Abstract
Analysis of bacterial transcriptomes have shown the existence of a genome-wide process of overlapping transcription due to the presence of antisense RNAs, as well as mRNAs that overlapped in their entire length or in some portion of the 5′- and 3′-UTR regions. The biological advantages of such overlapping transcription are unclear but may play important regulatory roles at the level of transcription, RNA stability and translation. In a recent report, the human pathogen Staphylococcus aureus is observed to generate genome-wide overlapping transcription in the same bacterial cells leading to a collection of short RNA fragments generated by the endoribonuclease III, RNase III. This processing appears most prominently in Gram-positive bacteria. The implications of both the use of pervasive overlapping transcription and the processing of these double stranded templates into short RNAs are explored and the consequences discussed.
Keywords: overlapping transcription, RNase III, RNA processing, bacteria, transcriptome
Pervasive Overlapping Transcription in Bacteria
Implementation of high-throughput RNA analysis techniques to the identification of the entire collection of RNA molecules (transcriptome) produced by a bacterial population has directed our view of RNA landscapes away from a protein-centric genome annotation. As with studies involving eukaryotic cells, the first bacterial transcriptomic studies also revealed the existence of a genome-wide process of overlapping transcription.1-15 To be clear overlapping transcription is defined as a process that generates overlapping sense/antisense RNAs from a genomic region. The resulting RNA transcripts show perfect complementarity at least in some portion of the length of the overlapping RNAs. There are at least four different mechanisms to generate overlapping transcription in bacteria (Fig. 1): (1) bona fide antisense RNAs (asRNA), RNA molecules that do not encoded for proteins and show complementarity with part of a gene, a complete gene or a group of genes; (2) 5′ overlapping UTRs between mRNAs of contiguous genes (head-to-head) that are transcribed in divergent directions; (3) 3′ overlapping UTRs between mRNAs of contiguous genes transcribed in convergent directions (tail-to-tail). In this case, the overlapping process can be caused by read-through of transcriptional terminator, the presence of anti-terminator elements or the location of the transcriptional terminators inside the contiguous gene; and (4) overlapping operons, genes that being located in the middle of an operon are transcribed in opposite direction to the rest of the operon.12,16 In this definition of overlapping transcription, we exclude short transcripts that are encoded at genomic locations distant from the RNAs they regulate and sharing only limited complementarities with their targets, because they are not produced from complementary strands of the same DNA region.
Figure 1. Processing of different types of overlapping transcripts by RNase III. Schematic representation of examples of different type of overlapping transcripts in bacteria. These include bona fide antisense RNAs (asRNAs), overlapping 5′ and 3′ untranslated regions (UTRs) of mRNA (mRNA) and overlapping operons. The sense/antisense RNA duplex are processed by RNase III to short RNA fragments (average of 20 nucleotides) that accumulate in similar amounts in both strands of all genome regions where these types of overlapping transcription are taking place.
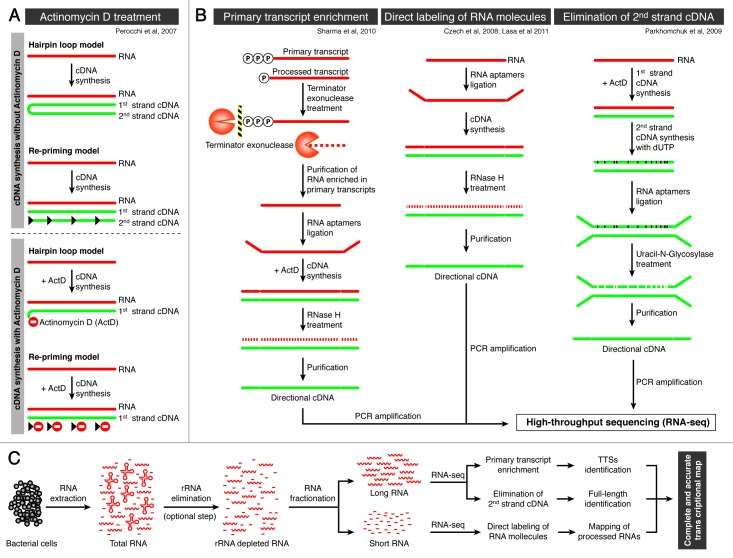
