Abstract
Editing by ADAR enzymes is essential for mammalian life. Still, knowledge of the spatio-temporal editing patterns in mammals is limited. By use of 454 amplicon sequencing we examined the editing status of 12 regionally extracted mRNAs from porcine developing brain encompassing a total of 64 putative ADAR editing sites. In total 24 brain tissues, dissected from up to five regions from embryonic gestation day 23, 42, 60, 80, 100 and 115, were examined for editing.
Generally, editing increased during embryonic development concomitantly with an increase in ADAR2 mRNA level. Notably, the Gria2 (GluR-B) Q/R site, reported to be ~100% edited in previous studies, is only 54% edited at embryonic day 23. Transcripts with multiple editing sites in close proximity to each other exhibit coupled editing and an extraordinary incidence of long-range coupling of editing events more than 32 kb apart is observed for the kainate glutamate receptor 2 transcript, Grik2. Our study reveals complex spatio-temporal ADAR editing patterns of coordinated editing events that may play important roles in the development of the mammalian brain.
Keywords: 5-HT2C receptor, ADAR, ADAR2, Blcap, GluR-B, Gria2, Htr2c, Pig, RNA editing
Introduction
RNA editing is a posttranscriptional mechanism employed by eukaryotes to generate increased transcript diversity from a limited sized genome. During editing an RNA molecule is chemically altered at the nucleotide base; the most prevalent form of editing in the central nervous system is the deamination of adenosines (A) to inosine (I), which is recognized as guanosine (G) by the cellular translation machinery,1 a reaction mediated by the ADAR1 and -2 enzymes (adenosine deaminases that act on RNA). ADAR1 is ubiquitously expressed from three different promoters, two of which are constitutively active, resulting in the same nuclear localized ADAR1 (cADAR1) and one interferon induced, resulting in a version of the ADAR1 enzyme (iADAR1) that is partially localized to the cytoplasm.2 ADAR2 is highly expressed in the nucleus of cells of the central nervous system but also found in other tissues.3 ADAR1 and ADAR2 seem to overlap in their editing specificity and may functionally overlap.4,5 An additional brain-specific nuclear ADAR, ADAR3, is thought to be nonfunctional.6 ADAR1 and -2 function as homodimers in vivo;7-9 however, heterodimerization of ADARs has been observed and could modulate editing activity.6,9,10
ADARs can promiscuously deaminate extended stretches of perfectly complementary dsRNA in a relatively sequence independent manner, such as the abundant fold-back structures found in untranslated regions. Yet, surprisingly few mRNA coding regions are edited by ADARs: In all animals studied only about 30 genes, predominantly neuroreceptors and channels have been shown to exhibit exonic ADAR editing.11 It has been suggested that one ADAR enzyme may edit a specific site and recruit additional ADARs to nearby sites by protein-protein interactions leading to multiple editing events along imperfect RNA duplexes,12 a phenomenon referred to as coupled editing. mRNA editing can have a great impact on protein function: in most of the 12 edited transcripts investigated in this study editing leads to altered amino acid incorporation during translation hereby potentially influencing the function of the encoded proteins. An exception is the ADAR2 mRNA where self-editing by the ADAR2 enzyme creates a new 3′ splice acceptor site in intron 1 upon editing of an AA dinucleotide to AI, which mimics the canonical AG found at 3′ splice sites. This leads to inclusion of additional 47 nucleotides at the 5′ end of exon 2 in the ADAR2 transcript (referred to as the ADAR2alt transcript) that due to premature termination produces a nonfunctional product.13,14 In mice, the importance of this editing system is underlined by the observation that ADAR1 and -2 knockouts (KO) are embryonic and perinatal lethal, respectively.15,16 ADAR2 KO mice die from severe seizures caused by the lack of editing at one specific site, the Q/R site in the AMPA glutamate receptor, Gria2, resulting in increased influx of Ca2+ ions. The Gria2 Q/R site is normally edited to approximately 100%,15,17,18 which has raised the question why this site is maintained as an A in the genome only to be edited to I. Although ADAR2 KO mice die after birth they go through normal embryonic development, hence the lack of the RNA editing at Gria2 Q/R does not perturb embryogenesis.15
In spite of intense research of the editing process the timing and function of mammalian editing during brain development are not well understood. Here we examine the spatio-temporal editing of porcine orthologs of 12 human and mouse ADAR editing regions, encompassing nearly all the transcripts currently known to be edited by ADAR enzymes. This is done in high depth using 454 sequencing on a total of 24 samples from five different brain regions and six time points ranging from embryonic day 23 (E23) to E115 (time of birth). The porcine brain resembles the human brain by several distinct features that make it an attractive model organism. The brains are similar in size, growth and anatomic complexity including convolutions of the cortex.19 Important for this study, the large size of the pig brain enables a more precise analysis and insight into RNA editing at very early developmental stages.
Results
The spatio-temporal pattern of A-to-I editing in the transcripts listed in Table 1 was analyzed during embryonic development of the porcine brain using the 454 next generation sequencing technology. The 12 selected transcripts, which previously have been described to be edited by ADAR enzymes, mostly encode neuroreceptors essential for neuron function. Three of the editing sites detected in Grik2 were amplified using two independent RT-PCR amplicons generated by the use of different primer sets, implying that the technique used for quantifying editing events is unbiased (Fig. S1).
Table 1. List of genes analyzed in this report.
| Gene name | Alternative name | Full name |
|---|---|---|
| Grik2 |
GluR6, GluK2 |
Glutamate receptor, ionotropic, kainate 2 |
| Gria2 |
GluR2, GluRB, GluA2 |
Glutamate receptor, ionotropic, ampa 2 |
| Gria3 |
GluR3, GluRC, GluA3 |
Glutamate receptor, ionotropic, ampa 3 |
| Kcna1 |
MK1, Kv1.1 |
Potassium channel, voltage-gated, shaker-related subfamily, member 1 |
| Htr2c |
5-Ht2c, 5-Htr2c |
Serotonin 5-ht-2c receptor |
| Gabra3 |
|
Gamma-aminobutyric acid receptor, α-3 |
| ADARb1 |
ADAR2 |
Adenosine deaminase, rna-specific, b1 |
| Flna |
Fln, Fln1 |
Filamin A |
| Blcap |
BC10 |
Bladder cancer-associated protein |
| Igfbp7 |
Mac25, Psf, Agm |
Insulin-like growth factor-binding protein 7 |
| Ednrb |
Etb |
Endothelin receptor type B |
| Cyfip2 | PIR121 | Cytoplasmic fmrp-interacting protein 2 |
Alternative names and full names are indicated.
