Abstract
In the nematode Caenorhabditis elegans (C. elegans), gene inactivation by RNA interference can achieve remarkable potency due to the amplification of initial silencing triggers by RNA-dependent RNA polymerases (RdRPs). RdRPs catalyze the biogenesis of an abundant species of secondary small interfering RNAs (siRNAs) using the target mRNA as template. The interaction between primary siRNAs derived from the exogenous double-stranded RNA (dsRNA) trigger and the target mRNA is required for the recruitment of RdRPs. Other genetic requirements for RdRP activities have not been characterized. Recent studies have identified the RDE-10/RDE-11 complex which interacts with the primary siRNA bound target mRNA and acts upstream of the RdRPs. rde-10 and rde-11 mutants show an RNAi defective phenotype because the biogenesis of secondary siRNAs is completely abolished. In addition, the RDE-10/RDE-11 complex plays a similar role in the endogenous RNAi pathway for the biogenesis of a subset of siRNAs targeting recently acquired, duplicated genes.
Keywords: C. elegans, rde-10, rde-11, RNAi, small interfering RNAs
Introduction
The RNAi pathway plays important roles in gene regulation, chromatin modification, and defense against parasitic genes including transposons and viruses. One of the most striking features of RNAi in Caenorhabditis elegans (C. elegans) is its ability to amplify the initial response by the activities of RNA-dependent RNA polymerases (RdRPs) (e.g., RRF-1 and EGO-1) which synthesize secondary small interfering RNAs (siRNAs).1-6 The initial RNAi response is elicited by exogenous long double-stranded RNAs (dsRNAs) which are processed by the endonuclease Dicer and the Argonaute protein RDE-1 into ~23 nt primary siRNAs.3,7-10 Primary siRNAs are incorporated into the RNA-induced silencing complex (RISC) through their association with RDE-1 to then downregulate target mRNA levels. Primary RISC and target mRNA interaction leads to the recruitment of RdRPs and de novo synthesis of secondary siRNAs using the target mRNA as a template.4-6 Secondary siRNAs are much more abundant than primary siRNAs and associate with a family of worm-specific Argonautes (WAGOs) to repress target gene expression at both transcriptional and post-transcriptional levels.3,11,12 The mechanisms of both the primary and secondary steps of RNAi are not well understood.
Endogenous siRNAs are predominantly 22 nt long with a guanine at the 5′ end in C. elegans.11,13 These 22G siRNAs are derived from at least half of all C. elegans genes and classified into either WAGO class or CSR-1 class depending on their biogenesis, function and interacting Argonautes. Endogenous 22G siRNAs are RdRP products, but unlike secondary siRNAs in the exogenous RNAi pathway, the biogenesis of the vast majority of endogenous 22G siRNAs does not seem to depend on a cognate primary siRNA. However, a small fraction of WAGO class 22G siRNAs is secondary to a family of primary 26G siRNAs whose biogenesis requires Dicer, the exonuclease ERI-1 and the RdRP RRF-3.14-18 Primary 26G siRNAs interact with Argonaute proteins ERGO-1 or ALG-3/4. The ERGO-1 class 26G siRNAs are expressed in oocytes and embryos, whereas downstream 22G siRNAs produced from 26G siRNA targets are enriched in somatic tissues.15,17,19 These siRNAs regulate recently acquired, duplicated genes.17,19 The ALG-3/4 class 26G and downstream 22G siRNAs are expressed in sperm cells and regulate spermatogenesis.15,18 It is not clear how the primary and secondary steps are connected in these endogenous RNAi pathways.