The possibility that well understood technical artifacts had the potential to generate pseudo-antisense transcription data prompted initial criticism.17 While these early uncertainties were partially justified and the signals detected from the antisense strand of highly expressed genes in the earliest studies were in many cases due to the DNA-dependent DNA polymerase activity of the reverse transcriptase,18 these technical issues have been solved in more recent studies. Examples of technical improvements included (Fig. 2): (1) cDNA synthesis performed in the presence of actinomycin D, which specifically inhibits the DNA-dependent DNA polymerase activity of the reverse transcriptase enzyme18; (2) enrichment of RNA samples for primary transcripts by use of terminator exonuclease treatment that degrades 5′P (processed transcript) but not 5′PPP (primary transcript) allowing for the identification of the transcript start and not the extent of the asRNA2; (3) direct labeling of 3′ and 5′ ends of the RNA molecules with adapters before cDNA synthesis, preserving the strand orientation of each RNA molecule12,19; and (iv) synthesis of the cDNA second strand in the presence of dUTP, which allows the selective removal of the strand with UNG (Uracil-N-Glycosylase) after ligation of 5′ and 3′ adaptors.20 The introduction of these technical modifications in the cDNA synthesis and labeling methodologies far from contradicting initial observations have confirmed that overlapping transcription is a very common process. Thus, the percentage of genes that have been associated with at least one antisense transcript in recent studies ranged from 13% in Bacillus subtilis,4 27% in Synechocystis PCC6803,10 30% in Anabaena,11 46% en Helicobacter pylori,2 to 49% in Staphylococcus aureus.12 The first impression of these data are that strong differences in the extent of overlapping transcription between bacteria exist. However, it is uncertain whether these differences reflect real biological differences or a combination of both biological and methodological bias. The answer to this question would require comparative transcriptomic studies using standardized protocols and computational tools.
Figure 2. Flowchart illustrating technical improvements used to preserve the polarity of RNA molecules for transcriptome analysis. (A) Hypothetical model proposed by Perocchi et al.18 showing how spurious second-strand synthesis can occur during reverse transcription and the mode of inhibition by Actinomycin D. During first-strand cDNA synthesis from RNA molecules by reverse transcription, unintended second-strand cDNA synthesis could occur using the first-strand cDNA as a template. (B) Methods to preserve the polarity of RNA molecules during preparation of libraries for RNA sequencing. Primary transcript enrichment: 5′-monophosphate dependent terminator exonuclease (TEX) specifically degrades RNAs with 5′ monophosphates (5′ P), while primary transcripts with a 5′ triphosphate (5′ PPP) or RNA with other termini are protected.2 Direct labeling of RNA molecules: RNA fraction is ligated to a linker in the 3′ end. After removal of non-ligated oligonucleotide, the RNA is ligated to 5′ RNA adaptor by using T4 RNA ligase. For the first strand synthesis of cDNA, an oligonucleotide complementary to the 3′ linker is used.12,19 Removal of second strand cDNA: After the first strand cDNA synthesis non-incorporated nucleotides are removed and dTTP is substituted by dUTP during the synthesis of the second strand. After ligation with a Y-shaped adaptor, the dUTP-containing strand is selectively removed with UNG (Uracil-N-Glycosylase), leaving the first cDNA strand intact.20 (C) Summary flowchart suggesting an experimental design to define a complete and accurate transcriptional map.
What mechanisms regulate the transcription of overlapping transcripts? The origin of the mRNAs participating in overlapping transcription involves promoters recognized by sigma factors. The few studies that have been done on bona-fide antisense transcripts reveal that they are transcribed from similar promoters as their sense counterparts.3,4 Therefore, it is conceivable that the sense and antisense transcripts are regulated via the same mechanisms. However, it is noteworthy that expression of antisense RNAs is for most genes lower than those of the corresponding sense transcript, suggesting that the promoters of antisense transcripts have evolved and modulated their strength to that of the sense transcript or that transcription process of sense and antisense transcripts is coordinated by unknown mechanisms.