In total 64 sites were edited > 1% in at least five tissues, which was set as cut-off value to avoid contributions from sequencing errors. Table S1 for full list of sequenced A-to-I editing percentages at all sites in all tissues examined.
Overall editing and ADAR2 mRNA level increase during embryonic development
We find a general increase in editing throughout embryonic brain development (Fig. 1), which is consistent with earlier observations of A-to-I editing during brain development in mice and humans.12,17,20 To investigate whether this gradual increase in editing reflects ADAR enzyme levels we analyzed ADAR1 and -2 mRNA levels by RT-qPCR (Fig. 1; Fig.S2). It revealed that the mean ADAR2 mRNA expression correlates positively with the observed A-to-I editing levels. ADAR2alt, which lacks editing activity, was only expressed at very low levels. We found that cADAR1 was > 10 times higher expressed than the partially cytoplasmic and interferon induced iADAR1 at all time points.
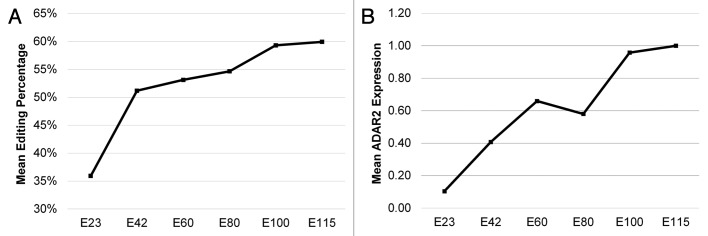
Figure 1. Mean editing and ADAR2 mRNA expression. (A) Mean editing of ADAR edited transcripts displaying at least 10% editing as a function of embryonic development. The mean editing of transcripts across all brain regions examined is indicated in percentage based on measurement at the indicated gestation time points. (B) The mean ADAR2 mRNA expression level relative to GAPDH averaged across all brain regions at the indicated gestation time points.
Among the 64 editing sites analyzed, 33 were edited more than 5% in at least one sample and were chosen for further comparison with ADAR mRNA levels by Pearson correlations (Table 2). In line with the inferred overall correlation between editing percentages and ADAR2 transcript levels (Fig. 1), we found good correlation between ADAR2 transcript and most of the specific targets (Table 2), many of which are known to be edited by ADAR2.4,15,21 The ADAR2alt transcript has an expression signature similar to ADAR2 (Fig. S2), consistent with the similar Pearson coefficients; however, the expression levels are much lower than for ADAR2. In contrast, both ADAR1 mRNAs appear not to correlate well with editing sites (Table 2).
Table 2. Correlation of ADAR transcript levels with editing percentages.
| Transcript | Site | iADAR1 | cADAR1 | ADAR2 | ADAR2alt | Reported |
|---|---|---|---|---|---|---|
| Grik2 |
I/V |
|
-0.44 |
0.74 |
0.83 |
ADAR215 |
| |
Y/C |
|
|
0.78 |
0.59 |
ADAR215 |
| |
G/G |
-0.63 |
|
|
|
|
| |
M/V |
|
|
0.64 |
0.75 |
|
| |
Q/R |
|
|
0.59 |
|
ADAR215 |
| Gria2 |
Q/R |
|
|
0.43 |
|
ADAR215 |
| |
+4 |
|
|
|
|
|
| |
Hotspot (+60) |
|
|
0.66 |
0.57 |
|
| Gria3 |
R/G |
|
|
|
|
|
| Kcna1 |
I/V |
|
-0.48 |
|
0.48 |
ADAR235 |
| Htr2c |
A |
|
-0.42 |
|
|
ADAR1 and -2 44 |
| |
B |
|
-0.46 |
|
|
ADAR1 and -2 15 |
| |
C' |
|
|
0.45 |
|
|
| |
C |
|
|
|
|
ADAR1 and -2 44 |
| |
D |
|
-0.46 |
|
|
ADAR215,44 |
| Adar2 |
-16 |
-0.43 |
-0.44 |
0.54 |
0.71 |
|
| |
-8 |
|
-0.44 |
0.61 |
0.85 |
|
| |
-4 |
-0.54 |
|
0.59 |
0.69 |
|
| |
-2 |
|
-0.50 |
0.49 |
0.66 |
|
| |
-1 |
|
-0.57 |
0.67 |
0.79 |
ADAR215 |
| |
+10 |
-0.41 |
-0.47 |
0.67 |
0.78 |
|
| |
+23 |
|
|
0.60 |
0.74 |
ADAR215 |
| |
+24 |
-0.60 |
|
0.51 |
0.68 |
ADAR215 |
| |
+28 |
|
-0.63 |
|
0.50 |
|
| Flna |
Q/R |
|
|
0.50 |
0.44 |
ADAR1 and -2 4 |
| |
K/R |
|
|
|
|
|
| Blcap |
5a |
|
|
0.41 |
|
|
| |
M/V |
|
0.51 |
|
|
|
| |
Y/C |
|
|
|
|
ADAR1 and -2 21 |
| |
Q/R |
|
0.47 |
|
|
ADAR1 and -2 21 |
| |
K/R |
-0.41 |
0.53 |
|
|
ADAR1 and -2 21 |
| Igfbp7 |
B (K/R) |
-0.46 |
|
0.62 |
0.52 |
|
| C (K/R) | -0.47 | 0.74 | 0.79 |
Pearson correlation coefficients between ADAR transcript levels (qPCR data) and A-to-I editing percentages (454 data) for correlations attaining a significant p value (< 0.05, values given in bold have p values < 0.005) are indicted. Only sites averaging > 9% editing in all tissues were used for this analysis. Values of 0.1–0.3 can be considered a small positive correlation, 0.3–0.5 as a medium correlation and 0.5–1.0 as a strong correlation. See text for description of ADAR variants.
Collectively, this is consistent with ADAR2 being the primarily editing enzyme of the 33 selected sites during porcine brain development.