A novel RDE-10/RDE-11 complex that bridges the primary and secondary steps of both exogenous and endogenous RNAi pathways was discovered from genetic screens and proteomic analyses of the gene products identified in those screens (Fig. 1).10,20 The RDE-10 and RDE-11 proteins are only conserved within nematodes. RDE-10 lacks known functional domains, whereas RDE-11 has a RING-type zinc-finger domain, a protein interaction domain that is also found in E3 ubiquitin-protein ligases. Several other RNAi pathway components, including RSD-2, the TUDOR domain protein RSD-6, the half-molecule ATP-binding cassette (ABC) transporter protein HAF-6, function together and/or share similar roles with RDE-10 and RDE-11 in the amplification of RNAi response.10,21,22
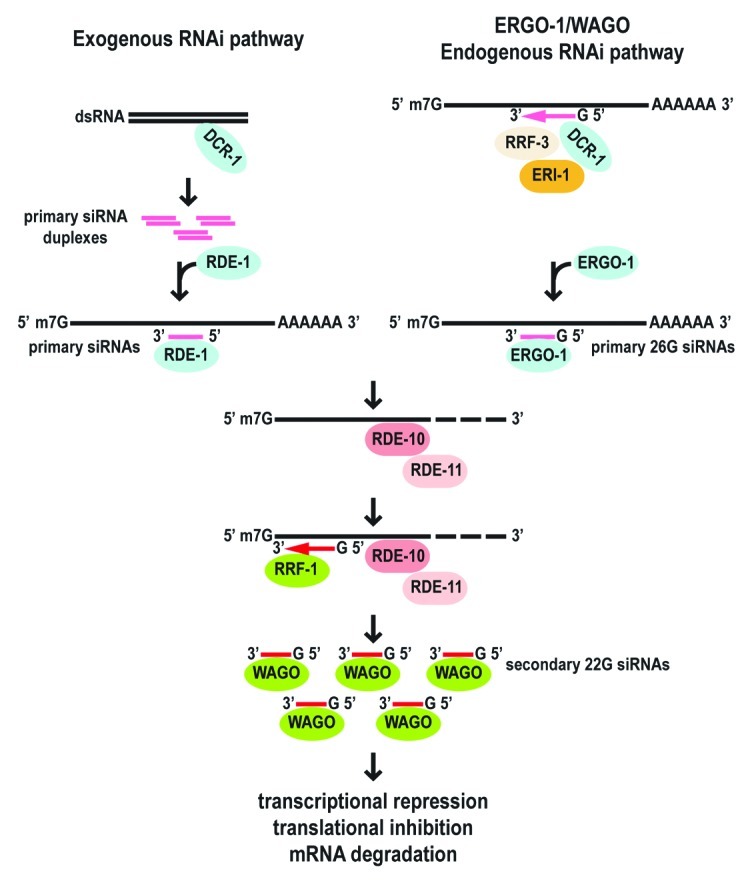
Figure 1. A model of the roles of the RDE-10/RDE-11 complex in both exogenous and endogenous RNAi pathways.
The Dosage-Sensitive RNAi-Defective Genes rde-10 and rde-11 are Required for Secondary siRNA Accumulation in the Exogenous RNAi Pathway
Mutations in the rde-10 and rde-11 genes cause a dosage-sensitive RNAi-defective phenotype: these mutants are resistant to low concentrations of exogenous dsRNAs, but are sensitive to high concentrations of dsRNAs.10 Mutations in several other RNAi pathway genes, including rsd-2, rsd-3, rsd-6 and haf-6, share the same phenotype.21,22 In contrast, rde-1 mutants are resistant to high concentrations of dsRNAs. One attractive model is that these dosage-sensitive RNAi-defective genes are dispensable for the biogenesis and activity of primary siRNAs, but essential for the amplification of RNAi response mediated by secondary siRNAs. In the presence of high concentrations of exogenous dsRNAs, enough primary siRNAs are generated to mount an RNAi response; therefore the function of these dosage-sensitive RNAi-defective genes is not required. However, if the initial trigger dsRNAs are introduced at low concentrations, then secondary siRNA accumulation is essential for a robust RNAi response. rde-1 mutants fail to generate both primary and secondary siRNAs and do not respond to exogenous dsRNAs even at very high concentrations.
This model was validated by comparing the levels of primary and secondary siRNAs between wild-type, rde-10 and rde-11 mutant animals exposed to exogenous dsRNAs by high-throughput sequencing.10,20 Primary siRNAs and their passenger strands bear 5′ monophosphates that are readily suitable for 5′ adaptor ligation for small RNA cDNA library generation.4,5 Secondary siRNAs are more abundant than primary siRNAs. They contain 5′ triphosphates which need to be trimmed to monophosphates by tobacco acid pyrophosphatase (TAP) prior to 5′ adaptor ligation.4,5 In non-TAP treated samples which only capture primary siRNAs and their passenger strands, the levels of total siRNAs derived from exogenous dsRNAs were similar among wild-type, rde-10 and rde-11 mutant animals. However, in TAP-treated samples which contain both primary and secondary siRNAs, siRNAs homologous to trigger dsRNAs were greatly reduced in rde-10 and rde-11 mutants compared with wild-type animals. These data indicate that rde-10, rde-11, and presumably other dosage-sensitive RNAi-defective genes are essential for the efficacy of exogenous RNAi by promoting secondary siRNA amplification.