Role of RNase III in Antisense Regulation
In a recent study devoted to analyzing the transcriptome of the human pathogen Staphylococcus aureus the total RNA sample was fractionated in long and short (< 50 nucleotides) RNA fractions. RNA sequencing of both fractions revealed a genome-wide process of overlapping sense/antisense RNA processing by the activity of double stranded endoribonuclease, RNase III.12 The end products of the process are a collection of short RNA fragments (20 nucleotides on average) that accumulate in every genome region where overlapping transcription is detected. Given that short RNA fragments originate from the digestion of overlapping transcripts, the total amount of short RNA fragments is similar in both strands and is proportional to the amount of double stranded RNA molecules.
This process of overlapping RNA digestion and production of a collection of short RNA molecules that are symmetrically distributed in both strands of the annotated genes is not exclusive of S. aureus and it also occurs in different Gram-positive bacteria such as Bacillus subtilis, Listeria monocytogenes and Enterococcus faecalis. In contrast, analysis of the transcriptome of the Gram-negative bacteria Salmonella enterica ser. Enteritidis using the same approach could not identify the collection of short RNA fragments. The absence of this process in Salmonella supports that a technical artifact does not generate the collection of short RNAs during the preparation of the RNA libraries. Different reasons can be envisioned to explain why the complement of short RNA fragments is not detected in Salmonella. It is possible that the size of the RNA fragments produced by RNase III enzyme of Salmonella are longer than 50 nucleotides, in which case the RNA fragments would be excluded from the RNA fraction used to prepare the short RNA libraries. Alternatively, overlapping transcripts might be processed by a different mechanism or the resulting short RNA molecules might be unstable in Gram-negative bacteria. Indeed, although fundamental principles govern RNA degradation in bacteria, significant differences have been also identified in the degradosome composition of Gram-positive and Gram-negative bacteria.21-23 Analysis of the short RNA fraction of other Gram-negative bacteria as well as isogenic mutants in different RNases would aid to clarify whether the digestion of overlapping transcripts occurs through different mechanisms in both types of bacteria.
Irrespective of the length of the sense/antisense complementarity region, the formation of the RNA duplexes between overlapping transcripts have been shown to affect the final amount of the protein encoded by the sense RNA in different ways. Examples have been described in which sense-antisense duplex formation results in the sense RNA degradation by RNases such as RNase III and RNase E, an endoribonuclease that cleaves single strand RNA molecules, thus lowering the amount of translatable sense RNA.24,25 Other interactions between overlapping transcripts have been shown to increase the amount of sense RNA coding protein since the duplex formation process protects the sense transcript from degradation or increases the likelihood that sense transcripts will be made at levels exceeding the amount degraded due to the formation of a double stranded substrate of RNase III.26,27 Finally, the overlapping of sense and antisense transcripts can inhibit the binding of the sense transcript to the ribosome and translation process.28
What is the Role of the Genome–Wide Overlapping Transcription Process?
The presence of a stable collection of short RNA fragments derived from the digestion of overlapping transcripts by RNase III demonstrates that both overlapping transcripts are present at the same time in the same cell. The observation of co-expression in the same cell is informative because current transcriptomic studies are performed with RNA purified from at least few millions of bacteria and without the RNase III results it is impossible to determine whether the expression of overlapping transcripts occurs in the same bacteria or it is mutually exclusive.
The question then arises as to what is the role of overlapping transcription and RNase III mediated digestion for bacterial gene regulation? Two possible alternatives may be considered to answer this question: short RNA fragments are residual non-functional products of the overlapping RNA digestion or such fragments are functional molecules with a specific role in gene regulation. With respect to the first possibility, our results support the hypothesis that overlapping transcription provides a simple mechanism to remove all those transcripts that are produced in response to transitory stimuli or escape the regular transcription repression process. For this purpose, the antisense transcript would establish the threshold level that the sense RNA have to reach in order to be translated, removing all the residual RNA molecules whose level are not enough to produce the minimal amount of protein required to be functional. It has been speculated that stochastic variations on transcriptional levels might be beneficial to enhance the phenotypic heterogeneity of the cells within a genetically uniform microbial population.29 However, if the transcription initiation process is more leaky than expected and all mRNAs are indiscriminately translated into protein, then, the cytoplasm will accumulate hundreds of unintended proteins in insufficient amounts to achieve their function. The presence of these proteins would have adverse effects in a particular environmental condition. Alternatively, we cannot exclude the possibility that the RNA transcripts resulting from the RNase III-mediated digestion process could be more stable or translate more efficiently than the primary transcripts.
