Abstract
Background
Biohydrogen from cyanobacteria has attracted public interest due to its potential as a renewable energy carrier produced from solar energy and water. Anabaena siamensis TISTR 8012, a novel strain isolated from rice paddy field in Thailand, has been identified as a promising cyanobacterial strain for use as a high-yield hydrogen producer attributed to the activities of two enzymes, nitrogenase and bidirectional hydrogenase. One main obstacle for high hydrogen production by A. siamensis is a light-driven hydrogen consumption catalyzed by the uptake hydrogenase. To overcome this and in order to enhance the potential for nitrogenase based hydrogen production, we engineered a hydrogen uptake deficient strain by interrupting hupS encoding the small subunit of the uptake hydrogenase.
Results
An engineered strain lacking a functional uptake hydrogenase (∆hupS) produced about 4-folds more hydrogen than the wild type strain. Moreover, the ∆hupS strain showed long term, sustained hydrogen production under light exposure with 2–3 folds higher nitrogenase activity compared to the wild type. In addition, HupS inactivation had no major effects on cell growth and heterocyst differentiation. Gene expression analysis using RT-PCR indicates that electrons and ATP molecules required for hydrogen production in the ∆hupS strain may be obtained from the electron transport chain associated with the photosynthetic oxidation of water in the vegetative cells. The ∆hupS strain was found to compete well with the wild type up to 50 h in a mixed culture, thereafter the wild type started to grow on the relative expense of the ∆hupS strain.
Conclusions
Inactivation of hupS is an effective strategy for improving biohydrogen production, in rates and specifically in total yield, in nitrogen-fixing cultures of the cyanobacterium Anabaena siamensis TISTR 8012.
Keywords: Anabaena siamensis, Heterocyst differentiation, HupS inactivation, Hydrogen production, Nitrogenase activity, Uptake hydrogenase
Introduction
The N2-fixing cyanobacterium Anabaena siamensis TISTR 8012, a novel strain isolated from rice paddy field in Thailand has been reported to have a high potential for hydrogen production with the ability to utilize sugars as substrate to produce hydrogen [1]. In Anabaena, there may be three enzymes directly involved in hydrogen metabolism. 1) Nitrogenase, a multiprotein enzyme complex consisting of two proteins, dinitrogenase (MoFe protein), encoded by nifD and nifK, and the dinitrogenase reductase (Fe protein), encoded by nifH. This enzyme catalyzes the reduction of atmospheric N2 to ammonia as well as the reduction of proton (H+) to hydrogen [2,3]. In the absence of the substrate N2, nitrogenase may exclusively catalyze hydrogen production. 2) Uptake hydrogenase, a heterodimeric enzyme with at least two subunits, HupS (small subunit) and HupL (large subunit). The large subunit, encoded by hupL, contains the active site, consisting of four conserved cysteine residues that are involved in the coordination of the metallic NiFe at center of the active site. The small subunit, encoded by hupS, contains three FeS clusters which have a function in transferring electrons from active site of HupL to the electron transport chain. The physiological function of the uptake hydrogenase is recycling of hydrogen produced by nitrogenase [2-5]. 3) Bidirectional hydrogenase, a heteropentameric, NAD+-reducing enzyme, encoded by hoxEFUYH. It consists of two protein complexes; hydrogenase (HoxY and HoxH) and a diaphorase unit (HoxE, HoxF and HoxU). The bidirectional hydrogenase is commonly found, though not universal, in both N2-fixing and non-N2-fixing cyanobacteria and catalyzes both consumption and production of molecular hydrogen [2,3,6].
In A. siamensis, an enhanced hydrogen production is mainly achieved through the nitrogenase enzyme [7]. However, the net hydrogen yield is lost due to the activity of the uptake hydrogenase. To overcome this, we engineered a hydrogen uptake deficient strain by interrupting hupS with an antibiotic resistance cassette. Previous studies have reported that N2-fixing cyanobacteria such as Nostoc punctiforme, Anabaena sp. strain PCC 7120, Anabaena variabilis and Nostoc sp. strain PCC 7942 with inactivated uptake hydrogenases show an ability to produce hydrogen at higher rate when compared to their corresponding wild type strains [8-12]. Interestingly, previous reports mainly focused on HupL inactivation since the active site of uptake hydrogenase is located in the large subunit. Therefore, we focused on HupS in A. siamensis TISTR 8012. The structural hupS and hupL genes of A. siamensis have been identified and sequenced [13]. hupS is located upstream of hupL and the predicted gene products for hupS and hupL consist of 320 and 531 amino acids, respectively. Their deduced amino acid sequences show higher than 90% and 80% similarity for HupS and HupL, respectively when compared to other cyanobacteria [13]. RT-PCR analysis revealed that hupS and hupL were co-transcribed with an enhanced transcription when the cells were grown under N2-fixing condition [13]. HupS and HupL of A. siamensis and other cyanobacteria need to go through a maturation process to become a fully functional enzyme [14].
Thus, in the present study we engineered a strain lacking a functional uptake hydrogenase (∆hupS) with the aim to enhance hydrogen production in A. siamensis TISTR 8012. In addition, the nitrogenase activity and transcript levels of genes involved in hydrogen metabolism and photosynthetic pathways in the ∆hupS strain were investigated. As expected, the ∆hupS strain was more efficient in hydrogen production under long term of light exposure than the wild type strain and the production could be prolonged for more than 72 h under light conditions.