The RDE-10 and RDE-11 Proteins Form a Complex in vivo that Interacts With Partially Degraded Target mRNAs
Consistent with rde-10 and rde-11 functioning at the same step of exogenous RNAi, their protein products form a tight complex as revealed by immunoprecipitation of epitope-tagged RDE-10 and tandem mass spectrometry analysis.10,20 The interaction between RDE-10 and RDE-11 proteins is direct as shown by immunoprecipitation using in vitro translated proteins.20 In addition, the RING-type zinc-finger domain of RDE-11 is required for its interaction with RDE-10 and proper function.20 Several previously identified small RNA pathway components co-purify with RDE-10, including RSD-2, a dosage-sensitive RNAi-defective gene product, and ERGO-1, a primary Argonaute associated with endogenous 26G siRNAs. Although ERGO-1 and RSD-2 were not at the top of the list of abundances in the purifications, their identifications by MS/MS occurred with good confidence.10 Based on their relatively low expressed sequence tag (EST) numbers in Wormbase, they are unlikely to be abundant contaminating proteins in the proteomic analysis. Further, some of the genes identified by the RDE-10 proteomic analysis are conserved and essential for viability and fertility, precluding their isolation from forward genetic screens. For proteins encoded by these genes, their physical interaction with RDE-10 can serve as an entry point to understand their roles in the RNAi pathway. Since the RDE-10/RDE-11 complex is dispensable for the biogenesis and activity of primary siRNAs, it presumably functions downstream of the DCR/ERI complex and primary RISC. The association of RDE-10 with ERGO-1, a primary Argonaute in one of the endogenous RNAi pathways, indicates that the RDE-10/RDE-11 complex is likely to function upstream of the RdRPs for secondary siRNA biogenesis. The primary Argonaute in the exogenous RNAi pathway, RDE-1, was not identified in the RDE-10 proteomics, possibly because affinity purifications were performed on animals without exposure to exogenous dsRNAs that engage RDE-1.
RDE-10 preferentially interacts with target mRNA during an RNAi response as shown by RNA co-immunoprecipitation assays followed by qRT-PCR.20 Although the levels of target mRNAs are significantly reduced due to degradation in wild-type animals treated with dsRNAs, their association with RDE-10 is greatly enriched. Consistent with the model that the RDE-10/RDE-11 complex functions downstream of RDE-1 but upstream of the RdRP RRF-1, this enrichment depends on rde-1, but not rrf-1. The recruitment of RDE-10 to target mRNA does not require RDE-11, however, the RDE-11 function is essential for the subsequent target mRNA degradation. The RDE-10/RDE-11 complex bound target mRNA degradation is initiated by 3′ deadenylation while the 5′ cap remains intact. A detectable downregulation of total target mRNA levels requires RRF-1 activity. RRF-1 was not identified in the RDE-10 proteomics, possibly because the interaction is transient. Alternatively, RRF-1 interacts directly with RDE-10/RDE-11 complex modified target mRNAs. Since the primary Argonaute RDE-1 is only known to degrade the passenger strand of primary siRNA duplex, while the secondary WAGO Argonautes do not possess the “slicer” endonuclease activity, the RNAi pathway likely employs other cellular machinery to degrade target mRNAs.3,11,23
The RDE-10/RDE-11 Complex Regulates the Biogenesis of a Subset of Endogenous siRNAs
A comprehensive analysis of the roles of rde-10 and rde-11 in endogenous small RNA pathways was performed by high-throughput sequencing of small RNA cDNA libraries derived from wild type, rde-10 and rde-11 mutant worms at different developmental stages.10 Small RNA samples were treated with TAP prior to 5′ adaptor ligation, therefore these libraries contain all known classes of small RNAs with either a 5′ monophosphate (e.g., miRNAs, piRNAs and 26G siRNAs) or a 5′ triphosphate (e.g., 22G siRNAs). The analysis was focused on small RNA features derived from coding genes, pseudogenes, transposons and non-annotated genomic regions that yielded ≥ 10 siRNA reads per million total small RNA reads (RPM) in wild type and were reduced by ≥ 67% in rde-10 and rde-11 mutants. Mutations in rde-10 and rde-11 do not cause major changes in any of the known classes of small RNAs, aside from a few dozen clusters of WAGO class 22G siRNAs that are greatly reduced. A significant fraction of these WAGO class 22G siRNAs are secondary to the ERGO-1 class 26G siRNAs and target duplicated genes.17,19 Similar to rde-10 and rde-11, the dosage-sensitive RNAi-defective genes rsd-2, rsd-6 and haf-6 also regulate the levels of 22G siRNAs targeting duplicated genes as shown by high-throughput sequencing and qRT-PCR. These results indicate that similar to their roles in the exogenous RNAi pathway, rde-10 and other dosage-sensitive RNAi-defective genes potentiate an RNAi response by promoting secondary siRNA accumulation in the endogenous RNAi pathway.