Developmental timing and tissue specific differences of mRNA editing
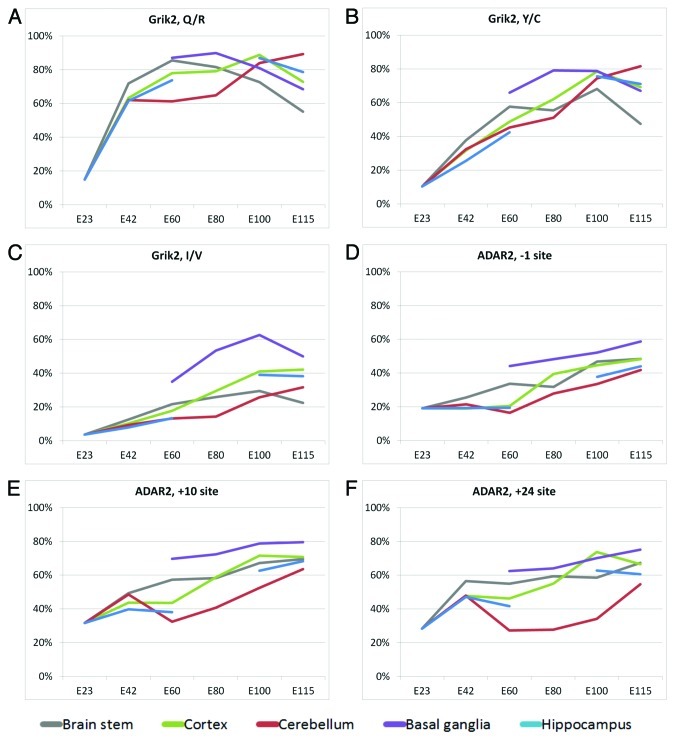
Despite the general increase in editing during development some editing sites peak at intermediate time points. The most prominent example of this is the Q/R editing site in Grik2 mRNA17 (Fig. 2A), which peaks at E60 for brain stem, E80 for basal ganglia and E100 for cortex and hippocampus (note that hippocampus was not examined at E80). Only cerebellum peaks at E115 for this site. The overall level of editing can also differ widely among different brain regions, as seen for the Grik2 I/V site (Fig. 2C) where the basal ganglia exhibits markedly higher editing levels than other tissues. This higher level of editing in basal ganglia is also seen in each of the three primary ADAR2 pre-mRNA self-editing sites (-1, +10 and +24) (Fig. 2D and E). Especially cerebellum is edited to a low extent in ADAR2 pre-mRNA and an unexpected editing pattern is observed for editing sites +10 and +24 where the editing level decreases from E42 to E60 where after it increases to the maximum level at E115.
Figure 2. Editing level for Grik2 editing sites (A-C) and the three primary ADAR2 self-editing sites (D-F) at different gestation time points. The color code refers to tissue type and is indicated below the figure. Note that only one tissue, whole forebrain, was dissected from E23 due to the small size of the embryo at this stage.
These findings cannot be explained by changes in ADAR transcript levels alone (Fig. S2).
Evaluation of early editing events
The larger size of the pig brain as opposed to e.g., the mouse brain enabled us to analyze editing events during mammalian brain development as early as 23 d after fertilization. Interestingly, we found that the Gria2 Q/R site, previously reported ~100% edited in all instances in adult human as well as embryonic and adult rat and mouse brain,17,22,23 exhibited a partial editing level of 54% at day 23 rising to ~100% at all subsequent time points (Fig. 3A); a finding confirmed by Sanger sequencing (data not shown). The low editing level of Gria2 Q/R at this time point is further highlighted by inspection of the Htr2c D site, which exhibits 60% editing (Fig. 3F); remarkably higher than the Gria2 Q/R site. The Htr2c D site, like the four other Htr2c sites exhibit a very stable level of editing throughout all of porcine embryonic development (Fig. 3B-F). This is in contrast to the previously reported increase of Htr2c editing in mouse during development.17 Whether this reflects species specific or experimental variation is not clear but the high read depth employed here (Table S3) ensures an accurate quantification of editing levels. Also, mapping our Htr2c amplicon sequence to the Sus scrofa genome 9.2 generates only one credible hit, attesting that the constant editing percentages of Htr2c sites are not contributions from multiple genomic locations.
Figure 3. Editing level for the Gria2 Q/R site (A) and all the 5 Htr2c sites (B-F) indicated in percentages. Note that only one tissue, whole forebrain, was dissected from E23.
Another significant editing site in the Gria2 amplicon, which is previously unreported, is located 84 nucleotides upstream of the Gria2 Q/R site displaying a remarkable 14% editing in E23 tissue but declining to only 0–4.6% at later time points. This may either represent a pig specific editing event or, more likely, reflect our ability to probe very early developmental stages in a mammalian brain (day 23 of 115 d of gestation) also signified by remarkably low editing of Gria2 Q/R.
Species-specific differences in editing
Unexpectedly, the I/M editing site, previously reported in Gabra3,24 is nearly completely unedited in porcine brain development (see Table S1 for all editing percentages). When specifically examining the expected Gabra3 editing site we observe only from 0.0% to 0.7% editing. This is unexpected since all other mammals examined display appreciable editing of this site.25 Importantly, the duplex representing the putative editing substrate, containing the exonic editing site, is essentially identical to that found in mouse. The most likely explanation is the absence of a specific structure in the downstream intron (Daniel C. et al., submitted.).
The transcripts Ednrb (edited at a Q/R site in human brain26) and Cyfip2 (edited at a K/E site in human and mouse brain4,27) show no editing in any of our porcine samples. However, Cyfip2 exhibits multiple low level editing sites.
Principle editing sites for ADAR enzymes
It has been suggested that ADAR enzymes may bind primarily to a high-affinity binding site in the RNA and subsequently trigger secondary editing events.12 Since several of the editing sites detected occur within single amplicons we are able to perform more detailed analysis of coupled editing of these sites. The site with highest affinity for ADAR enzyme is likely to be edited before sites with lower affinity on the same amplicon, particularly at the early time points when ADAR enzyme concentration is limited. Therefore the site with highest percentage of singular editing at E23 and E42 is viewed as the principal editing site of the amplicon. The complete list of singular editing for every tissue is shown in Table S2. In the following the mean percentage of singular editing at E23 and E42 tissues will be used to define the principal editing site.
The Igfbp7 B site28 is clearly the principal site in this amplicon with 45.6% editing. The same is evident for the Grik2 Q/R site, which has the most pronounced singular editing at 32.1%. Other amplicons are less obvious, such as the Blcap21 amplicon where the Y/C site, with only 8.3% singular editing, is the principal site with all adjacent sites reaching only 1.6% editing level. Similarly, the principal editing site in the ADAR2 amplicon appears to be position +24 with 11.1% singular editing vs. 5.4% at the +10 site. Finally, the Htr2c site D also shows 11.1% singular editing with the C site significantly lower at 5.4%.
