Abstract
Studies of miRNA association with Argonaute (AGO) proteins in mammalian cells have indicated lack of bias toward particular AGO. However, to our knowledge, the use of quantitative methods for studying miRNA association with different AGOs has not been reported so far. In this work we compared the total miRNA content in AGO1 and AGO2 immunoprecipitates obtained from MCF7 adenocarcinoma cells using TaqMan Low Density miRNA Arrays and successfully verified selected miRNAs with qPCR. For most of the miRNA species AGO1 and AGO2 profiles were well correlated, however, some miRNAs demonstrated consistent biases toward one of the Argonautes. Furthermore, miRNAs which were predominantly AGO2-associated derived mostly from sense strands of the corresponding pre-miRNAs while the majority of AGO1 biased miRNAs originated from antisense strands of the pre-miRNAs. Additionally, we show that circulating miRNA in human blood plasma can be immunoprecipitated with both AGO1 and AGO2 antibody. However, unlike in cell lysates, AGO1 and AGO2 associated miRNA profiles in plasma did not correlate, indicating that many cell types contribute to circulating miRNA (given that expression of AGO proteins is tissue specific). Furthermore, AGO-specific miRNA profiles in blood cells differed significantly from miRNAs profiles in plasma indicating that most circulating miRNAs are likely to derive from non-blood cells. Since circulating miRNAs hold great promise as biomarkers for numerous cancers and other diseases, we hypothesize that AGO-specific miRNA profiles might add an additional dimension to circulating miRNA-based diagnostics.
Keywords: Argonaute, blood, circulating, miRNA, plasma
Introduction
MicroRNAs (miRNAs) are small (19–24 nt) non-coding RNAs involved in post-transcriptional regulation in living cells by targeted hydrolysis of mRNAs or its translation inhibition.1 The maturation of miRNA starts with processing of primary non-coding transcript (pri-miRNAs) into 60–70 nt pre-miRNA hairpins by nuclease DROSHA/DGCR8.2,3 Upon transport into the cytoplasm pre-miRNAs are further cut by RNase III-like enzyme DICER1 into miRNA/miRNA* duplexes.4,5 Finally one strand of the miRNA/miRNA* duplex is loaded onto an Argonaute (AGO) protein within RNAi induced silencing complex (RISC).6-8 An Argonaute associated miRNA binds to complementary regions within targeted mRNA what leads to either AGO-mediated endonuclease cleavage of the mRNA or a reduction in translation efficiency.9
The regularity of miRNA sorting into distinct AGO proteins in mammalian cells is poorly understood. Early reports indicated that miRNAs are randomly sorted to individual Argonautes.10,11 However, RNA-sequencing of AGO1, AGO2 and AGO3 associated miRNAs revealed that some miRNAs can have a certain bias toward particular Argonaute.12,13 Furthermore, it remains unclear whether the average efficacy of miRNA loading is equal between different AGO proteins. To our knowledge application of qPCR-based methods for addressing differential miRNA sorting across distinct human AGO proteins were not reported so far. In this study we employed TaqMan Low Density miRNA Arrays and individual qPCR miRNA assays to profile miRNA following immunoprecipitation (IP) of AGO1 and AGO2 proteins from lysates of MCF7 human breast adenocarcinoma cells and MCF10a mammary epithelial cells. Our findings confirm that no significant bias in miRNA association toward particular AGO exists for most of the miRNAs. However, several miRNAs clearly demonstrated preferential association with certain AGO protein. We further discuss the possible mechanisms and implications of differential AGO sorting of certain miRNAs.
Extracellular miRNAs have recently been detected in human blood plasma and other body fluids as nuclease resistant entities and proved themselves as promising biomarkers for broad spectrum of human diseases including cancer.14-20 Furthermore, hundreds of publications regarding diagnostic potential of circuiting miRNA have appeared in the past four years indicating a significant interest in this emerging field. However, the cellular origin of circulating miRNA in the blood plasma remains largely unclear. More importantly, there is little information whether blood cells contribution to the plasma miRNAs is significant and whether miRNAs from blood cells may mask cancer-derived miRNAs.