Results and discussion
The confirmation on a complete segregation of a ∆hupS strain of Anabaena siamensis
After transformation of recombinant plasmid (pRLhupSNm) into cells of Anabaena siamensis TISTR 8012 (Figure 1), recombinant colonies were selected on BG11 plate containing 25 ug mL-1 of neomycin and transferred to BG11 broth containing antibiotic at the same concentration before analyzing for complete segregation using colony PCRs. To ensure complete segregation of ∆hupS cells, colony PCRs were performed by using a primer pair specific to hupS as shown in Figure 2A. The results show that PCR products obtained from different recombinant colonies after two weeks did not show complete segregation whereas the completely segregated recombinant strains were found after 4 weeks of cultivation (Figure 2B). A completely segregated recombinant strain was selected for further analysis.
Figure 1.
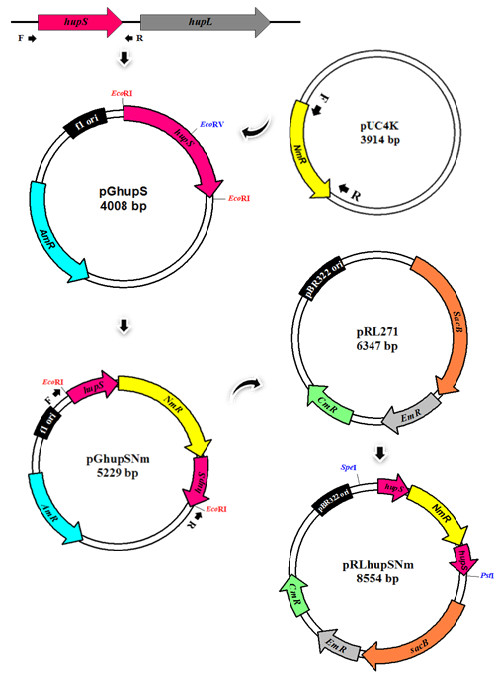
Strategy for the construction of a recombinant plasmid containing an interruption in hupS (∆hupS). Details as described in Materials and methods.
Figure 2.
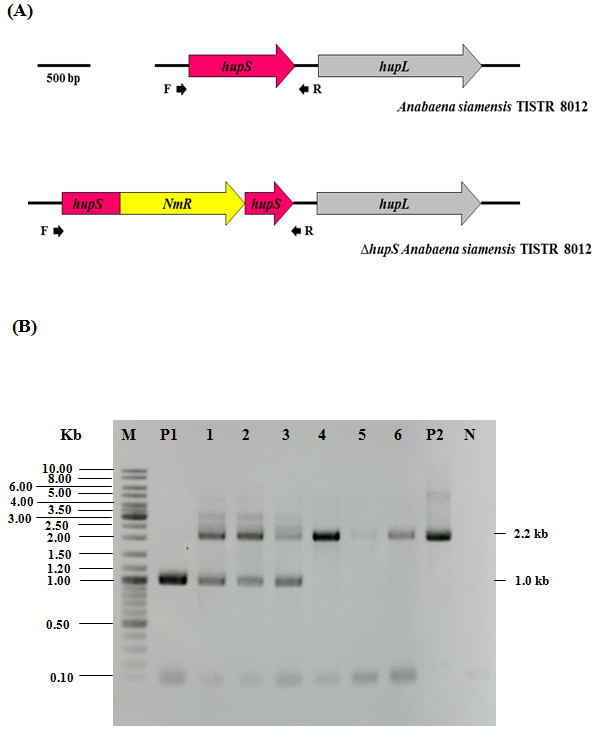
Engineering of a ∆hupS strain of Anabaena siamensis TISTR 8012. (A) Physical map of hupSL in wild type A. siamensis TISTR 8012 and an engineered strain lacking a functional uptake hydrogenase (∆hupS) created by interrupting hupS with neomycin antibiotic resistant cassette. (B) Confirmation of complete segregation of ∆hupS was performed by using colony PCRs and analyzed by 0.8% agarose gel electrophoresis. Primer pairs specific to hupS was used; Lane M: GeneRulerTM DNA ladder (Fermentas), Lane P1: Positive control (PCR product using genomic DNA of wild type as template), Lanes 1–3: PCR products of recombinant colonies cultured in BG11 medium in which the addition of antibiotic for 2 weeks did not show complete segregation, Lanes 4–6: PCR products of recombinant colonies cultured in BG11medium in which the addition of antibiotic for 4 weeks did show complete segregation, Lane P2: Positive control (PCR product using pRLhupSNm plasmid as template), Lane N: Negative control using H2O as template.
Effect of hupS inactivation on hydrogen production, growth rate and heterocyst differentiation
The physiological characterization of the ∆hupS strain of A. siamensis TISTR 8012 was investigated by comparision with the corresponding wild type strain. A. siamensis TISTR 8012 wild type and ∆hupS strain were grown in media with combined N-source (BG11) and without N-source (BG110). Samples were taken to measure the optical density of cell culture every three days of cultivation. The results showed that even though the growth rate of the wild type and ∆hupS strains had a similar pattern in both media the ∆hupS strain grew slightly slower than the wild type strain (Figure 3A, B), suggesting that HupS inactivation had only minor effects on cell growth. This is in agreement with earlier observations using other filamentous cyanobacterial strains [8-12]. Moreover, under N2-fixing condition there was no discernible difference in physiological morphology between the ∆hupS and wild type strains when observed under the Scanning Electron Microscope (SEM) (Figure 3C,D) and the heterocyst frequency in the ∆hupS filaments gradually increased with time in a similar manner to that in the wild type (data not shown).
Figure 3.
