Abstract
Many short β-peptides adopt well-defined conformations in organic solvents, but specialized stabilizing elements are required for folding to occur in aqueous solution. Several different strategies have been developed to stabilize the 14-helical secondary structure in water, and here we provide a direct comparison of three such strategies. We have synthesized and characterized β-peptide heptamers in which either a salt bridge between side chains or a covalent link between side chains or a few cyclically constrained residues have been incorporated to promote 14-helicity. The incorporation of a salt bridge does not generate significant 14-helicity in water, according to CD and 2D NMR data. In contrast, incorporation of either a lactam bridge between side chains or cyclic residues results in stable 14-helices in water. The β-peptides featuring trans-2-aminocyclohexane-carboxylic acid (ACHC) residues show the highest 14-helical backbone stability, with hardly any sensitivity to pH or ionic strength. The β-peptides featuring a side chain-to-side chain cyclization show lower 14-helical backbone stability and higher sensitivity to pH and ionic strength, but an increased order between the side chains because of the cyclization.
Keywords: β-peptides, 14-helix, cyclic peptides, foldamers
Introduction
Foldamers are oligomers that can adopt predictable secondary structures.[1] Foldamers that display stable conformations in aqueous solution provide attractive scaffolds for development of biologically active molecules.[2] β-Peptide foldamers have received considerable attention in this respect.[3] These β-amino acid oligomers are typically straightforward to prepare, and they provide access to a variety of ordered backbone conformations. Among the regular β-peptide conformations, the 14-helix has been most intensively studied, in terms of sequence-stability relationships and biological applications.[2,4] The 14-helix is defined by formation of backbone hydrogen bonds i, i-2 C=O--H-N that encompasses a 14-atom ring. 14-Helical β-peptides have been shown to bind to specific proteins[5] and to display antibacterial[6] and antifungal[7] activities. Most β-peptides studied to date have been composed partially or entirely of β3-amino acid residues because protected β3-amino acids are readily available in enantiopure form from the analogous α-amino acids via the Arndt-Eistert process.[4] β-Peptides containing exclusively β3residues generally fold to the 14-helix in organic solvents, but special design features are required to enable 14-helical folding in water.[2,4]
The first clear evidence for 14-helix formation in water was obtained for β-peptides containing residues with a six-membered ring constraint, such as trans-2-aminocyclohexanecarboxylic acid (ACHC).[8] These constrained residues have a much larger 14-helix propensity than do β3-residues. However, it has subsequently been shown that alternative design strategies, based entirely on β3-residues, can generate 14-helicity in water. Both Seebach et al.[9] and Cheng and DeGrado[10] showed that 14-helicity is promoted in aqueous solutions by i, i+3 spacing of β3-residues bearing oppositely charged side chains. The 14-helix has approximately three residues per turn; therefore, this i, i+3 spacing allows intrahelical salt bridge formation. Initial all-β3 designs featured charge-charge interactions at two of the three helical faces of the 14-helix; however, subsequent studies have shown that residues with side chain branching adjacent to the backbone, such as β3-homovaline (β3-hVal), can replace some of the charged side chains.[11] Covalent linkage of two side-chains at β-amino acid positions i and i+3 via a disulfide bridge was shown to lead to the stabilization of the 14-helix in methanol.[12] Covalent linkage of two side-chains at β-amino acid positions i and i+3 via amide bonds, as an alternative to ion pairing, resulted in the promotion of 14-helicity in water.[13]
It would be valuable to know the relative helix-stabilizing efficacies of the different strategies outlined above. However, published examples do not allow the necessary comparisons because of differences in β-amino acid composition and β-peptide length. We therefore decided to prepare and analyze a homologous set of β-peptide heptamers (Figure 1) that would enable us to assess the relative extents of 14-helix promotion among salt bridging, covalent side chain linkage and ACHC incorporation, in a consistent β3-amino acid context. Although it is not yet possible to quantify the population of the 14-helix (or any other β-peptide secondary structure) in a given β-peptide, we find that a combination of two-dimensional nuclear magnetic resonance (2D NMR) and circular dichroism (CD) measurements allows us to draw clear qualitative distinctions among the 14-helix-stabilizing strategies.
Figure 1.