In the case of 5′ and 3′ overlapping UTRs, the consequences of the digestion could be different for the 5′ divergent overlapping UTRs or for the 3′ convergent overlapping UTRs. Regardless of the specific consequences, digestion of overlapping UTRs would allow coordination of the expression of neighboring genes. This finding is in line with the idea that distribution of bacterial genes within the genomes is not random.30 Thus, to the deeply rooted concept that genes encoding proteins of the same metabolic pathway are clustered together (operons) to facilitate the regulation of their expression, such a second regulatory level coordinating the expression of adjacent transcription units should be considered when investigating bacterial gene regulation. Needless to say that overlapping transcription between adjacent genes also has important consequences when phenotypes associated with insertion or deletion mutants are investigated. Additional thoughts related with the function of the overlapping RNA digestion process are the binding kinetics between overlapping transcripts and the digestion rate of the RNA duplex. Extensive experimental efforts would be necessary to uncover how these factors affect to the overlapping RNA digestion process.
Concerning the possibility that short RNA fragments may fulfill a function by themselves, the average size of the RNA fragments generated by overlapping RNA digestion is 20–22 nucleotides depending on the bacterial species in which they are generated. The size and double stranded structure of the fragments is similar to that of the eukaryotic microRNAs (miRNAs). miRNAs are produced by the successive actions of two RNase III enzymes, Drosha and Dicer, in precursor RNA molecules. Following their processing, one strand of the miRNAs is loaded into a ribonucleoprotein complexes, which key component is the Argonaute (AGO) protein. Then, miRNA-AGO complex interact with their mRNA target by based pairing and direct the inactivation of target RNAs by mRNA degradation or translational arrest and heterochromatin formation.31-34
The existence of a miRNA-based regulatory mechanism in prokaryotic cells was not considered due to the absence of the required machinery to generate the miRNAs and more importantly to the absence of argonaute-like proteins. However, very recently a highly conserved protein (SMc01113/YbeY) sharing structural homology with the MID domain of the Argonaute protein has been described.35 YbeY protein is required for maturation of bacterial 5S, 16S and 23S rRNAs and it seems to facilitate the establishment of interactions between small RNA and the mRNA targets, in a similar way to Hfq protein.35,36 Furthermore, structural and docking analysis suggests that YbeY could contribute catalytically, like an RNase, to RNA cleavage after binding to a guide RNA. Thus, it is tempting to speculate that similarly to what happens in eukaryotes, binding of one strand of the short RNA to the MID domain of YbeY protein can facilitate the mRNA target recognition and subsequently affect the mRNA stability or translation efficiency. YbeY and RNase III are both required for correct maturation of rRNAs and double knockout of both genes encoding these proteins caused a strong defect on bacterial growth.36 This overlap in function and sharing of components between RNA processing and rRNA maturation constitute an additional difficulty for understanding the biological relevance of short RNA molecules because it is puzzling to distinguish whether phenotypes associated to the absence of these enzymes are due to defects on ribosomal maturation or functions related with post-transcriptional gene regulation. A definitive strategy to answer this question and demonstrate the functionality of the short RNAs would require the depletion of the short RNA pool or trans-complementation of the bacterial cell with a collection of short RNA molecules, two challenging approaches that warrants methodological developments.
Perspectives and Unresolved Issues Associated With Sense and Antisense Transcription
The mechanisms through which overlapping transcription can affect sense RNA expression are diverse and are thought to be primarily based on direct interactions between sense/antisense transcripts. It is intriguing to consider what determines the accessibility of the sense/antisense RNA duplex to RNase III, or why all RNA duplexes are not degraded by RNase III-like activities.
If sense and antisense transcripts are being transcribed at the same locus within the same cell it is possible that transcriptional interference may play a role in their regulation.37-39 Several mechanisms have been proposed for transcriptional interference including the collision between both RNA polymerase complexes and removal of the transcription initiation complex by the continuous passing of the elongation RNA polymerase complex in the opposite strand have been proposed. In addition, the fate of both sense and antisense transcripts depend upon several factors including: the availability of complementary sequences to interact given that nascent RNAs are immediately bound and coated with a variety of RNA binding proteins, the affinity with which both RNA molecules will interact depending on the length of complementarity of the molecules, and the local and global folding predictions that may further decide the possible annealing fates of both RNAs.