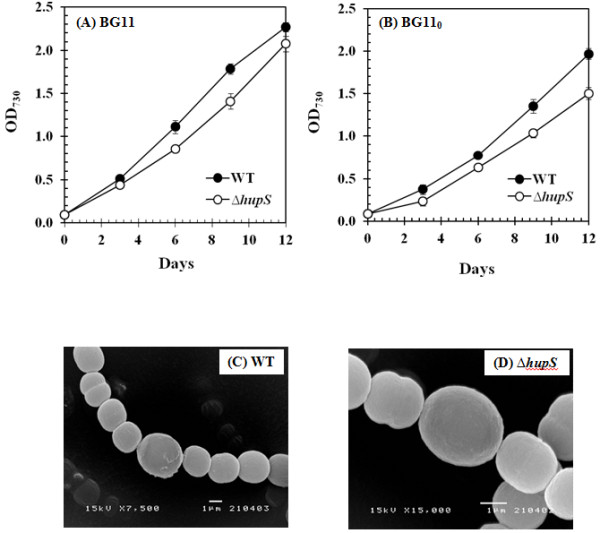
Characterization of wild type and ∆hupS strains of Anabaena siamensis TISTR 8012. Comparison of growth rate between wild type and the ∆hupS strains when cells were grown in either BG11 (A) or BG110 (B) medium, means ± S.D. (n=3). Error bars are included in the graphs, some may be smaller than the symbols used. The morphology of wild type (C) and ∆hupS strain (D) of A. siamensis TISTR 8012 cells observed under Scanning Electron Microscope (SEM) grown without the addition of N-source (BG110 medium).
Furthermore, the transcription levels of key genes involved in heterocyst differentiation, ntcA and hetR, were examined under N2-fixing conditions by RT-PCR. The transcription factor NtcA encoded by ntcA, is a key transcriptional factor required for the activation of many genes involved in nitrogen and carbon metabolism [15,16]. In addition, NtcA is also required for the development and function of mature heterocysts. HetR, a serine-type protease encoded by hetR, is expressed early during heterocyst differentiation and is crucial to the differentiation process [17]. Mutation in the hetR inhibits early steps in the formation of heterocysts while over-expression of hetR gives rise to multiple heterocysts [18]. Both NtcA and HetR are auto-regulated [19,20]. The experiments revealed no discernible differences in either ntcA or hetR transcript levels between ∆hupS and wild type strains after 72 h cultivation (Figure 4A). This suggests that HupS is not essential for heterocyst differentiation.
Figure 4.
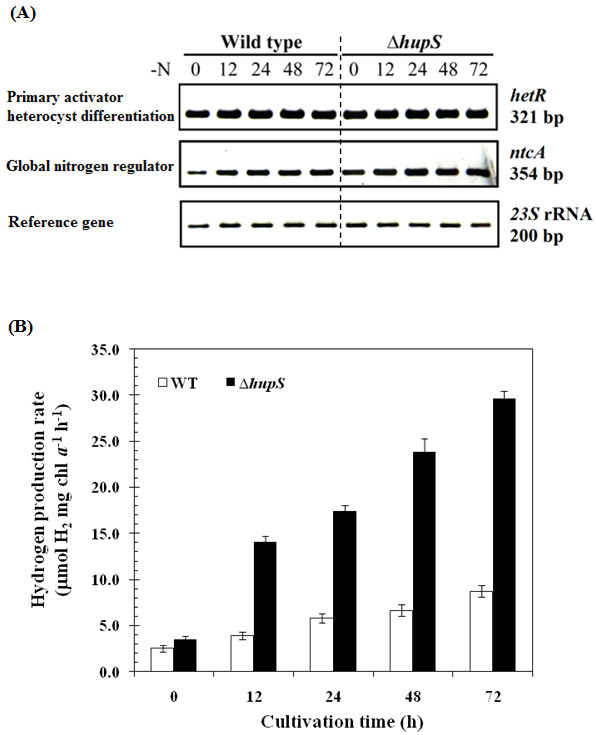
Effect of HupS inactivation on genes transcription and hydrogen production. Comparison of transcription levels of hetR and ntcA genes (A) and hydrogen production (B) between wild type and ∆hupS strains of Anabaena siamensis TISTR 8012 when grown in BG110 medium for various times at 40 μEm-2s-1. Hydrogen production rate was determined under continuous illumination of 40 μEm-2s-1 and anaerobic condition for 12 h. Means ± S.D. (n=3).
When analyzing hydrogen production, the wild type and ∆hupS strains of A. siamensis TISTR 8012 were grown under N2-fixing conditions (BG110 medium) for 12, 24, 48, and 72 h, respectively. Hydrogen production was then determined under continuous illumination of 40 μEm-2s-1 and anaerobic condition for 12 h. Interestingly, the ∆hupS strain produced hydrogen at a significantly higher rate than that of the wild type (Figure 4B). The maximum hydrogen production rate of the ∆hupS strain was 29.7 μmol H2 mg chl a-1h-1 when grown in BG110 medium for 72 h, which is almost 4-folds higher than that observed in the wild type under normal growth condition. These results demonstrate that inactivation of hupS is an effective strategy for improving cyanobacterial photobiological hydrogen production in A. siamensis TISTR 8012. Similar observations have earlier been made in other filamentous cyanobacterial strains [8-12].