Previous studies have indicated that extracellular miRNAs in blood plasma co-immunoprecipitates with anti-AGO2 antibody and remain stable for long time after cell death due to the unusual stability of AGO2 protein in nuclease-rich environment.21,22 The fact that expression of the four human AGO proteins is cell type and tissue specific led us to hypothesize the existence of distinct miRNA-AGO profiles in human extracellular body fluids. In consistence with our hypothesis, we observed a dramatic lack of correlation between AGO1 and AGO2 associated miRNA profiles in blood plasma indicating that different tissues and cells contribute to the extracellular miRNA content. Furthermore, we had shown that AGO-specific miRNA profiles in blood cells and plasma do not correlate for most of the miRNAs. Whether AGO-specificity of miRNA profiles might add additional value for biomarkers for diseases and/or organs damage remains to be elucidated.
Results
AGO1 and AGO2 miRNA profiles in cultured cells
To estimate patterns of miRNA association with AGO1 and AGO2 proteins we used monoclonal anti-AGO1 and anti-AGO2 antibodies to immunoprecipitate the corresponding Argonauts from lysates of MCF7 breast adenocarcinoma cells and analyzed the miRNA content in each immunoprecipitate using TaqMan Low Density miRNA Arrays (TLDA) from Applied Biosystems. Both antibodies recognized the corresponding proteins specifically on western blots and did not show any cross reactivity with other AGOs (data not shown). Moreover, successful use of the same anti-AGO1 and anti-AGO2 clones in coIP experiments has been consistently reported.13,23 Well proven antibodies for AGO3 and AGO4 proteins were not available at the time when the work was performed. More importantly, AGO3 and AGO4 mRNAs and proteins are expressed in dramatically lower level comparing to AGO1 and AGO2 in many cell types including MCF7 cells (Fig. S1). Therefore, we concentrated our analyses only on AGO1 and AGO2 proteins.
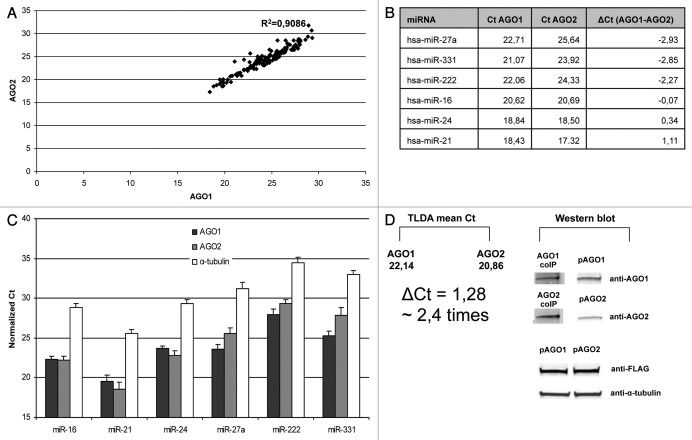
According to TLDA results AGO1 and AGO2 miRNA profiles generally showed good correlation in MCF7 cell line (Fig. 1A; Fig. S1). Out of 667 different miRNA species on the array 141 miRNAs were consistently detected in both biological replicates. Criteria for any miRNA being “detected” were: (1) Ct < 32 in either AGO1 or AGO2 immunoprecipitate; (2) no or at least 4 Ct values higher signals on negative control). Such cut-off parameters were chosen because we experienced difficulties in detecting miRNA with Ct > 32 in pre-amp TLDA using individual TaqMan miRNA qPCR assays. Out of 141 miRNAs 14 miRNAs were biased toward AGO1 [ΔCt (AGO1-AGO2) ≤ -1], 8 miRNAs were preferentially associated with AGO2 [ΔCt (AGO1-AGO2) ≥ 1] and 119 miRNAs were detected in approximately equal amounts in both AGO1 and AGO2 immunoprecpitates [-1 ≤ ΔCt (AGO1-AGO2) ≤ 1] (Table 1; Fig. S2). We have further verified several miRNA species using individual TaqMan qPCR miRNA assays (for miR-16, miR-21, miR-24, miR-27a, miR-222 and miR-331–3p) (Fig. 1B). The rationale for choosing these miRNA species for were: (1) relatively low Ct values on pre-amp TLDA (17–25); (2) miR-27a, miR-222 and miR-331 were significantly enriched in AGO1 immunoprecipitate; (3) miR-21 was among AGO2-biased miRNAs while miR-16 and miR-24 were approximately equally AGO1 and AGO2 dispersed (see criteria above). The difference in AGOs association of six selected miRNAs was consistently observed in three independent replicates not only in MCF7 but also in MCF10a (Fig. 1C; Fig. S3). This indicated that the observed differences in AGO1–AGO2 miRNA profiles were not MCF7 cell line specific.