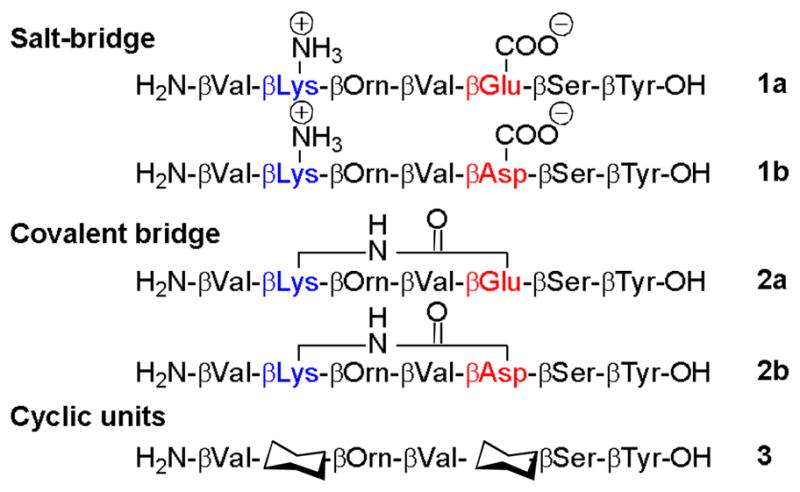
β-Peptides featuring three different strategies for 14-helix stabilization in water; salt bridge formation between side-chains (1a and 1b), covalent bridge formation between side chains (2a and 2b), and cyclic residues (3), with the cyclohexyl symbol representing trans-2-aminocyclohexanecarboxylic acid.
Results and Discussion
Design and synthesis of the β-peptides
A β-peptide composed of seven residues can form two full turns of the 14-helix containing up to five intramolecular backbone hydrogen bonds. We chose positions 2 and 5 to incorporate the different helix-stabilizing elements, and kept the other five positions constant in order to isolate the impact of the variable elements on 14-helicity. At positions 1 and 4 β3-hVal residues were incorporated, because a side chain branch point adjacent to the backbone appears to promote 14-helicity.[11a,14] Position 7 is β3-hTyr; the UV absorbance of the aromatic side chain is useful for HPLC purification and concentration determination. Hydrophilic residues were placed at positions 3 (β3-hOrn) and 6 (β3-hSer) to promote aqueous solubility (Figure 1).
β-Peptides 1a and 1b contain complementary basic and acidic side chains at positions 2 and 5, respectively, which allows formation of a salt bridge in the 14-helical conformation. Placing the basic residue near the N-terminus and the acidic residue near the C-terminus leads to a more favourable interaction of the charged side chains with the 14-helix dipole than would the alternative arrangement.[10] The β3-hOrn was placed next to the β2-hLys in order to generate the most favorable charge-charge interactions in β-peptides 1a and 1b;[10, 15a] placement of the ornithine at position 6 would possibly have resulted in significant interference with the acidic residue at position 5. The acidic residue at position 5 was varied between β3-hAsp and β3-hGlu in order to determine whether the length of the tether between the carboxylate and the backbone influences the contribution of ion pair formation to helix stability. The basic lysine residue at position 2 has been replaced previously by an ornithine, without significant changes in the stability of the helix.[13] β-Peptides 2a and 2b were formed from precursors with the sequences of 1a and 1b, respectively, but in 2a–b the side chains from residues at positions 2 and 5 have been linked covalently via amide formation. In β-peptide 3, pre-organized ACHC residues occupy positions 2 and 5. ACHC has a stronger 14-helix propensity than does any β3-residue.[8,15]
All β-peptides were synthesized according to previously described protocols on solid-phase, taking into consideration the specific differences of the β-amino acids and the on-bead side chain cyclization.[13,16] After purification by preparative RP-HPLC, the expected products, identified by mass spectrometry, were isolated in 7–35% yield, with > 99% purity (see Supporting Information).