The first insights into the enzymes involved in regulation of overlapping transcription and the function of this conserved biological process are emerging. Due to their simplicity and feasibility for genetic manipulation, investigations with bacteria can provide clues to understanding of the function of overlapping transcription in eukaryotic cells. However, in order to fulfil this mission at least two methodological difficulties associated with the particularity of overlapping transcription process needs to be solved. One difficulty inherent to the double stranded DNA structure is how to genetically manipulate one of the strands without perturbing the expression of the complementary overlapping strand. The second difficulty is the necessity of evaluating the results of the experiments at single cell level, which implies development of specific reporter tools.
Finally, with reference to many topics it is often said that “size matters.” An important lesson that emerges from these studies is that bacterial short RNA fraction deserves much more attention than has been paid to date and only the combination of long and short RNA fractions together with complementary sequencing strategies, as it is shown in Figure 2C, can provide the complete and accurate landscape of bacterial transcriptomes.
Acknowledgments
A.T.-A. is recipient of “Ramon y Cajal” contracts from the Spanish Ministry of Science and Innovation. This research was supported by grants ERA-NET Pathogenomics (PIM2010EPA-00606), BIO2008-05284-C02, BIO2011-30503-C02 and BFU2011-23222 from Spanish Ministry of Economy and Competitiveness.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/21167
References
- 1.Güell M, van Noort V, Yus E, Chen W-H, Leigh-Bell J, Michalodimitrakis K, et al. Transcriptome complexity in a genome-reduced bacterium. Science. 2009;326:1268–71. doi: 10.1126/science.1176951. [DOI] [PubMed] [Google Scholar]
- 2.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–5. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 3.Wurtzel O, Sapra R, Chen F, Zhu Y, Simmons BA, Sorek R. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010;20:133–41. doi: 10.1101/gr.100396.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicolas P, Mäder U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, et al. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science. 2012;335:1103–6. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- 5.Mendoza-Vargas A, Olvera L, Olvera M, Grande R, Vega-Alvarado L, Taboada B, et al. Genome-wide identification of transcription start sites, promoters and transcription factor binding sites in E. coli. PLoS One. 2009;4:e7526. doi: 10.1371/journal.pone.0007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dornenburg JE, Devita AM, Palumbo MJ, Wade JT. Widespread antisense transcription in Escherichia coli. MBio. 2010;1 doi: 10.1128/mBio.00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filiatrault MJ, Stodghill PV, Bronstein PA, Moll S, Lindeberg M, Grills G, et al. Transcriptome analysis of Pseudomonas syringae identifies new genes, noncoding RNAs, and antisense activity. J Bacteriol. 2010;192:2359–72. doi: 10.1128/JB.01445-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vijayan V, Jain IH, O’Shea EK. A high resolution map of a cyanobacterial transcriptome. Genome Biol. 2011;12:R47. doi: 10.1186/gb-2011-12-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidtke C, Findeiss S, Sharma CM, Kuhfuss J, Hoffmann S, Vogel J, et al. Genome-wide transcriptome analysis of the plant pathogen Xanthomonas identifies sRNAs with putative virulence functions. Nucleic Acids Res. 2012;40:2020–31. doi: 10.1093/nar/gkr904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitschke J, Georg J, Scholz I, Sharma CM, Dienst D, Bantscheff J, et al. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci U S A. 2011;108:2124–9. doi: 10.1073/pnas.1015154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitschke J, Vioque A, Haas F, Hess WR, Muro-Pastor AM. Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. Proc Natl Acad Sci U S A. 2011;108:20130–5. doi: 10.1073/pnas.1112724108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lasa I, Toledo-Arana A, Dobin A, Villanueva M, de los Mozos IR, Vergara-Irigaray M, et al. Genome-wide antisense transcription drives mRNA processing in bacteria. Proc Natl Acad Sci U S A. 2011;108:20172–7. doi: 10.1073/pnas.1113521108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilms I, Overlöper A, Nowrousian M, Sharma CM, Narberhaus F. Deep sequencing uncovers numerous small RNAs on all four replicons of the plant pathogen Agrobacterium tumefaciens. RNA Biol. 2012;9:9. doi: 10.4161/rna.17212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Høvik H, Yu W-H, Olsen I, Chen T. Comprehensive transcriptome analysis of the periodontopathogenic bacterium Porphyromonas gingivalis W83. J Bacteriol. 