Figure 1. Association of miRNAs with endogenous AGO1 and AGO2 proteins in MCF7 cells. (A) Correlation plot showing Ct values of miRNA signals detected in anti-AGO1 and anti-AGO2 pellets using TaqMan Low Density miRNA Arrays (TLDA). Note, the high correlation coefficient indicate low bias in binding toward AGO1 and AGO2 proteins for most miRNAs. (B) Ct values of the miRNA species on TLDA which were selected for verification with individual qPCR assays. (C) Individual TaqMan miRNA Assays of miR-16, miR-21, miR-24, miR-27a, miR-331 and miR-222 in anti-AGO1, anti-AGO2 and anti-α-tubulin (control) immunoprecipitates from MCF7 cells. Spiked in cel-miR-39 was used as control for isolation efficiency in all samples. Data are presented as Ct values normalized on both cel-miR-39 and the mean Ct of [miR-16, miR-21, miR-24, miR-27a, miR-331 and miR-222] within each sample (quantile normalization). Each bar represents mean + SD of three independent experiments. (D) Estimation of average miRNA association with each AGO protein. The means of Ct values of miRNAs in either anti-AGO1 or anti-AGO2 samples detected on the array (left) were compared with the relative content of AGO1 and AGO2 proteins detected by western blot (right) in the same immunopellets. To account for the difference in anti-AGO1 and anti-AGO2 antibody efficiency on western blot, equal amounts of recombinant FLAG-AGO1 (pAGO1) and FLAG-AGO2 (pAGO2) proteins were applied on the SDS gel. Note, the content of AGO1 protein in the anti-AGO1 coIP pellet was lower as compared with AGO2 protein in anti-AGO2 coIP pellet (~2,5 times as analyzed by densitometry). Accordingly, the observed difference in the mean Ct values between AGO1 and AGO2 coIP pellets corresponded to approximately 2,5 times difference in the total miRNA content.
Table 1. TaqMan Low Density miRNA Arrays data on miRNAs significantly enriched in either anti-AGO1 or anti-AGO2 coIP pellets.
| miRNA | Ct AGO1 | Ct AGO2 | ΔCt (AGO1-AGO2) | Pre-miRNA strand |
|---|---|---|---|---|
| hsa-miR-27a-4373287 |
22,7108326 |
25,6429944 |
-2,932161805 |
3p |
| hsa-miR-331–3p-4373046 |
21,0664601 |
23,9169649 |
-2,850504805 |
3p |
| hsa-miR-7–1*-4381118 |
25,6456706 |
28,4489174 |
-2,803246805 |
3p |
| hsa-miR-223–4395406 |
28,9383351 |
31,6983179 |
-2,759982805 |
3p |
| hsa-miR-222–4395387 |
22,0658651 |
24,3264319 |
-2,260566805 |
3p |
| hsa-miR-671–3p-4395433 |
26,4881046 |
28,4872449 |
-1,999140305 |
3p |
| hsa-miR-744–4395435 |
24,7962121 |
26,4697534 |
-1,673541305 |
5p |
| hsa-miR-30e*-4373057 |
23,5356746 |
24,9752734 |
-1,439598805 |
3p |
| hsa-miR-526b*-4395494 |
29,2333581 |
30,6153529 |
-1,381994805 |
3p |
| hsa-miR-30a*-4373062 |
24,9439171 |
26,2166564 |
-1,272739305 |
3p |
| hsa-miR-301b-4395503 |
27,3119716 |
28,5231709 |
-1,211199305 |
3p |
| hsa-miR-328–4373049 |
23,9407611 |
25,0871804 |
-1,146419305 |
3p |
| hsa-miR-101–4395364 |
26,6729960 |
27,8188929 |
-1,145896804 |
3p |
| hsa-miR-345–4395297 |
22,6739915 |
23,7664224 |
-1,092430804 |
5p |
| |
|
|
|
|
| hsa-miR-200b-4395362 |
23,3568091 |
22,2788469 |
1,077962195 |
3p |
| hsa-miR-7–4378130 |
27,8013806 |
26,7211454 |
1,080235195 |
5p |
| hsa-miR-21–4373090 |
18,4337241 |
17,3231909 |
1,110533195 |
5p |
| hsa-miR-20b-4373263 |
24,2134021 |
23,0771789 |
1,136223195 |
5p |
| hsa-miR-20a-4373286 |
20,6647581 |
19,5261814 |
1,138576695 |
5p |
| hsa-miR-590–5p-4395176 |
24,7511536 |
23,6107949 |
1,140358695 |
5p |
| hsa-miR-106a-4395280 |
19,8540316 |
18,6838149 |
1,170216695 |
5p |
| hsa-miR-17–4395419 | 19,7704171 | 18,4849974 | 1,285419695 | 5p |
Upper row–miRNAs that are preferentially AGO2 associates are SENSE miRNAs. Lower row, miRNAs that are preferentially AGO1 associated are ANTISENSE miRNAs.