Sustained hydrogen production and enhanced nitrogenase activity under long term of light exposure in the ∆hupS strain
We have previously reported that hydrogen production of wild type cells of A. siamensis TISTR 8012 decreased when the cells during the production phase were subject to long duration of high light intensity exposure (200 μE m-2s-1) with the consequence of high uptake hydrogenase activity [1]. Therefore, the ∆hupS strain was grown under N2-fixing condition followed by measuring light dependent hydrogen production for various times up to 96 h. Interestingly, the ∆hupS strain showed much higher hydrogen production under long term light exposure than the wild type strain (Figure 5A). The hydrogen production rate in the ∆hupS strain continued to increase even after exposure to light longer than 12 h and the production could be prolonged up to 96 h under high light conditions. The nitrogenase activity of the ∆hupS strain was also investigated under the same conditions. The ∆hupS strain showed increased nitrogenase activity upon incubation up to 96 h with maximum activity detected after 24 h (Figure 5B). In contrast, the wild type strain had about 2–3 fold lower nitrogenase activity than the ∆hupS strain. A previous study in A. variabilis mutant strain AVM13 in which hupL was interrupted showed no difference of nitrogenase activity when compared to wild type strain. Moreover, the higher hydrogen production by the AVM13 strain under N2-fixing conditions was not sustainable and a dramatically decreased hydrogen production was detected after 30 h incubation [8]. Our results demonstrate that the ∆hupS strain of A. siamensis TISTR 8012 has a high potential for hydrogen production and an ability for long term, sustained hydrogen production under light exposure.
Figure 5.
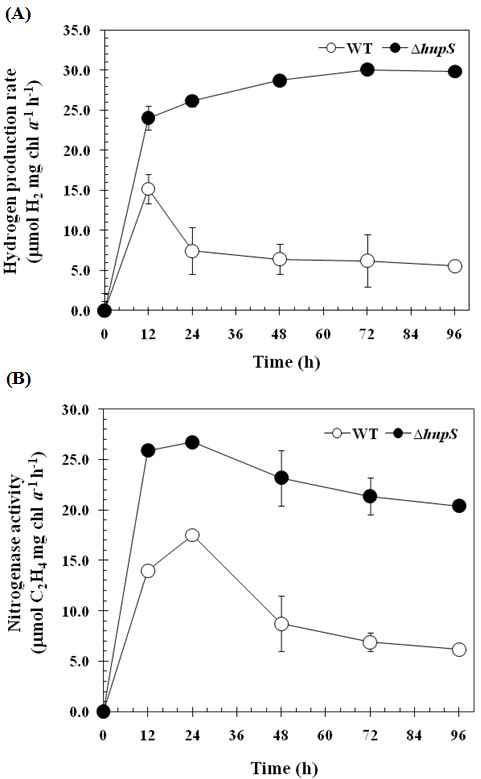
Effect of HupS inactivation on the sustainability of hydrogen production and nitrogenase activity. Comparison of hydrogen production (A) and nitrogenase activity (B) between wild type and ∆hupS strains of Anabaena siamensis TISTR 8012 was done after growth in BG110 medium for 4 days at 40 μEm-2s-1. The collected cells were determined for hydrogen production and nitrogenase activity after incubation for various times under continuous illumination of high light intensity at 200 μE m-2s-1 and anaerobic condition. Means ± S.D. (n=3).
Transcription levels of genes related to hydrogen metabolism and photosynthesis in wild type and ∆hupS
The ∆hupS strain was examined for the relative transcript levels of genes encoding proteins involved in hydrogen metabolism and the photosynthesis pathway in order to provide important information for the regulation of hydrogen metabolism as affected by hupS inactivation. Significantly enhanced nifD but not hoxH transcript levels under N2-fixing conditions were observed in the ∆hupS strain (Figure 6A). This indicates that the increased hydrogen production in the ∆hupS strain may largely be due to higher nitrogenase activity with little or no contribution from the bidirectional hydrogenase. However, since the uptake hydrogenase which is one of the enzymes to supply electrons to the nitrogen fixation process is inactivated, the nitrogenase is likely to receive ATP and reducing equivalents from other pathways. The transcript levels of psbA, encoding the D1 protein of photosystem II, and fdxH, encoding a heterocyst-specific ferredoxin mediating electron transport to the nitrogenase in heterocysts, were significantly up-regulated in the ∆hupS strain (Figure 6B).
Figure 6.
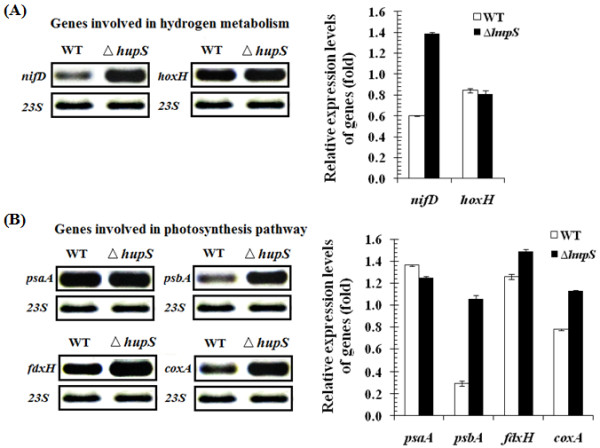
Transcript analyses of genes involved in hydrogen metabolism (A) and photosynthesis pathway (B). RT-PCR using total RNA isolated from wild type and ∆hupS cells grown in BG110 medium without N-source for 12 h. The PCR amplification using cDNAs of respective genes were performed using specific primers. 23S rRNA was used as control when determining the relative level of the respective transcript and the intensities of the PCR generated DNA fragments were determined by using GeneTools program.
This may suggest that electrons and ATP needed for hydrogen production in the ∆hupS strain of A. siamensis TISTR 8012 can be obtained from the electron transport chain associated with the photosynthetic oxidation of water of photosystem II in the vegetative cells. In addition, there was no change observed in the transcription level of the psaA encoding the core protein of photosystem I (Figure 6B). It should be noted that the source of electron transfer to nitrogenase could arise from not only vegetative cells but also from within the heterocysts. Previously, proteins involved in the oxidative pentose phosphate pathway have been reported to be more abundant in heterocysts of a hydrogen uptake deficient strain of Nostoc punctiforme[21].