Comparisons via 2D NMR spectroscopy
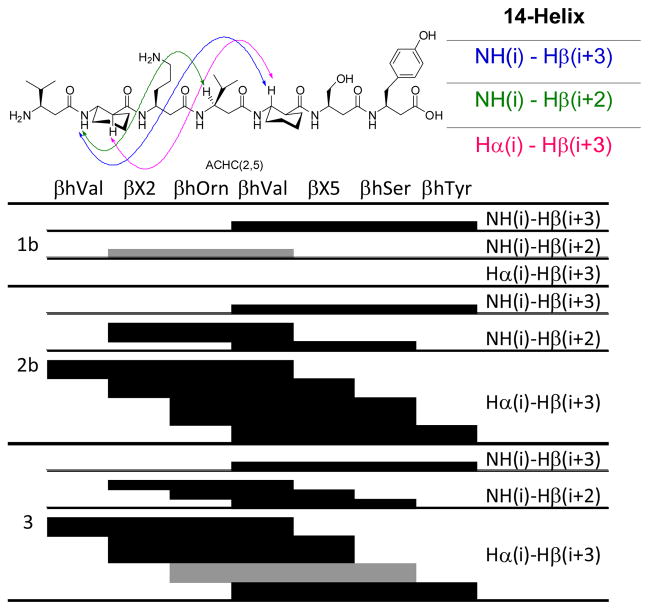
2D NMR provides the strongest spectroscopic evidence for helical folding in solution, via the observation of NOEs between protons from residues that are not sequentially adjacent. Because 2D NMR analysis is labor-intensive, we analyzed only a subset of our β-peptides in this way, 1b, 2b and 3. Each was examined at a concentration of 1–2 mM in a 10 mM phosphate buffered 9:1 H2O/D2O mixture at pH 7.4 and 10°C. TOCSY data (see Supporting Information) were used for a complete assignment of proton chemical shifts. ROESY data (see Supporting Information) were evaluated to identify medium-range NOEs that indicate β-peptide folding. Three NOE patterns involving backbone protons are diagnostic for 14-helix formation: (i) between an amide proton and the proton at the β-position two residues toward the C-terminus [NH(i)-CβH(i+2)], (ii) between an amide proton and the proton at the β-position three residues toward the C-terminus [NH(i)-CβH(i+3)], and (iii) between the axial proton at the α-position and the proton at the β-position three residues toward the C-terminus [CαH(i)-CβH(i+3)] (Figure 2, top).
Figure 2.
(Top) 14-Helix characteristic NOE interactions, illustrated with β-peptide 3. (Bottom) Summary of observed NOE correlations characteristic of a 14-helix (thickness of the bars reflects intensities of the connectivities; gray color expresses ambiguously assigned NOE).
β-Peptide 1b, stabilized via a salt-bridge, showed mainly sequential and intraresidue NOEs; few of the characteristic medium-range NOEs were detected (Figure 2). The resonances for 1b displayed considerable overlap, especially those corresponding to the amide protons of β3-hLys2 and β3-hAsp5 (see Supporting Information). Poor proton resonance dispersion is commonly observed when the extent of folding is low. The NH(β3-hLys2)-CβH(β3-hVal4) NOE might have been present but could not be assigned unambiguously because of resonance overlap.
Side chain-cyclized β-peptide 2b showed a larger dispersion of the amide signals compared to 1b, indicating a higher degree of folding of the β-peptide. Even though 2b displayed some resonance overlap we could nevertheless assign three important medium-range NOEs involving amide protons and all four of the possible CαH(i)-CβH(i+3) NOEs, for residue pairs 1/4, 2/5, 3/6 and 4/7 (Figure 2 and Supporting Information). Among these latter four NOEs, the correlation between β3-hOrn3 and β3-hSer6 was the strongest, which suggests that the bridged residues are particularly well ordered. Many other medium-range NOEs were observed involving side chain protons of 2b. These medium-range side chain to side chain and side chain to backbone NOEs were typically weaker than the inter-backbone medium-range NOEs, but could be unambiguously assigned and provide important information concerning the conformational order of the side chains. Medium-range NOEs involving the side chains were observed for the residue couples 2/5, 3/6 and 4/7 (Table 1). The two protons at the δ-position of β3-hLys2 could be differentiated in the spectra, suggesting a high degree of order in this lactam bridged side-chain. The CδH and CεH protons of β3-hLys2 were found to correlate with the backbone of β3-hAsp5. Comparable side-chain to backbone medium-range NOE correlations were also observed for the β3-hOrn3 - β3-hSer6 couple, which is not connected via a covalent bridge. The β3-hVal4 - β3-hTyr7 couple featured, in addition to a side-chain to backbone medium-range NOE, side-chain to side-chain medium-range NOEs. The aliphatic protons of β3-hVal4 featured medium-range correlations with the aromatic protons of β3-hTyr7. The combination of both the typical backbone medium-range NOE pattern for a 14-helix with the additionally observed medium-range correlations involving the side-chains provides strong evidence for a well ordered 14-helix structure for 2b.
Table 1.