The only previous study of singular editing events was performed in mouse Htr2c, ADAR2 and Grik2 transcripts;12 The principal sites detected for mouse Htr2c and ADAR2 transcripts appears to be in agreement with the porcine samples however, the study by Ensterö et al. did not include the Q/R site in the Grik2 amplicon, which we find to be the predominate site in this region.12
Coupling of editing events
It has been suggested that the singular editing of the principal editing site may function as nucleation point for additional ADAR editing events.
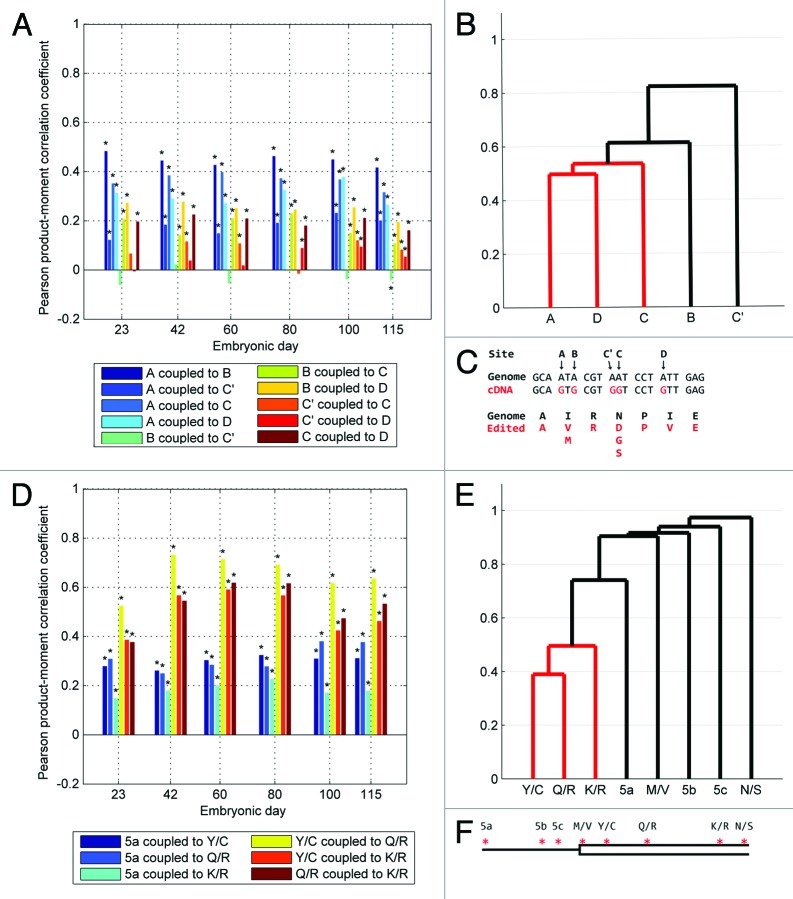
To address this issue we analyzed the coupling between individual sites and displayed it by graphing the Pearson correlation coefficient and a representative group analysis dendrogram. Generally, we find that edited sites positively couple with each other inside the same amplicon. E.g. most sites in the Htr2c amplicon display some degree of coupling, signified by statistically significant positive Pearson coefficients with the notable exception that the C' editing site has a generally low Pearson coefficient when comparing to the other sites (Fig. 4A). The uncoupling of the C' site is also evident from the group analysis (Fig. 4B). Other tissues yielded similar results (Figs. S3 and S4).
Figure 4. Coupling of editing events for Htr2c and Blcap transcripts. (A) Pearson correlation coefficients for coupling between all editing sites on the Htr2c amplicon in cortex. See legend for color codes. Black asterisks denote significance compared with bonferroni corrected critical p values. (B) Group analysis of all cortex E115 Htr2c editing sites. (C) The genomic nucleotide sequence immediately surrounding the editing sites in Htr2c (A, B, C', C and D) and the sequence of the corresponding cDNA with edited nucleotides depicted in red. The genomically coded amino acid sequence is shown in black and the resulting amino acids after editing are shown in red. (D) Pearson correlation coefficients for coupling between the predominant editing sites on the Blcap amplicon in cortex. See legend for color codes. Black asterisks denote significance compared with bonferroni corrected critical p values. (E) Group analysis of cortex E115 Blcap editing sites. (F) Schematic drawing (not to scale) of the editing sites (red asterisks) in Blcap showing the three editing sites in 5′UTR region (thin line; 5a, 5b, 5c) and the five editing sites inside the coding region (open box) detected in this study. Note that only one tissue, whole forebrain, was dissected from E23.
For the Blcap sites high Pearson correlation coefficients are observed for coupled editing, especially between the three primary open reading frame (ORF) sites (Y/C, Q/R and K/R) (Fig. 4D and E) The amplicon contains eight editing sites but due to low overall editing levels the 5b, 5c, M/V and N/S sites were omitted from the Pearson coefficient calculations. Group analysis confirmed strong coupling between the three ORF sites and less between the remaining sites. Similar results were observed for editing sites in Blcap in other tissues (Figs. S5 and S6).
Sequencing of the amplicon spanning the intron-exon junction of ADAR2 enabled the detection of an editing event that potentially could alter splicing of the ADAR2 pre-mRNA. Multiple spurious editing events presumably of little importance are detected at low levels in the intronic section of this amplicon. Three highly edited sites stand out as being coupled; the -1 site, which is the actual site conferring alternate splicing in the edited form, the +10 site and the +24 site (Fig. 5A). The Pearson coefficients for the cortex clearly indicated coupled editing between the three sites (-1, +10 and +24) which was confirmed by group analysis (Fig. 5B). The same scenario was observed in other tissues (Figs. S7 and S8).
Figure 5. Coupling of editing events for ADAR2 and Grik2 transcripts. (A) Pearson correlation coefficients for coupling between the predominant self-editing sites on the ADAR2 amplicon in cortex. See legend for color codes. Black asterisks denote significance compared with bonferroni corrected critical p values. (B) Group analysis of all cortex E115 ADAR2 self-editing sites. (C) Schematic drawing (not to scale) of the editing sites detected in ADAR2 (red asterisks). The entire depicted region is intronic in the unedited pre-mRNA but editing of the -1 site causes inclusion of the open boxed section in the downstream exon. (D) Pearson correlation coefficients for coupling between all editing sites on the Grik2 amplicon in cortex. See legend for color codes. Black asterisks denote significance compared with bonferroni corrected critical p values. (E) Group analysis of all cortex E115 Grik2 editing sites. (F) Schematic drawing (not to scale) of the editing sites in Grik2 showing the five editing sites (red asterisks) detected in this study. I/V and Y/C sites are separated from G/G, M/V and Q/R sites by a 32 kb intron. Note that only one tissue, whole forebrain, was dissected from E23.
