. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Z, Tang Y. Genome-wide analysis of enzyme structure-function combination across three domains of life. Protein Pept Lett. 2007;14:291–297. doi: 10.2174/092986607780090775. [DOI] [PubMed] [Google Scholar]