Medium-range side chain NOEs observed among the four different i, i+3 residue pairs of 2b and 3 in water.
| β3-hVal1/β3-hVal4 | β3-hLys2/β3-hAsp5 | β3-hOrn3/β3-hSer6 | β3-hVal4/β3-hTyr7 | |
|---|---|---|---|---|
| 2b | CδH(2)-CβH(5) | CγH(3)-CβH(6) | CδH(4)-CβH(7) | |
| Cδ′H (2)-CβH(5) | Cγ′H(3)-CβH(6) | CδH(4)-CεH(7) | ||
| CεH(2)-CβH(5) | CδH(3)-CβH(6) | CδH(4)-CζH(7) | ||
| CεH(2)-NH(5) | CγH(4)-CεH(7) | |||
| 3 | CδH(1)-CβH(4) | CδH(4)-CβH(7) | ||
| CδH(4)-CεH(7) | ||||
| CγH(4)-CβH(7) | ||||
| CγH(4)-CεH(7) |
The six amide protons of β-peptide 3, featuring the cyclic residues, showed excellent dispersion between 8.1 and 7.6 ppm, which led to well-resolved TOCSY and ROESY data and indicates a high degree of folding,. A large number of NOEs diagnostic for the 14-helix were observed for 3; including four correlations in the amide region (Figure 2). Three of the four possible CαH(i)-CβH(i+3) NOEs were observed; the NOE between CαH(Orn3)-CβH(Ser6) could not be unambiguously assigned due to overlap between the CβH’s of β3-hOrn3 and β3-hSer6. The highest intensity was found for a connectivity at an internal position, CαH(ACHC2)-CβH(ACHC5). Compared to 2b, few medium-range NOEs involving protons at side-chains were observed. The medium-range side chain NOEs observed were limited to the residue pairs 1/4 and 4/7 (Table 1). The absence of NOE’s involving the side-chains of residue couple 2/5 could result from the restrained nature of the cyclic β-amino acid residue at these positions. However, the absence of side-chain to backbone medium-range NOE correlations for the β3-hOrn3 - β3-hSer6 pair is striking as this pair is the same in 2b and 3. The lower number of medium-range NOEs involving side-chains in 3 compared to 2b suggests a more dynamic character of the side-chains of 3, despite the more rigid backbone.
For all of the three β-peptides examined via 2D NMR, no NOE inconsistent with the 14-helix conformation was detected. β-Peptides 2b and 3 featured many NOEs characteristic for the 14-helix, with additional medium-range NOEs involving the side-chains. Overall the NMR results indicate that β-peptides 2b and 3 have well-populated 14-helical conformations in water. β-Peptide 1b, which could form a single intra-helical salt bridge, does not seem to display significant 14-helicity, in contrast for example to long β-peptides that feature two sequential salt-bridges.[17] The use of cyclic residues as biasing elements (3) could preorganize the backbone to promote a 14-helix in which the side chains have a certain degree of freedom. The use of a covalent-bridge appears to have an additional stabilizing effect on the conformations of the side chains, as suggested by the higher number of observed medium-range NOEs involving side chain protons for 2b relative to 3.
Comparisons via circular dichroism spectroscopy
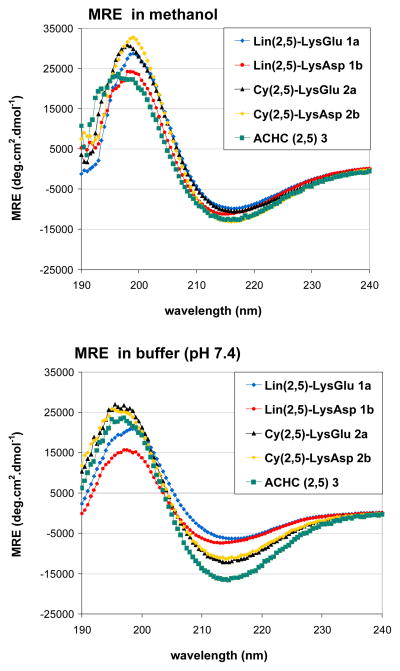
Circular dichroism (CD) measurements were performed in order to compare the extent of 14-helical folding among the five β-peptides discussed here. Figure 3 shows the CD signatures of each β-peptide in methanol and in aqueous 10 mM sodium-phosphate buffer, pH 7.4 (all samples were evaluated at 20 °C). In methanol each heptamer displays the characteristic CD pattern of a 14-helix, with a minimum around 215 nm. The mean residue ellipticity (MRE) at 215 nm can be used as a very approximate basis for comparing the extent of 14-helical folding among different β-peptides.[2]
Figure 3.