2012;194:100–14. doi: 10.1128/JB.06385-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georg J, Voss B, Scholz I, Mitschke J, Wilde A, Hess WR. Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol Syst Biol. 2009;5:305. doi: 10.1038/msb.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–6. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 17.Johnson JM, Edwards S, Shoemaker D, Schadt EE. Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 2005;21:93–102. doi: 10.1016/j.tig.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 18.Perocchi F, Xu Z, Clauder-Münster S, Steinmetz LM. Antisense artifacts in transcriptome microarray experiments are resolved by actinomycin D. Nucleic Acids Res. 2007;35:e128. doi: 10.1093/nar/gkm683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, et al. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009;37:e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, et al. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol Rev. 2010;34:883–923. doi: 10.1111/j.1574-6976.2010.00242.x. [DOI] [PubMed] [Google Scholar]
- 22.Carpousis AJ. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu Rev Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- 23.Condon C. Maturation and degradation of RNA in bacteria. Curr Opin Microbiol. 2007;10:271–8. doi: 10.1016/j.mib.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 24.Krinke L, Wulff DL. RNase III-dependent hydrolysis of lambda cII-O gene mRNA mediated by lambda OOP antisense RNA. Genes Dev. 1990;4(12A):2223–33. doi: 10.1101/gad.4.12a.2223. [DOI] [PubMed] [Google Scholar]
- 25.Lee E-J, Groisman EA. An antisense RNA that governs the expression kinetics of a multifunctional virulence gene. Mol Microbiol. 2010;76:1020–33. doi: 10.1111/j.1365-2958.2010.07161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011;43:880–91. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stazic D, Lindell D, Steglich C. Antisense RNA protects mRNA from RNase E degradation by RNA-RNA duplex formation during phage infection. Nucleic Acids Res. 2011;39:4890–9. doi: 10.1093/nar/gkr037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawano M, Aravind L, Storz G. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol Microbiol. 2007;64:738–54. doi: 10.1111/j.1365-2958.2007.05688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dubnau D, Losick R. Bistability in bacteria. Mol Microbiol. 2006;61:564–72. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- 30.Lawrence JG. Gene organization: selection, selfishness, and serendipity. Annu Rev Microbiol. 2003;57:419–40. doi: 10.1146/annurev.micro.57.030502.090816. [DOI] [PubMed] [Google Scholar]
- 31.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 32.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–20. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cernilogar FM, Onorati MC, Kothe GO, Burroughs AM, Parsi KM, Breiling A, et al. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature. 2011;480:391–5. doi: 10.1038/nature10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakama M, Kawakami K, Kajitani T, Urano T, Murakami Y. DNA-RNA hybrid formation mediates RNAi-directed heterochromatin formation. Genes Cells. 2012;17:218–33. doi: 10.1111/j.1365-2443.2012.01583.x. [DOI] [PubMed] [Google Scholar]
- 35.Pandey SP, Minesinger BK, Kumar J, Walker GC. A highly conserved protein of unknown function in Sinorhizobium meliloti affects sRNA regulation similar to Hfq. Nucleic Acids Res. 2011;39:4691–708. doi: 10.1093/nar/gkr060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davies BW, Köhrer C, Jacob AI, Simmons LA, Zhu J, Aleman LM, et al. Role of Escherichia coli YbeY, a highly conserved protein, in rRNA processing. Mol Microbiol. 2010;78:506–18. doi: 10.1111/j.1365-2958.2010.07351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shearwin KE, Callen BP, Egan JB. Transcriptional interference--a crash course. Trends Genet. 2005;21:339–45. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomason MK, Storz G. Bacterial antisense RNAs: how many are there, and what are they doing? Annu Rev Genet. 2010;44:167–88. doi: 10.1146/annurev-genet-102209-163523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Georg J, Hess WR. cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev. 2011;75:286–300. doi: 10.1128/MMBR.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]