The inactivation of HupS of A. siamensis TISTR 8012 (∆hupS) resulted in a significant up-regulation of coxA encoding the cytochrome c oxidase subunit I which is present in vegetative cells only [22]. The increase of CoxA activity would lower the level of O2 in vegetative cells resulting in less inhibition of bidirectional hydrogenase, leading to enhanced hydrogen production. Nevertheless, to further explore the effect of HupS inactivation on cell metabolism, the global protein expression level should be investigated.
Growth competition of wild type and ∆hupS strains in a mixed culture
The ability of the ∆hupS strain of A. siamensis TISTR 8012 to compete with the wild type in a mixture without the addition of an antibiotic was studied using a molecular method to analyze the relative abundances of the two strains in a single sample. To demonstrate the possibility to use this method to quantify the relative abundance of the wild type and ∆hupS strains, axenic cultures of wild type and ∆hupS strains were mixed in known proportions before performing colony PCRs using primers specific to hupS. Obtained DNA fragments were analyzed by 0.8% agarose gel electrophoresis, their band intensities were quantified resulting in a standard calibration curve (Figure 7A). The method was then applied to an experiment analyzing the growth of the wild type and ∆hupS strains in a mixed culture for 120 h. After around 50 h in the 50:50 mixed culture, the wild type started to grow on the relative expense of the ∆hupS strain (Figure 7B), in agreement with an earlier observation using a ∆hupL strain of Anabaena/Nostoc PCC 7120 [23]. The mixed culture of wild type and ∆hupS strains reached a ratio of 60:40 after 96 h. It is worth noting that the ∆hupS strain may grow in antibiotic free medium without reverting back to wild type. However, the long term stability of the engineered strain should be further established in long term, large scale hydrogen production facilities.
Figure 7.
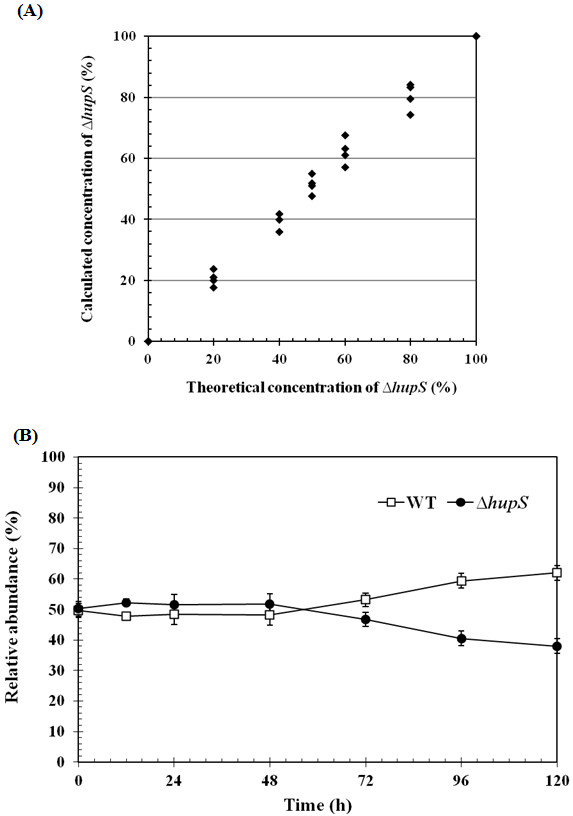
The ability of the ∆hupS strain to compete with the wild type strain analyzed using a molecular method to determine the relative abundances of the strains in mixed cultures. (A) Standard curve, theoretical and calculated % of ∆hupS strain with respect to wild type in mixed cultures. (B) The relative abundance of wild type and ∆hupS strains. The relative abundance at time = 0 was set at 50% (ratio of wild type to ∆hupS strain was 1:1).
Conclusions
This study further explores the potential for solar based biohydrogen production using a purpose designed engineered cyanobacterial strain, ∆hupS, using a general strategy outlined elsewhere [24]. We demonstrate that inactivation of hupS encoding the small subunit of the uptake hydrogenase in the cyanobacterium Anabaena siamensis TISTR 8012 results in long term sustainable, light dependent hydrogen production with enhanced nitrogenase activity and only minor effects on cell growth and heterocyst differentiation when compared with the wild type strain. The ∆hupS strain was found to compete well with the wild type up to 50 h in a mixed culture. However, the effects of HupS inactivation on general and specific metabolism as well as the long term stability of the ∆hupS strain warrant further investigations.
Materials and methods
Strain and growth conditions
The N2-fixing cyanobacterium Anabaena siamensis TISTR 8012 cells were grown in 50 mL of BG110 medium (without N-source) and BG11 medium containing 18 mM NaNO3 as N-source [25], both media were buffered with 20 mM HEPES-NaOH (pH 7.5). For ∆hupS strain, cells were grown in either BG11 or BG110 media containing 25 μg mL-1 neomycin antibiotic. The initial cell concentration was adjusted to an OD730 of 0.1 and cultures were incubated aerobically under continuous illumination of 40 μEm-2s-1 with cool white fluorescent lamps from two sides on a rotatory shaker at 160 rpm and 30°C. The growth rate was monitored by measuring the optical density of the culture at 730 nm with a spectrophotometer. The total amount of chlorophyll a (chl a) was determined spectrophotometrically at 665 nm in 90% (v/v) methanol extracts [26]. For the morphological study, A. siamensis TISTR 8012 cells were observed under Scanning Electron Microscope, SEM (JEOL model JSM-5410LV, Japan).