CD spectra of β-peptides 1–3 in methanol and phosphate buffer (pH 7.4, 20°C) normalized to Mean Residue Ellipticity (MRE).
The CD spectra of β-peptides 1–3 in methanol display similar MRE values around 215 nm, which suggests that these five β-peptides display similar extents of 14-helicity in this helix-stabilizing solvent. Changing from methanol to aqueous buffer, however, results in significant differences among the β-peptides in terms of MRE at 215 nm, which suggest differences in the favorability of 14-helical folding among these molecules. β-Peptides 1a and 1b feature MRE (215 nm) values of around −7000 deg cm2 dmol−1, with the minimum shifted to higher wavelength for 1a. β-Peptides 2a and 2b feature MRE (215 nm) values of around −11000 deg cm2 dmol−1, which is similar to the MRE values displayed by these β-peptides in methanol. β-Peptide 3 features an MRE (215 nm) value of around −16000 deg cm2 dmol−1; this minimum is the most intense among all of the β-peptides analyzed in aqueous buffer. The CD minimum at 215 nm for 3 in water is modestly but significantly more intense than that of 3 in methanol. Overall, the data agree well with previous observations,[15a] and they suggest that only 2a, 2b and 3 display significant 14-helicity in water.
Analysis of the CD data suggests that β-peptides 1a and 1b, which can form an intra-14-helical salt-bridge, show the least 14-helicity in aqueous solution among the five heptamers we examined. The small MRE (215 nm) in water for a heptameric β-peptide stabilized by one salt-bridge is in line with previous results.[18] The length of the side-chains, i.e. β-peptides 1a vs. 2b, does not result in drastic changes for these two β-peptides, but has a more fine-tuning effect. The more intense minima near 215 nm for β-peptides 2a, 2b and 3 relative to 1a and 1b allow us to conclude that either i, i+3 side chain linkage or the use of preorganized ACHC residues significantly promotes 14-helical folding relative to simple all-β3 sequences that can form a potentially stabilizing internal salt bridge between side chain groups. The length of the lactam bridges between β-peptides 2a and 2b differs by one CH2 group and the effect on helix stability is minimal. Larger changes within this bridging element, however, do result in significant changes in the helix stability.[13] β-Peptide 3 features the most intense CD signal at 215 nm, which might indicate that the ACHC-based strategy is most effective among those examined here for 14-helix stabilization.
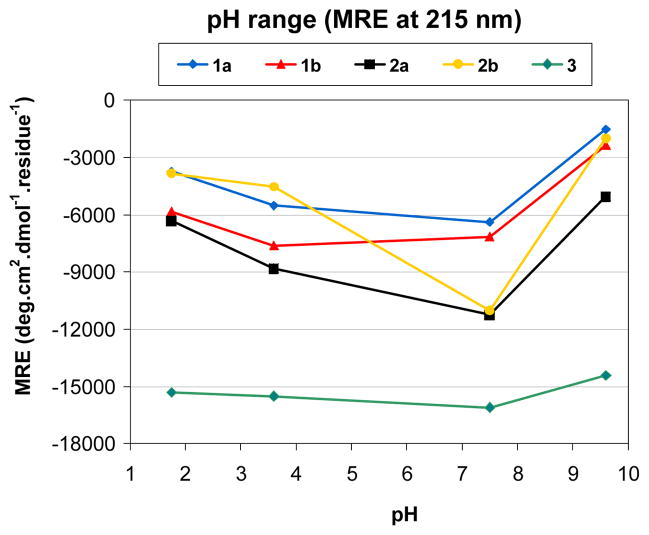
In order to evaluate the stability of the 14-helix of β-peptides 1–3 under different environmental conditions, CD spectroscopy measurements were performed at different pH values (Figure 4). In addition to the data displayed in Figure 3 at pH 7.4, data were acquired at pH 1.8, 3.6, and 9.6.
Figure 4.
Effect of pH on the stability of the 14-helix, expressed in MRE at 215 nm at four different pHs.