The Dosage-Sensitive RNAi-Defective Genes are Unlikely to Regulate the Spreading of the Silencing Signals
In C. elegans, the gene silencing activity of the exogenous RNAi trigger is able to spread throughout the organism beyond the initial site of delivery.24 The sid-1 gene encodes a conserved transmembrane protein which serves as a channel for the transport of dsRNAs.25-27 The rsd-2, rsd-6, and haf-6 genes have been speculated to regulate the intercellular and intracellular trafficking of RNA silencing signals as well based on the following observations: 1) these mutants are resistant to ingested dsRNAs targeting germline expressed genes, but are sensitive to the same dsRNAs injected directly into the gonads; 2) RSD-2 and RSD-6 associate with endoplasmic reticulum, while HAF-6 is a plasma membrane protein.21,22,28 The current model suggests that feeding these mutants with E.coli that produce dsRNAs fails to trigger an RNAi response because this method delivers low concentrations of dsRNAs. In addition, the rsd-2, rsd-6, and haf-6 genes regulate the levels of a subset of WAGO class 22G siRNAs.10 In contrast, mutations in sid-1 gene did not cause decrease of several abundant representative endogenous siRNAs10(C. Zhang and G. Ruvkun, unpublished data). Therefore, rsd-2, rsd-6, and haf-6 are unlikely to be spreading mutants. It would be intriguing to identify the protein interactors of membrane-associated RNAi pathway components RSD-2, RSD-6 and HAF-6. They may associate with other RNAi pathway factors or other membrane proteins whose functions have been previously characterized. These initial studies will help direct further in-depth characterization of these proteins.
The Biological Importance of rde-10, rde-11 and Other Dosage-Sensitive RNAi-Defective Genes
The rde-10, rde-11, rsd-2, and rsd-6 genes promote secondary siRNA accumulation in both exogenous and endogenous RNAi pathways. They are only conserved among Caenorhabditis species. We speculate that these genes are important for nematodes to adapt to their natural habitats where the physical and biological conditions are subject to dramatic fluctuation. The RNAi pathway is required for antiviral defense in C. elegans.29,30 rde-10, rde-11 and other dosage-sensitive RNAi-defective genes presumably enhance the potency of the antiviral response. Although endogenous 22G siRNAs are derived from half of all C. elegans genes, rde-10, rde-11, rsd-2, and rsd-6 are specifically required for the accumulation of a small number of WAGO class 22G siRNAs.10,11,13,31 A significant fraction of these 22G siRNAs function downstream of ERGO-1-class 26G siRNAs to silence recently acquired, duplicated genes. Most of these duplicated genes are poorly conserved and lack indication of function or expression.19 They share extensive sequence homology, and often times are at close proximity in the genome with each other.19 Gene duplication occurs at a relatively high frequency in C. elegans and may be acquired by viral infection or transposon mobilization.19,32,33 The dosage-sensitive RNAi-defective genes and other components in the ERGO-1/WAGO endogenous RNAi pathway effectively silence majority of these genes, thus this pathway may contribute to the optimal survival, fitness and fecundity of C. elegans. RdRPs, which are required for secondary siRNA biogenesis in nematodes, have also been identified in fungi and plants. These organisms may have functional homologs of C. elegans dosage-sensitive RNAi-defective genes that are essential for secondary siRNA biogenesis and effective genome surveillance.
Acknowledgments
We thank Sylvia Fischer and Taiowa Montgomery for helpful comments. This work was supported by a grant to G.R. from National Institute of Health (GM44619).