Construct of recombinant plasmid and conjugative gene transfer
The strategy for construction of recombinant plasmid containing target gene interruption could be divided into three steps as shown in Figure 1. The first step, the hupS gene sequence information in A. siamensis TISTR 8012 was obtained from the NCBI database, accession number AY152844. hupS was amplified from extracted genomic DNA of A. siamensis TISTR 8012 cells by using specific primers, HupSF2 (gcatgcatgactaacgtactctggct) and HupSR2 (gcatgcgtctccattcccattaccta). The obtained hupS PCR product was purified and ligated into the pGEM-T easy vector (Promega), creating pGhupS plasmid. The second step, the MluI fragment containing a neomycin (NmR) resistant cassette gene from pUC4K vector was modified blunt-ending and then inserted into EcoRV site within the hupS gene of the pGhupS plasmid to produce pGhupSNm plasmid. In the last step, a hupSNm fragment from pGhupSNm plasmid was amplified by using specific primers and then cloned into pRL271 vector to produce pRLhupSNm plasmid. pRLhupSNm plasmid functions as a cargo plasmid suitable to be transferred into A. siamensis TISTR 8012 cell. All plasmids were checked and confirmed by sequencing.
The cargo plasmid, pRLhupSNm was transformed into E. coli HB101 carrying the helper plasmid pRL623 and transferred to A. siamensis TISTR 8012 cell with the help of the conjugative plasmid pRL443 as shown in Table 1 by using the triparental mating method [27]. Single recombinant exconjugant colonies were selected on BG11 plate containing neomycin antibiotic at concentration of 25 μg mL-1. To ensure the complete segregation, obtained gene knockout was analyzed by colony PCRs.
Table 1.
Plasmids used in this study
| Plasmids | Relevant characteristic(s) | Source / Reference |
|---|---|---|
| pGEM-T easy |
Cloning vector, Apr lacZ′, mcs |
Promega |
| pRL271 |
Cloning vector carrying sacB, Em and Cm |
GenBank accession #L05081 |
| pUC4K |
Source of Nm cassette |
Amersham |
| pRL632 |
Helper plasmid carrying metylates AvaI, AvaII and AvaIII sites |
[27] |
| pRL443 |
Conjugative plasmid, Km spontaneous mutant of RK2 |
[27] |
| pGhupS |
pGem-T easy vector contained hupS |
This study |
| pGhupSNm |
Nm cassette inserted into EcoRV site within hupS of pGhupS |
This study |
| pRLhupSNm | Cloning vector, Apr lacZ′, mcs | This study |
Hydrogen production determination
The cells were harvested and resuspended in 5 mL medium in a 13 mL of glass vial, then sealed with a rubber septum and a proper screw lid. The vial was bubbled with argon gas for 15 min to eliminate oxygen and incubated under different conditions at 30°C before determining hydrogen production. After 12 h incubation, a 400 μL of head space gas sample was withdrawn from the vial with a gas tight syringe and the hydrogen gas was analyzed by a gas chromatograph (Peri-chrom PR2100, France) with a Molecular Sieve 5A 60/80 mesh column equipped with a thermal conductivity detector and argon as the carrier gas. The hydrogen production rate was expressed as μmol H2 mg chl a-1 h-1.
Nitrogenase activity determination
In vivo nitrogenase activity was measured using the acetylene-reduction assay. In the absence of N2, the enzyme catalyzes the conversion of acetylene (C2H2) to ethylene (C2H4) gas. The reaction was carried out in a glass vial by incubation of the cells suspension (2 mL) with 1 mL of 10% (v/v) acetylene (C2H2) balanced in argon. The ethylene (C2H4) production was detected by using a Gas Chromatograph with a Porapak Q, 50/80 mesh column equipped with a flame ionization detector (Shimadzu, Japan). Enzyme activity was expressed as μmol C2H4 mg chl a-1 h-1.
Transcription analysis
The total RNA was extracted from cells in each condition by using the TRI Reagent® and treated with DNase (Fermentas) for DNA digestion. The treated RNA (1 μg) was converted to single stranded cDNA with the iScriptTM cDNA Synthesis Kit (Bio-RAD), according to the manufacturer’s instruction. RT-PCR amplifications using cDNAs of the respective genes were performed using corresponding primers. Negative controls for the RT-reaction were RT-PCR on DNaseI treated RNA without RT-enzyme. Negative controls for the PCR reactions were PCR amplification without cDNA added and positive controls were performed with genomic DNA using the corresponding primers. All primers used are listed in Table 2. The PCR conditions consisted of 95°C for 3 min, followed by 30 cycles of 95°C for 15 sec, 50°C for 20 sec and 72°C for 20 sec, and then a final extension at 72°C for 3 min. The PCR product was analyzed by 1.0% (w/v) agarose gel electrophoresis.
Table 2.