Significant pH-dependent differences in 14-helical folding were observed among the five heptamers. β-Peptides 1a and 1b show a substantial decrease in 14-helicity upon increasing the pH from 7.4 to 9.6, suggesting that little or no 14-helix population remains at high pH. Decreasing the pH from 7.4 to 1.8 has a smaller effect on 14-helix population. β-Peptides 2a and 2b feature a relatively larger pH dependence of the 14-helicity than do 1a and 1b, with higher apparent population of the helical conformation at neutral pH for series 2 than for series 1. For both series the destabilizing effect at high and low pH could result from interactions between the helix macrodipole and the charged states at the N- and/or C-termini. Increase of the pH results in deprotonation of the backbone ammonium at the N-terminus and loss of the stabilizing interaction between the positive terminal charge and the helix macrodipole; comparable effects result from protonation of the C-terminal carboxylate at low pH. 14-Helical folding of β-peptide 3, however, shows very little susceptibility to changes in pH. This behavior is surprising since terminal charge-helix macrodipole interactions should be comparable for 3 and for the other β-peptides in this series. This insensitivity to pH suggests that the ACHC residues in 3 are so highly preorganized for the 14-helical conformation that the presence or absence of terminal charge-helix macrodipole interactions has no effect on the extent of helix formation. This interpretation, in turn, raises the possibility that 3 approaches complete population of the 14-helical conformation in aqueous solution, a conclusion that is consistent with the results obtained for other short ACHC-containing β-peptides, featuring similar MRE values.[15a]
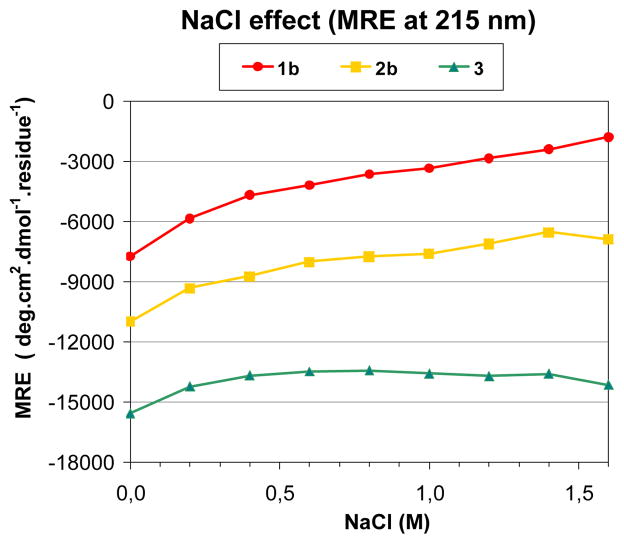
We used CD spectroscopy to examine the effect of changing ionic strength (analyzed by varying NaCl concentration between 0 and 1.6 M) on the extent of 14-helicity in pH 7.4 aqueous buffer (Figure 5). β-Peptide 1b shows a strong and 2b a mild decrease in the CD intensity at 215 nm upon increasing NaCl concentrations, which suggests that salt destabilizes the 14-helical conformation. The effect of NaCl is particularly pronounced for 1b, with a decrease of 77% in the intensity at 215 nm. This large effect presumably arises because the intra-helical salt bridge available to 1b promotes the formation of the small population of 14-helix detected in the absence of NaCl, and the intramolecular electrostatic attraction is screened as ions are added to the solution. β-Peptide 2b apparently retains significant 14-helicity even at 1.6 M NaCl, since the intensity at 215 nm has declined by only 37% relative to aqueous buffer without any salt. In this case the decrease may arise from ionic screening of the charge-macrodipole interactions. The CD spectra of β-peptide 3 show an overall loss of only 9% of the intensity at 215 nm. This trend supports the view that a small number of ACHC residues strongly promote 14-helix formation.
Figure 5.
Effect of [NaCl] on the 14-Helix, expressed in MRE at 215 nm.
Conclusion
We have conducted the first direct comparison of three distinct strategies that have been developed to stabilize the 14-helix secondary structure among β-peptides. Our results suggest that incorporation of a small proportion of ACHC residues (two out of seven, in our system) represents the most effective way to promote 14-helicity in water. In addition, covalent side chain linkage of appropriate length is effective at supporting the 14-helix conformation in aqueous solution, but is more susceptible to changes in the environment. On the other hand, non-covalent side chain linkage, via a single ion pair formation, exerts only a weak helix-promoting effect in aqueous solution. This last observation presumably explains the use of multiple ion pairing interactions that have been necessary to stabilize 14-helical conformations in other systems.[9–11,17] The necessity of multiple ion pairs limits the number of residue positions that can be used to achieve desired functions, such as binding to a target biomolecule. Moreover, the use of ion pairing for 14-helix stabilization creates a high level of dependence on environmental conditions (such as pH and ionic strength). The findings presented here show that incorporation of either a lactam bridge or a small proportion of preorganized ACHC residues results in short, helix-forming β-peptide sequences that offer great flexibility for incorporation of functionally important side chains on a rigid backbone structure.