A trend entailing lower coupling at later time points was observed (Fig. 5A; Figs. S7 and S8) even though the editing percentages are higher at the later time points (Table S1). This could possibly be due to the higher concentration of ADAR enzymes at later time points. As suggested by Öhman group,12 editing of low affinity sites may become limited in competition with high affinity sites under low ADAR enzyme concentrations; however, high concentrations of ADAR enzyme may promote initiation of editing at suboptimal editing sites. Such an effect is likely to be most pronounced in a substrate like the ADAR2 transcript where many potential suboptimal initiating sites are present.
The Grik2 amplicon sequenced in this study contains three highly and two less edited ADAR editing sites. The Pearson correlation coefficients for coupling between each of these sites in the Grik2 transcript shows coupling between the three highly edited sites, I/V, Y/C and Q/R (Fig. 5D). Remarkably, his region contains a large intron of 32 kb, which, based on the fact that the intron constitutes part of the recognition site for editing, presumably remains unspliced when editing occur. As a consequence the Q/R site is situated more than 32 kb from the I/V and Y/C sites (Fig. 5F) but still functionally coupled based on the Pearson coefficients and the group analysis (Fig. 5D and E), a peculiarity recently also observed in humans.29 Similar coupling scheme was observed in other tissues for the Grik2 region (Figs. S9 and S10).
Tertiary structure prediction
In the situation where an amplicon contains multiple editing sites and where the editing complementary sequence (ECS) is known it is possible to model the tertiary structure for the editing region. Inspection of the predicted tertiary structure for Blcap, Htr2c and ADAR2 transcripts corroborates the model that editing of coupled sites by ADAR enzymes proceeds predominantly via sequential editing along the same side of the imperfect stem of the target transcript (Fig. 6).12 However the Htr2c C site seems to be an exception to this trend.
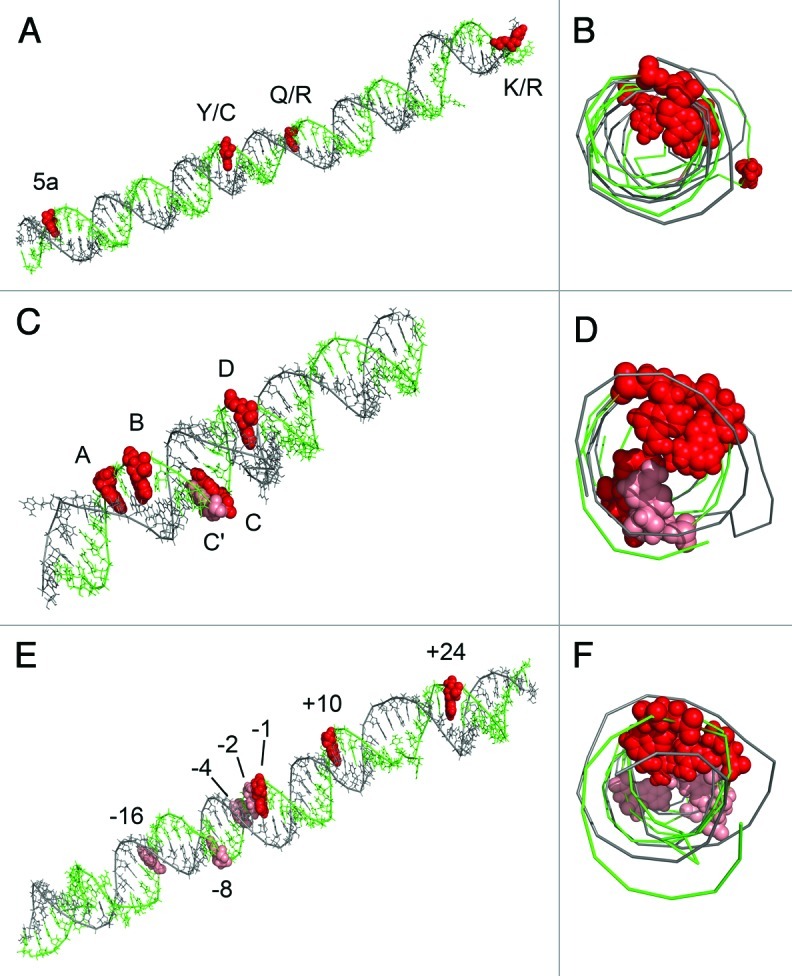
Figure 6. Tertiary modeling of the edited regions of the Blcap (A and B), Htr2c (C and D) and ADAR2 (E and F) pre-mRNAs with intronic ECS. The tertiary models are shown from the side (A, C and E) and head-on (B, D and F). Red spheres are editing sites displaying coupling. Light red spheres are uncoupled editing sites.
Discussion
Examining ADAR editing by deep sequencing provided a detailed picture of the embryonic editing patterns that confirms most known specific targets of ADAR editing but also reveals novel sites. In particular, the combined spatial and temporal resolution of editing during development of the porcine embryo has yielded novel insight into tissue specific fluctuations of editing levels. Confirming previous observations, we found that the general editing level increases during development; however, notable exceptions exist such as the sites on Grik2 displaying transiently elevated editing. Our data also confirmed that the Q/R site of Gria2, one of the most studied editing sites, is near 100% edited during most developmental stages.15,17,18 However, here we report that the Gria2 Q/R site is edited in only 54% at the earliest time point (E23) which may be a consequence of lower levels of ADAR2 at early developmental stages. Another interesting observation is the editing of Htr2c transcripts, which remains remarkably stable throughout embryonic development; in contrast to the increased editing seen for Htr2c transcripts over embryonic development in mouse whole brain;17 a comparison between the secondary structures of the Htr2c pre-mRNAs from pig and mouse reveals that the distal region from a conserved edited structure (red box in Fig. S11) contains a more elongated stem in the pig (right black box in Fig. S11). This structure could potentially increase ADAR enzyme recruitment and editing of the adjacent duplex in pig. The fact that the Htr2c D site is 60% edited at the E23 stage suggests that ADAR2 levels are significant at this point in development and that the editing of Gria2 Q/R may be actively precluded. An alternative explanation is that Gria2 expression at E23 may occur in a subset of brain cells expressing low levels of ADAR2. Functional importance of the partial editing of Gria2 Q/R in the earliest development also provides a potential evolutionary explanation for retaining a genomic adenosine at this position rather than changing to a guanosine in the genome.