Primers used in RT-PCR reactions
| Primer | Sequence 5′to 3 | Target of primer pair | PCR product, bp |
|---|---|---|---|
| ASnifDF1 |
tcgtattcggtggtgacaaa |
nifD |
204 |
| ASnifDR1 |
gagacacaccacggaaacct |
|
|
| AShoxHF1 |
gaatccgtctgcgtcaattt |
hoxH |
284 |
| AShoxHR1 |
gcaaatgtccgtcgtaggtt |
|
|
| 23F |
gctaagcgatgtaccgaagc |
23S rDNA |
200 |
| 23R |
taacccagagtggacgaacc |
|
|
| PsaAF1 |
ctgttgaaaggtgtattgtt |
psaA |
489 |
| PsaAR1 |
aggagctaccttcagtttat |
|
|
| PsbAF1 |
gcacattcaactttatgatt |
psbA |
390 |
| PsbAR1 |
ccaaaattgagttattgaag |
|
|
| FdxHF1 |
atggctagctaccaagttag |
fdxH |
299 |
| FdxHR1 |
ttaagcaaggtacggttctt |
|
|
| CoxAF1 |
gcgagattacttcagtttta |
coxA |
426 |
| CoxAR1 |
atccaaataccttctcctac |
|
|
| NtcAF1 |
cgagtctactttcttttgaa |
ntcA |
354 |
| NtcAR1 |
aaaatcacgacagagaatta |
|
|
| HetRF |
ggatgaccggacatttgcac |
hetR |
321 |
| HetRR | ccataagcgatcgcaagagg |
Determination of the relative abundance of wild type and ∆hupS strain of Anabaena siamensis TISTR 8012 in a mixed culture
Axenic cultures of A. siamensis TISTR 8012 wild type and the ∆hupS strain were mixed in known proportions. Colony PCRs were performed using primers specific to hupS and analyzed by 0.8% agarose gel electrophoresis. The sizes of the obtained PCR product were approximately 1.0 kb and 2.2 kb representing hupS of the wild type and hupS interrupted with neomycin resistant cassette gene of the engineered strain, respectively. The intensities of the PCR bands were compared within each lane and calculated by using GeneTools program (SynGene, USA) to detect the presence and the relative abundance of the wild type and ∆hupS strain in a single sample. Using this protocol a standard curve was generated and shown as percent of ∆hupS with respect to wild type strain. For competition experiments, the axenic cultures of wild type and ∆hupS strains of A. siamensis TISTR 8012 were mixed together in a 50 mL growth flask in a ratio of 1:1 based on OD730 measurements and incubated under growth conditions. The relative abundance of the wild type versus the ∆hupS strain was then calculated during the experiment.
Competing interest
The authors declare that they have no competing interests.
Authors' contributions
WK, PL and AI designed the study, analyzed the data and wrote the manuscript. WK performed the experiments. All authors read and approved the final manuscript.
Contributor Information
Wanthanee Khetkorn, Email: p_wanthane@hotmail.com.
Peter Lindblad, Email: Peter.Lindblad@kemi.uu.se.
Aran Incharoensakdi, Email: aran.i@chula.ac.th.
Acknowledgements
This work was supported by the Royal Golden Jubilee Ph.D. program (PHD/0147/2549), the 90th Anniversary of Chulalongkorn University Fund (Ratchadaphiseksomphote Endowment Fund) to W. Khetkorn and A. Incharoensakdi, the Swedish Energy Agency, and the Swedish Research Links program (project 348-2009-6486). The research grants by Commission on Higher Education, Thailand (CHE) (the university staff development consortium), the Thai government SP2 (TKK2555) and the National Research University Project, CHE (FW0659A) to A. Incharoensakdi is also acknowledged. W. Khetkorn also thanks the Graduate School of Chulalongkorn University for providing post-doctoral Fellowship.
References
- Khetkorn W, Lindblad P, Incharoensakdi A. Enhanced biohydrogen production by the N2-fixing cyanobacterium Anabaena siamensis strain TISTR 8012. Int J Hydrogen Energy. 2010;35:12767–12776. doi: 10.1016/j.ijhydene.2010.08.135. [DOI] [Google Scholar]
- Tamagnini P, Leitao E, Oliveira P, Ferreira D, Pinto F, Harris DJ, Heidorn T, Lindblad P. Cyanobacterial hydrogenase: diversity, regulation and applications. FEMS Microbiol Rev. 2007;31:692–720. doi: 10.1111/j.1574-6976.2007.00085.x. [DOI] [PubMed] [Google Scholar]
- Tamagnini P, Axelsson R, Lindberg P, Oxelfelt F, Wunschiers R, Lindblad P. Hydrogenase and hydrogen metabolism of cyanobacteria. Microbiol Mol Rev. 2002;66:1–20. doi: 10.1128/MMBR.66.1.1-20.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houchins JP, Burris RH. Light and dark reactions of the uptake hydrogenase in Anabaena 7120. Plant Physiol. 1981;68:712–716. doi: 10.1104/pp.68.3.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad P, Sellstedt A. Occurrence and localization of an uptake hydrogenase in the filamentous heterocystous cyanobacterium Nostoc PCC 73102. Protoplasma. 1990;159:9–15. doi: 10.1007/BF01326630. [DOI] [Google Scholar]
- Kentemich T, Casper M, Bothe H. The reversible hydrogenase in Anacystis nidulans is a component of the cytoplasmic membrane. Naturwissenschaften. 1991;78:559–560. doi: 10.1007/BF01134448. [DOI] [Google Scholar]
- Khetkorn W, Baebprasert W, Lindblad P, Incharoensakdi A. Redirecting the electron flow towards the nitrogenase and bidirectional Hox-hydrogenase by using specific inhibitors results in enhanced H2 production in the cyanobacterium Anabaena siamensis TISTR 8012. Bioresource Technol. 2012;118:265–271. doi: 10.1016/j.biortech.2012.05.052. [DOI] [PubMed] [Google Scholar]