Supplementary Material
Acknowledgments
This work was supported by a Sofja Kovalevskaja Award of the Alexander von Humboldt Foundation and the BMBF to L.B, by NIH grant GM56414 and the UW-Madison NSEC, and the Spanish Ministerio de Educación y Ciencia (Postdoctoral Fellowship to E.V.). We thank Bernhard Griewel for recording 2D-NMR spectra.
Footnotes
Supporting information for this article is available on the WWW under http://www.chembiochem.org or from the author.
References
- 1.a) Gellman SH. Acc Chem Res. 1998;31:173–180. [Google Scholar]; b) Kirshenbaum K, Zuckermann RN, Dill KA. Curr Opin Struct Biol. 1999;9:530–535. doi: 10.1016/S0959-440X(99)80075-X. [DOI] [PubMed] [Google Scholar]; c) Hill DJ, Mio MJ, Prince RB, Hughes TS, Moore JS. Chem Rev. 2001;102:3892–4011. doi: 10.1021/cr990120t. [DOI] [PubMed] [Google Scholar]
- 2.a) Cheng RP, Gellman SH, DeGrado WF. Chem Rev. 2001;101:3219–3232. doi: 10.1021/cr000045i. [DOI] [PubMed] [Google Scholar]; b) Seebach D, Hook DF, Glattli A. Biopolymers. 2006;84:23–37. doi: 10.1002/bip.20391. [DOI] [PubMed] [Google Scholar]; c) Goodman CM, Choi S, Shandler S, DeGrado WF. Nat Chem Biol. 2007;3:252–262. doi: 10.1038/nchembio876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Werder M, Hauser H, Abele S, Seebach D. Helv Chim Acta. 1999;82:1774–1783. [Google Scholar]; b) Gademann K, Ernst M, Hoyer D, Seebach D. Angew Chem Int Ed. 1999;38:1223–1226. doi: 10.1002/(SICI)1521-3773(19990503)38:9<1223::AID-ANIE1223>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]; c) Porter EA, Wang X, Lee HS, Weisblum B, Gellman SH. Nature. 2000;404:565. doi: 10.1038/35007145. [DOI] [PubMed] [Google Scholar]; d) Rueping M, Mahajan Y, Sauer M, Seebach D. ChemBioChem. 2002;3:257–259. doi: 10.1002/1439-7633(20020301)3:2/3<257::AID-CBIC257>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]; e) Potocky TB, Menon AK, Gellman SH. J Biol Chem. 2003;278:50188–50194. doi: 10.1074/jbc.M308719200. [DOI] [PubMed] [Google Scholar]; f) Gelman MA, Richter S, Cao H, Umezawa N, Gellman SH, Rana TM. Org Lett. 2003;5:3563–3565. doi: 10.1021/ol034977v. [DOI] [PubMed] [Google Scholar]; g) English EP, Chumanov RS, Gellman SH, Compton T. J Biol Chem. 2006;281:2661–2667. doi: 10.1074/jbc.M508485200. [DOI] [PubMed] [Google Scholar]
- 4.a) Seebach D, Overhand M, Kuhnle FNM, Martinoni B, Oberer L, Hommel U, Widmer H. Helv Chim Acta. 1996;79:913–941. [Google Scholar]; b) Seebach D, Beck AK, Bierbaum DJ. Chem Biodiversity. 2004;1:1111–1239. doi: 10.1002/cbdv.200490087. [DOI] [PubMed] [Google Scholar]