The previously undetected editing site 84 nucleotides upstream of the Gria2 Q/R site is also of special developmental interest, based on the notion that this site is most highly edited at E23. Editing of this site could play a unique function in early development.
Editing levels of the examined transcripts vary, not only across time, but also across tissues. Whether these variations stem from control of ADAR editing or target accessibility during development is not clear. Although the overall editing level of the transcripts examined here may, to some degree, be explained by variations in ADAR2 expression, simply correlating editing percentages of specific editing sites with the levels of ADAR transcripts present in the tissues does not explain editing variations sufficiently well. Control of ADAR enzyme activity could be mediated by other factors such as limited availability of IP6, a co-factor crucial for correct ADAR protein folding and activity,30 or by other protein factors such as the positive regulator, Pin1, which interacts with ADAR2 enzyme promoting nuclear localization and stability of ADAR2 or the negative regulator, WWP2, which binds to ADAR2 and catalyzes ubiquitination and subsequent degradation of the enzyme.31
ADAR2 seems most active in basal ganglia based on the elevated editing of Grik2 and self-editing by ADAR2 in this tissue (Fig. 2B-F); the latter leading to increased production of the alternatively spliced ADAR2 transcript variant (Fig. S2D). Interestingly, the increased basal ganglia ADAR2 editing to some extent reflects an overall higher level of ADAR2 mRNA expression in basal ganglia (Fig. S2C). Previous studies have not been able to correlate ADAR expression levels to editing efficiency,17,32,33 but also lack the same resolution in the overall data set.
Essentially all amplicons examined displayed some level of coupling of individual editing sites, corroborating that editing of a principal editing site promotes additional editing events at adjacent sites. The tertiary modeling of the Blcap, Htr2c and ADAR2 transcripts with ECS regions (Fig. 6), shows that, with the exception of the Htr2c C site, the coupled editing sites accumulate at one side of the RNA duplex, which is in accordance with the model proposed for Htr2c and ADAR mRNAs in mouse, where that coupling is suggested to involve cooperative binding of ADAR enzymes through protein-protein interaction.12 An alternative explanation could be that A-to-I editing induces rearrangements in the RNA secondary structure which lead to the revelation of new editing targets nearby. No definite conclusions can be drawn about the mechanism behind the observed coupled editing, especially in the light of the Grik2 transcript. Here the coupling occurs across a large intron, which provides the ECS essential for editing.34 The question remains how such distant exons are coupled. One explanation may be that long-range RNA structure bridges the exons and enables ADAR enzymes to oligomerize from one exon to another and stimulate editing. Alternatively, the coupling could originate from co-transcriptional events where the nature of the elongating polymerase may control editing or sequestration of the pre-mRNA in putative “editing bodies” in the nucleus where higher concentrations of ADAR enzymes exist. It is also possible that the Q/R ECS is required for editing of the I/V and Y/C sites, which currently have no known ECS, and that the availability of this motif is depending on Q/R editing.
The long-range coupling of editing sites seen in Grik2 hints at an unexplored area of RNA interactions occurring at long distances in pre-mRNA. The example presented here is a special case where a long-range interaction in the pre-mRNA can be examined by RT-PCR amplification after splicing in the mRNA. Many other RNA interactions over long sequence distances are likely to exist; however, discovery of such interactions is experimentally highly challenging.
Generally, exonic ADAR editing sites have an intronic ECS present in an adjacent intron; however, this is not the case for ADAR2 mediated editing of the mRNA transcribed from the intronless voltage-gated potassium channel, Kcna1, gene. This transcript forms a short hairpin structure entirely within the exon, which is a substrate for ADAR2.35 The only other known example of an exonic ECS is seen in the GABAA receptor subunit α3, Gabra3, which has been shown to be site-selectively edited by ADAR enzymes.24 Editing of Gabra3 is widely conserved so it is intriguing that Gabra3 is not appreciably edited (< 1%) in any of the porcine samples examined in our study, in particular in light of the conserved substrate stem. However, a likely explanation was provided in a recent study, which demonstrated the need for a seemingly unrelated downstream intronic stem-loop structure for efficient editing of Gabra3, an element which is absent in porcine Gabra3 pre-mRNA (Daniel et al., submitted.).
The importance of ADAR editing is emphasized by the severe phenotypes of ADAR-1 and -2 knockouts15,16 consistent with the observation that editing is known to alter the kinetics and ion flux through the edited receptors and channels.15,36-38 The spatio-temporal editing pattern of several mRNAs and coupling between editing sites in porcine embryonic brain tissues uncovered in this study emphasize the importance of tight control of ADAR enzymatic activity. Revelation of this intrinsic network in more detail in the future will require a more detailed study at single cell resolution and should include the entire transcriptome.
Materials and Methods
RNA Extraction and RT-PCR
All procedures involving animals described in present study were reviewed and approved by the Danish Experimental Animal Inspectorate (“Rådet for Dyreforsøg”), Danish Ministry of Justice. Brain sections were dissected from pig embryos, snap frozen on dry ice and subsequently transferred to RNAlater-ICE (Ambion). High molecular weight RNA (≥ 200 nt.) was purified using the MirVana kit (Ambion). Samples were DNase treated using the TURBO DNA-free Kit (Ambion) and first strand cDNA was prepared with gene specific primers using M-MLV Reverse Transcriptase kit (Invitrogen). All primer sequences will be supplied by the corresponding author upon request.
PCR amplification using primers against A-to-I editing regions of the mRNAs listed in Table 1 was done using HotStarTaq Plus (Qiagen) and products were purified using Ultra-Sep Gel Extraction Kit (E.Z.N.A.).
454 amplicon sequencing
Purified RT-PCR products were prepared for 454 amplicon sequencing by use of GS FLX Titanium Rapid Library Preparation Kit (Roche) and RL-MID tags (Roche) to allow multiplexed sequencing. 454 amplicon sequencing was performed followed by sequence determination per manufacturer’s instructions. See Table S4 for the genomic sequence of each RT-PCR amplicon.