- Happe T, Schutz K, Bohme H. Transcriptional and mutational analysis of the uptake hydrogenase of the filamentous cyanobacterium Anabaena variabilis ATCC 29413. J Bacteriol. 2000;182:1624–1631. doi: 10.1128/JB.182.6.1624-1631.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg P, Schutz K, Happe T, Lindblad P. A hydrogen-producing, hydrogenase-free mutant strain of Nostoc punctiforme ATCC 29133. Int J Hydrogen Energy. 2002;27:1291–1296. doi: 10.1016/S0360-3199(02)00121-0. [DOI] [Google Scholar]
- Masukawa H, Mochimaru M, Sakurai H. Disruption of the uptake hydrogenase gene, but not of the bi-directional hydrogenase gene, leads to enhanced photobiological hydrogen production by the nitrogen-fixing cyanobacterium Anabaena sp. PCC 7120. Appl Microbiol Biotechnol. 2002;58:618–624. doi: 10.1007/s00253-002-0934-7. [DOI] [PubMed] [Google Scholar]
- Carrasco CD, Holliday SD, Hansel A, Lindblad P, Golden JW. Heterocyst-specific excision of the Anabaena sp. strain PCC 7120 hupL element requires xisC. J Bacteriol. 2005;187:6031–6038. doi: 10.1128/JB.187.17.6031-6038.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino F, Ikeda H, Masukawa H, Sakurai H. High photobiological hydrogen production activity of a Nostoc sp. PCC 7422 uptake hydrogenase-deficient mutant with high nitrogenase activity. Mar Biotechnol. 2007;9:101–112. doi: 10.1007/s10126-006-6035-3. [DOI] [PubMed] [Google Scholar]
- Phunpruch S, Baebprasert W, Thongpeng C, Incharoensakdi A. Nucleotide sequencing and transcriptional analysis of uptake hydrogenase genes in the filamentous N2-fixing cyanobacterium Anabaena siamensis. J Appl Phycol. 2006;18:713–722. doi: 10.1007/s10811-006-9077-z. [DOI] [Google Scholar]
- Bock A, King PW, Blokesch M, Posewitz MC. Maturation of hydrogenases. Adv Microb Physiol. 2006;51:1–71. doi: 10.1016/s0065-2911(06)51001-x. [DOI] [PubMed] [Google Scholar]
- Frias JE, Flores E, Herrero A. Requirement of the regulatory protein NtcA for the expression of nitrogen assimilation and heterocyst development genes in the cyanobacterium Anabaena sp. PCC 7120. Mol Microbiol. 1994;14:823–832. doi: 10.1111/j.1365-2958.1994.tb01318.x. [DOI] [PubMed] [Google Scholar]
- Wei TF, Ramasubramanian TS, Golden JW. Anabaena sp. strain PCC 7120 ntcA gene required for growth on nitrate and heterocyst development. J Bacteriol. 1994;176:4473–4482. doi: 10.1128/jb.176.15.4473-4482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Dong Y, Zhao J. HetR homodimer is a DNA-binding protein required for heterocyst differentiation, and the DNA-binding activity is inhibited by PatS. Proc Natl Acad Sci USA. 2004;2004(101):4848–4853. doi: 10.1073/pnas.0400429101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buikema WJ, Haselkorn R. Expression of the Anabaena hetR gene from a copper-regulated promoter leads to heterocyst differentiation under repressing condition. Proc Natl Acad Sci USA. 2001;98:2729–2734. doi: 10.1073/pnas.051624898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black TA, Cai YP, Wolk CP. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol Microbiol. 1993;9:77–84. doi: 10.1111/j.1365-2958.1993.tb01670.x. [DOI] [PubMed] [Google Scholar]
- Muro-Pastor AM, Valladares A, Flores E, Herrero A. Mutual dependence of the expression of the cell differentiation regulatory protein HetR and the global nitrogen regulator NtcA during heterocyst development. Mol Microbiol. 2002;44:1377–1385. doi: 10.1046/j.1365-2958.2002.02970.x. [DOI] [PubMed] [Google Scholar]
- Ekman M, Ow SY, Holmqvist M, Zhang X, Wangenen JV, Wright PC, Stensjö K. Metabolic adaptations in a H2 producing heterocyst-forming cyanobacterium: potentials and implications for biological engineering. J Proteome Res. 2011;10:1772–1784. doi: 10.1021/pr101055v. [DOI] [PubMed] [Google Scholar]
- Valladares A, Herrero A, Pils D, Schmetterer G, Flores E. Cytochrome c oxidase genes required for nitrogenase activity and diazotrophic growth in Anabaena sp. PCC 7120. Mol Microbiol. 2003;47:1239–1249. doi: 10.1046/j.1365-2958.2003.03372.x. [DOI] [PubMed] [Google Scholar]
- Lindblad P, Christensson K, Lindberg P, Pinto F, Tsygankov A. Photoproduction of H2 by wildtype Anabaena PCC 7120 and a hydrogen uptake deficient mutant: from laboratory experiments to outdoor culture. Int J Hydrogen Energy. 2002;27:1271–1281. doi: 10.1016/S0360-3199(02)00111-8. [DOI] [Google Scholar]
- Lindblad P, Lindberg P, Oliveira P, Stensjö K, Heidorn T. Design, engineering, and construction of photosynthetic microbial cell factories for renewable solar fuel production. Ambio. 2012;41(Suppl 2):163–168. doi: 10.1007/s13280-012-0274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. doi: 10.1099/00221287-111-1-1. [DOI] [Google Scholar]
- MacKinney G. Absorption of light by chlorophyll solutions. J Biol Chem. 1941;140:315–322. [Google Scholar]
- Elhai J, Wolk CP. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 1988;167:747–754. doi: 10.1016/0076-6879(88)67086-8. [DOI] [PubMed] [Google Scholar]