- 5.a) Werder M, Hauser H, Abele S, Seebach D. Helv Chim Acta. 1999;82:1774–1783. [Google Scholar]; b) Kritzer JA, Lear JD, Hodsdon ME, Schepartz A. J Am Chem Soc. 2004;126:9468–9469. doi: 10.1021/ja031625a. [DOI] [PubMed] [Google Scholar]; c) Kritzer JA, Luedtke NW, Harker EA, Schepartz A. J Am Chem Soc. 2005;127:14584–14585. doi: 10.1021/ja055050o. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Stephens OM, Kim S, Welch BD, Hodsdon ME, Kay MS, Schepartz A. J Am Chem Soc. 2005;127:13126–13127. doi: 10.1021/ja053444+. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Kritzer JA, Stephens OM, Guarracino DA, Reznik SK, Schepartz A. Bioorg Med Chem. 2005;13:11–16. doi: 10.1016/j.bmc.2004.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Kritzer JA, Hodsdon ME, Schepartz A. J Am Chem Soc. 2005;127:4118–4119. doi: 10.1021/ja042933r. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Murray JK, Farooqi B, Sadowsky JD, Scalf M, Freund WA, Smith LM, Chen J, Gellman SH. J Am Chem Soc. 2005;127:13271–13280. doi: 10.1021/ja052733v. [DOI] [PubMed] [Google Scholar]
- 6.a) Hamuro Y, Schneider JP, DeGrado WF. J Am Chem Soc. 1999;121:12200–12201. [Google Scholar]; b) Raguse TL, Porter EA, Weisblum B, Gellman SH. J Am Chem Soc. 2002;124:12774–12785. doi: 10.1021/ja0270423. [DOI] [PubMed] [Google Scholar]; c) Liu D, DeGrado WF. J Am Chem Soc. 2001;123:7553–7559. doi: 10.1021/ja0107475. [DOI] [PubMed] [Google Scholar]; d) Epand RF, Raguse TL, Gellman SH, Epand RM. Biochemistry. 2004;43:9527–9535. doi: 10.1021/bi049414l. [DOI] [PubMed] [Google Scholar]
- 7.Karlsson AJ, Pomerantz WC, Weisblum B, Gellman SH, Palecek SP. J Am Chem Soc. 2006;128:12630–12631. doi: 10.1021/ja064630y. [DOI] [PubMed] [Google Scholar]
- 8.Appella DH, Barchi JJ, Durell SR, Gellman SH. J Am Chem Soc. 1999;121:2309–2310. [Google Scholar]
- 9.Arvidsson PI, Rueping M, Seebach D. Chem Commun. 2001:649–650. [Google Scholar]
- 10.Cheng RP, DeGrado WF. J Am Chem Soc. 2001;123:5162–5163. doi: 10.1021/ja010438e. [DOI] [PubMed] [Google Scholar]
- 11.a) Hart SA, Bahadoor ABF, Matthews EE, Qiu XYJ, Schepartz A. J Am Chem Soc. 2003;125:4022–4023. doi: 10.1021/ja029868a. [DOI] [PubMed] [Google Scholar]; b) Kritzer JA, Tirado-Rives J, Hart SA, Lear JD, Jorgensen WL, Schepartz A. J Am Chem Soc. 2005;127:167–178. doi: 10.1021/ja0459375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Jacobi A, Seebach D. Helv Chim Acta. 1999;82:1150–1172. [Google Scholar]; b) Rueping M, Jaun B, Seebach D. Chem Commun. 2000:2267–2268. [Google Scholar]
- 13.Vaz E, Brunsveld L. Org Lett. 2006;8:4199–4202. doi: 10.1021/ol061349f. [DOI] [PubMed] [Google Scholar]
- 14.Raguse TL, Lai JR, Gellman SH. Helv Chim Acta. 2002;85:4154–4164. [Google Scholar]
- 15.a) Raguse TL, Lai JR, Gellman SH. J Am Chem Soc. 2003;125:5592–5593. doi: 10.1021/ja0341485. [DOI] [PubMed] [Google Scholar]; b) Lee M, Raguse TL, Schinnerl M, Pomerantz WC, Wang X, Wipf P, Gellman SH. Org Lett. 2007;9:1801–1804. doi: 10.1021/ol070511r. [DOI] [PubMed] [Google Scholar]
- 16.Murray JK, Gellman SH. Org Lett. 2005;7:1517–1520. doi: 10.1021/ol0501727. [DOI] [PubMed] [Google Scholar]
- 17.Guarracino DA, Chiang HR, Banks TN, Lear JD, Hodsdon ME, Schepartz A. Org Lett. 2006;8:807–810. doi: 10.1021/ol0527532. [DOI] [PubMed] [Google Scholar]
- 18.Norgren AS, Arvidsson PI. Org Biomol Chem. 2005;3:1359–1361. doi: 10.1039/b503237g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.