Editing site detection
The editing frequencies seen in Table S1 are made by an unbiased detection of editing sites performed with the SNP detection software designed for 454 read data called Swap454.39 The following parameters were used for mapping to reference sequences: minimum quality of non-reference nucleotide set to 20 (MIN_QUAL = 20) and minimum quality of neighboring nucleotide set to 15 (NQ = 15). The following parameters were used to call polymorphisms: Minimum 1% of accepted reads are allowed to differ from reference sequence (MIN_RATIO = 0.01) and at least two reads must be different than reference (MIN_READS = 2).
Detailed analysis of amplicons with multiple sites
Suitable amplicons with multiple editing sites (Igfbp7, Grik2, Blcap, ADAR2, Htr2c) were examined in detail. First sequences from 454 amplicon sequencing were mapped onto the reference sequence of each amplicon using the BLAST-Like Alignment Tool (BLAT)40 to make multiple alignments. Only reads covering all editing sites in each amplicon were kept and file sizes were reduced by only retaining data from sites previously detected as A-to-I (sequenced as G) edited by Swap454, denoting A as zero and G as one. Singular editing was investigated by inspecting these files for reads only having a single editing event. Coupling of editing sites was investigated by two different approaches, Pearson’s correlation coefficients and group analysis: Pearson’s correlation coefficients and associated p values were calculated using the Matlab function “corrcoef” on the files generated by BLAT and graphs were generated with a custom Matlab program. Group analysis was performed with the Matlab functions “pdist” to calculate parwise distances using the Jaccard method and “linkage” to create agglomerative hierarchical cluster trees based on the output from “pdist.” Cluster trees were then visualized as dendrograms using Matlab.
Tertiary structure prediction
For Blcap, Htr2c and ADAR2 transcripts RNAfold41 was used to fold secondary structure of edited region and ECS region joined by a GAAA tetraloop. The pipeline MC-sym42 was then used to predict tertiary structure of RNA duplexes: Sequence and secondary structure from RNAfold were entered in the MC-sym script generator and the GAAA tetraloop was manually removed from the MC-sym script. The script was used to generate 3D RNA models, which were screened as suggested by the makers of MC-sym. The best 3D model was visualized in Pymol.43
Quantitative PCR of ADARs
qPCR primers specific for porcine gene sequences were designed for cADAR1, iADAR1, ADAR2, ADAR2alt and GAPDH. Primer sets all span an intron-exon boundary and generate PCR product of ~100 nucleotides. The efficiencies of all primers were tested prior to use. All primer sequences will be supplied by the author upon request. qPCR was done in triplicate for all primers on all tissues and pipetting was done using the automated pipetting system, epMotion 5075 (Eppendorf) in 384 well format. The LightCycler 480 II system (Roche) was used for qPCR run. ADAR transcript levels are reported as expression relative to GAPDH.
Supplementary Material
Acknowledgments
We would like to thank Anders Lade Nielsen and Arne Lund Jørgensen (Department of Human Genetics, University of Aarhus, Denmark) for facilitating the supply of embryonic porcine brain tissues. Also we would like to thank Claus Bus for technical assistance during RNA preparation. This work was supported by the Lundbeck Foundation; and EU-FP7 SIROCCO Project (LSHG-CT-2006–037900).
Glossary
Abbreviations:
- A-to-I
adenosine to inosine
- ADAR
adenosine deaminases that act on RNA
- ADAR2alt
ADAR2 variant with out-of-frame alternative splicing
- cADAR1
constitutively expressed ADAR1, nuclear localization
- iADAR1
interferon induced ADAR1, partially cytoplasmic localization
- E23
embryonic day 23
- ECS
editing complementary sequence
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/21082
References
- 1.Paul MS, Bass BL. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J. 1998;17:1120–7. doi: 10.1093/emboj/17.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.George CX, Samuel CE. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc Natl Acad Sci U S A. 1999;96:4621–6. doi: 10.1073/pnas.96.8.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melcher T, Maas S, Herb A, Sprengel R, Seeburg PH, Higuchi M. A mammalian RNA editing enzyme. Nature. 1996;379:460–4. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- 4.Riedmann EM, Schopoff S, Hartner JC, Jantsch MF. Specificity of ADAR-mediated RNA editing in newly identified targets. RNA. 2008;14:1110–8. doi: 10.1261/rna.923308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horsch M, Seeburg PH, Adler T, Aguilar-Pimentel JA, Becker L, Calzada-Wack J, et al. Requirement of the RNA-editing enzyme ADAR2 for normal physiology in mice. J Biol Chem. 2011;286:18614–22. doi: 10.1074/jbc.M110.200881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen CX, Cho DS, Wang Q, Lai F, Carter KC, Nishikura K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 2000;6:755–67. doi: 10.1017/S1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poulsen H, Jorgensen R, Heding A, Nielsen FC, Bonven B, Egebjerg J. Dimerization of ADAR2 is mediated by the double-stranded RNA binding domain. RNA. 2006;12:1350–60. doi: 10.1261/rna.2314406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho DS, Yang W, Lee JT, Shiekhattar R, Murray JM, Nishikura K. Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J Biol Chem. 2003;278:17093–102. doi: 10.1074/jbc.M213127200. [DOI] [PubMed] [Google Scholar]
- 9.Chilibeck KA, Wu T, Liang C, Schellenberg MJ, Gesner EM, Lynch JM, et al. FRET analysis of in vivo dimerization by RNA-editing enzymes. J Biol Chem. 2006;281:16530–5. doi: 10.1074/jbc.M511831200. [DOI] [PubMed] [Google Scholar]
- 10.Cenci C, Barzotti R, Galeano F, Corbelli S, Rota R, Massimi L, et al. Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J Biol Chem. 2008;283:7251–60. doi: 10.1074/jbc.M708316200. [DOI] [PubMed] [Google Scholar]
- 11.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–49. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ensterö M, Daniel C, Wahlstedt H, Major F, Öhman M. Recognition and coupling of A-to-I edited sites are determined by the tertiary structure of the RNA. Nucleic Acids Res. 2009;37:6916–26. doi: 10.1093/nar/gkp731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orlandi C, Barbon A, Barlati S. Activity Regulation of Adenosine Deaminases Acting on RNA (ADARs) Mol Neurobiol. 2011 doi: 10.1007/s12035-011-8220-2. [DOI] [PubMed] [Google Scholar]