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/21246
References
- 1.Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr Biol. 2000;10:169–78. doi: 10.1016/S0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- 2.Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, et al. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–76. doi: 10.1016/S0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 3.Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, et al. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–57. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 4.Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–7. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 5.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–4. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 6.Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007;26:5007–19. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–9. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–71. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–32. doi: 10.1016/S0092-8674(00)81644-X. [DOI] [PubMed] [Google Scholar]
- 10.Zhang C, Montgomery TA, Fischer SE, Garcia SM, Riedel CG, Fahlgren N, et al. The Caenorhabditis elegans RDE-10/RDE-11 Complex Regulates RNAi by Promoting Secondary siRNA Amplification. Curr Biol. 2012;22:881–90. doi: 10.1016/j.cub.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu W, Shirayama M, Conte D, Jr., Vasale J, Batista PJ, Claycomb JM, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 2009;36:231–44. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, et al. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science. 2008;321:537–41. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–34. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 15.Han T, Manoharan AP, Harkins TT, Bouffard P, Fitzpatrick C, Chu DS, et al. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2009;106:18674–9. doi: 10.1073/pnas.0906378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gent JI, Lamm AT, Pavelec DM, Maniar JM, Parameswaran P, Tao L, et al. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol Cell. 2010;37:679–89. doi: 10.1016/j.molcel.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, et al. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc Natl Acad Sci U S A. 2010;107:3582–7. doi: 10.1073/pnas.0911908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conine CC, Batista PJ, Gu W, Claycomb JM, Chaves DA, Shirayama M, et al. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2010;107:3588–93. doi: 10.1073/pnas.0911685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer SE, Montgomery TA, Zhang C, Fahlgren N, Breen PC, Hwang A, et al. The ERI-6/7 helicase acts at the first stage of an siRNA amplification pathway that targets recent gene duplications. PLoS Genet. 2011;7:e1002369. doi: 10.1371/journal.pgen.1002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang H, Zhang Y, Vallandingham J, Li H, Florens L, Mak HY. The RDE-10/RDE-11 complex triggers RNAi-induced mRNA degradation by association with target mRNA in C. elegans. Genes Dev. 2012;26:846–56. doi: 10.1101/gad.180679.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tijsterman M, May RC, Simmer F, Okihara KL, Plasterk RH. Genes required for systemic RNA interference in Caenorhabditis elegans. Curr Biol. 2004;14:111–6. doi: 10.1016/j.cub.2003.12.029. [DOI] [PubMed] [Google Scholar]
- 22.Sundaram P, Echalier B, Han W, Hull D, Timmons L. ATP-binding cassette transporters are required for efficient RNA interference in Caenorhabditis elegans. Mol Biol Cell. 2006;17:3678–88. doi: 10.1091/mbc.E06-03-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steiner FA, Okihara KL, Hoogstrate SW, Sijen T, Ketting RF. RDE-1 slicer activity is required only for passenger-strand cleavage during RNAi in Caenorhabditis elegans. Nat Struct Mol Biol. 2009;16:207–11. doi: 10.1038/nsmb.1541. [DOI] [PubMed] [Google Scholar]
- 24.Jose AM, Hunter CP. Transport of sequence-specific RNA interference information between cells. Annu Rev Genet. 2007;41:305–30. doi: 10.1146/annurev.genet.41.110306.130216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shih JD, Hunter CP. SID-1 is a dsRNA-selective dsRNA-gated channel. RNA. 2011;17:1057–65. doi: 10.1261/rna.2596511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shih JD, Fitzgerald MC, Sutherlin M, Hunter CP. The SID-1 double-stranded RNA transporter is not selective for dsRNA length. RNA. 2009;15:384–90. doi: 10.1261/rna.1286409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456–9. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]
- 28.Han W, Sundaram P, Kenjale H, Grantham J, Timmons L. The Caenorhabditis elegans rsd-2 and rsd-6 genes are required for chromosome functions during exposure to unfavorable environments. Genetics. 2008;178:1875–93. doi: 10.1534/genetics.107.085472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Félix MA, Ashe A, Piffaretti J, Wu G, Nuez I, Bélicard T, et al. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 2011;9:e1000586. doi: 10.1371/journal.pbio.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rechavi O, Minevich G, Hobert O. Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans. Cell. 2011;147:1248–56. doi: 10.1016/j.cell.2011.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang C, Montgomery TA, Gabel HW, Fischer SE, Phillips CM, Fahlgren N, et al. mut-16 and other mutator class genes modulate 22G and 26G siRNA pathways in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2011;108:1201–8. doi: 10.1073/pnas.1018695108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–5. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 33.Lipinski KJ, Farslow JC, Fitzpatrick KA, Lynch M, Katju V, Bergthorsson U. High spontaneous rate of gene duplication in Caenorhabditis elegans. Curr Biol. 2011;21:306–10. doi: 10.1016/j.cub.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]