- 14.Slavov D, Gardiner K. Phylogenetic comparison of the pre-mRNA adenosine deaminase ADAR2 genes and transcripts: conservation and diversity in editing site sequence and alternative splicing patterns. Gene. 2002;299:83–94. doi: 10.1016/S0378-1119(02)01016-8. [DOI] [PubMed] [Google Scholar]
- 15.Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q, Khillan J, Gadue P, Nishikura K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–8. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- 17.Wahlstedt H, Daniel C, Ensterö M, Öhman M. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 2009;19:978–86. doi: 10.1101/gr.089409.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higuchi M, Single FN, Köhler M, Sommer B, Sprengel R, Seeburg PH. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993;75:1361–70. doi: 10.1016/0092-8674(93)90622-W. [DOI] [PubMed] [Google Scholar]
- 19.Lind NM, Moustgaard A, Jelsing J, Vajta G, Cumming P, Hansen AK. The use of pigs in neuroscience: modeling brain disorders. Neurosci Biobehav Rev. 2007;31:728–51. doi: 10.1016/j.neubiorev.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 20.Kawahara Y, Ito K, Sun H, Ito M, Kanazawa I, Kwak S. Regulation of glutamate receptor RNA editing and ADAR mRNA expression in developing human normal and Down’s syndrome brains. Brain Res Dev Brain Res. 2004;148:151–5. doi: 10.1016/j.devbrainres.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 21.Galeano F, Leroy A, Rossetti C, Gromova I, Gautier P, Keegan LP, et al. Human BLCAP transcript: new editing events in normal and cancerous tissues. Int J Cancer. 2010;127:127–37. doi: 10.1002/ijc.25022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paschen W, Hedreen JC, Ross CA. RNA editing of the glutamate receptor subunits GluR2 and GluR6 in human brain tissue. J Neurochem. 1994;63:1596–602. doi: 10.1046/j.1471-4159.1994.63051596.x. [DOI] [PubMed] [Google Scholar]
- 23.Bernard A, Khrestchatisky M. Assessing the extent of RNA editing in the TMII regions of GluR5 and GluR6 kainate receptors during rat brain development. J Neurochem. 1994;62:2057–60. doi: 10.1046/j.1471-4159.1994.62052057.x. [DOI] [PubMed] [Google Scholar]
- 24.Ohlson J, Pedersen JS, Haussler D, Öhman M. Editing modifies the GABA(A) receptor subunit alpha3. RNA. 2007;13:698–703. doi: 10.1261/rna.349107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daniel C, Wahlstedt H, Ohlson J, Björk P, Öhman M. Adenosine-to-inosine RNA editing affects trafficking of the gamma-aminobutyric acid type A (GABA(A)) receptor. J Biol Chem. 2011;286:2031–40. doi: 10.1074/jbc.M110.130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanoue A, Koshimizu TA, Tsuchiya M, Ishii K, Osawa M, Saeki M, et al. Two novel transcripts for human endothelin B receptor produced by RNA editing/alternative splicing from a single gene. J Biol Chem. 2002;277:33205–12. doi: 10.1074/jbc.M203972200. [DOI] [PubMed] [Google Scholar]
- 27.Levanon EY, Hallegger M, Kinar Y, Shemesh R, Djinovic-Carugo K, Rechavi G, et al. Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res. 2005;33:1162–8. doi: 10.1093/nar/gki239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gommans WM, Tatalias NE, Sie CP, Dupuis D, Vendetti N, Smith L, et al. Screening of human SNP database identifies recoding sites of A-to-I RNA editing. RNA. 2008;14:2074–85. doi: 10.1261/rna.816908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silberberg G, Lundin D, Navon R, Öhman M. Deregulation of the A-to-I RNA editing mechanism in psychiatric disorders. Hum Mol Genet. 2012;21:311–21. doi: 10.1093/hmg/ddr461. [DOI] [PubMed] [Google Scholar]
- 30.Macbeth MR, Schubert HL, Vandemark AP, Lingam AT, Hill CP, Bass BL. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science. 2005;309:1534–9. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marcucci R, Brindle J, Paro S, Casadio A, Hempel S, Morrice N, et al. Pin1 and WWP2 regulate GluR2 Q/R site RNA editing by ADAR2 with opposing effects. EMBO J. 2011;30:4211–22. doi: 10.1038/emboj.2011.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Connell MA, Gerber AP, Zukin RS, Zukin RS, Paupard M-C. Patterns of developmental expression of the RNA editing enzyme rADAR2. Neuroscience. 2000;95:869–79. doi: 10.1016/s0306-4522(99)00431-5. [DOI] [PubMed] [Google Scholar]
- 33.Hang PN, Tohda M, Matsumoto K. Developmental changes in expression and self-editing of adenosine deaminase type 2 pre-mRNA and mRNA in rat brain and cultured cortical neurons. Neurosci Res. 2008;61:398–403. doi: 10.1016/j.neures.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Herb A, Higuchi M, Sprengel R, Seeburg PH. Q/R site editing in kainate receptor GluR5 and GluR6 pre-mRNAs requires distant intronic sequences. Proc Natl Acad Sci U S A. 1996;93:1875–80. doi: 10.1073/pnas.93.5.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhalla T, Rosenthal JJ, Holmgren M, Reenan R. Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nat Struct Mol Biol. 2004;11:950–6. doi: 10.1038/nsmb825. [DOI] [PubMed] [Google Scholar]
- 36.Lomeli H, Mosbacher J, Melcher T, Höger T, Geiger JR, Kuner T, et al. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–13. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 37.Olaghere da Silva UB, Morabito MV, Canal CE, Airey DC, Emeson RB, Sanders-Bush E. Impact of RNA editing on functions of the serotonin 2C receptor in vivo. Front Neurosci. 2010;4:26. doi: 10.3389/neuro.23.001.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niswender CM, Copeland SC, Herrick-Davis K, Emeson RB, Sanders-Bush E. RNA editing of the human serotonin 5-hydroxytryptamine 2C receptor silences constitutive activity. J Biol Chem. 1999;274:9472–8. doi: 10.1074/jbc.274.14.9472. [DOI] [PubMed] [Google Scholar]
- 39.Brockman W, Alvarez P, Young S, Garber M, Giannoukos G, Lee WL, et al. Quality scores and SNP detection in sequencing-by-synthesis systems. Genome Res. 2008;18:763–70. doi: 10.1101/gr.070227.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–64. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36(Web Server issue):W70-4. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–5. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 43.Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010, http://www.citeulike.org/user/oteri/article/9562772
- 44.Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, et al. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–8. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.