Both AGO1 and AGO2 proteins were detectable in the immunoprecipitates obtained from MCF7 cells on western blot (Fig. 1D). To estimate relative levels two Argonautes it their corresponding immunoprecipitates we used equal amounts of FLAG-AGO1 and FLAF-AGO2 proteins to account for the difference in anti-AGO1 and anti-AGO2 staining efficiency. The precise quantification of miRNA binding to AGO1 and AGO2 (e.g., using mass spectrometry) was beyond the scope of this paper. However, the content of AGO1 protein in anti-AGO1 precipitates was approximately 2.5 times lower as compared with AGO2 protein in anti-AGO2 pellets (as analyzed by quantification of bands intensities relative to reference controls on western blot). In the same IP samples the means of TLDA Ct values were 22.1 and 20.9 for AGO1 and AGO2 respectively, what corresponds to approximately 2.4 times difference in miRNA content between the immunoprecipitates. We, therefore, conclude that the efficacy of miRNAs binding to the AGO1 and AGO2 proteins was approximately equal.
According to the previous report, several miRNA species analyzed after immunoprecipitatation of exogenously overexpressed epitope-tagged human AGO proteins did not show any bias toward either AGO1 or AGO2.10 In our hands the differences in miRNAs association with exogenously overexpressed AGO1 and AGO2 were not as evident as for endogenous AGOs when the content of miR-16, miR-21, miR-24, miR-27a, miR-222 and miR-331–3p was analyzed in anti-FLAG immunoprecipitates obtained from MCF7 cells overexpressing FLAG-AGO1 and FLAG-AGO2 proteins (data not shown). However, as the expression of exogenous Argonautes was drastically higher (8–7 Ct values less; ~256–128 times more) as compared with endogenous Argonauts what might be far beyond the physiological conditions.
We next addressed the question whether any particularities in miRNA structure can determine preferential binding to AGO1 and AGO2 proteins. Surprisingly, miRNA which were significantly biased toward AGO1 protein (see criteria above) derived mostly from the antisense strand of the corresponding pre-miRNAs while AGO2-biased miRNAs were mostly from sense strands (Table 1; Fig. S2). Thus, 7 out of 8 Ago2–biased miRNAs with ΔCt (AGO1-AGO2) > 1 were 5p-miRNAs, while among 14 AGO1-biased miRNAs [ΔCt (AGO1-AGO2) < -1] 12 derived from 3′-strands of the corresponding pre-miRNAs (3p-miRNAs). Since those miRNA species which showed no bias toward one of the AGO were both 5p and 3p miRNAs, the sense-antisense strand selectivity was not the only requirement for preferential AGO1 or AGO2 association.
AGO1 and AGO2 miRNA profiles in blood plasma
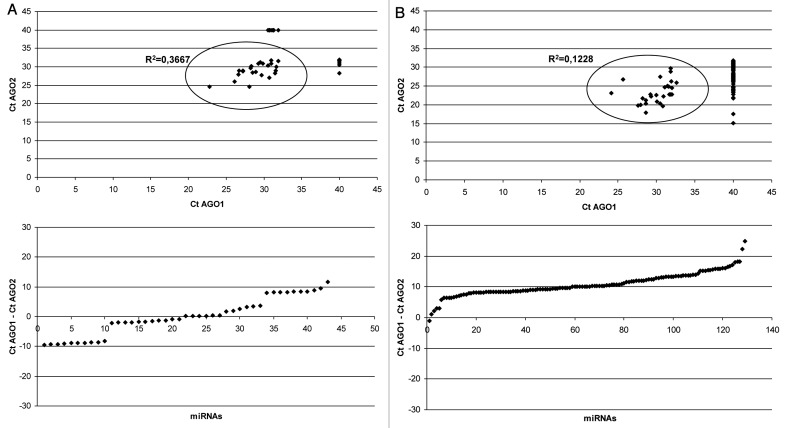
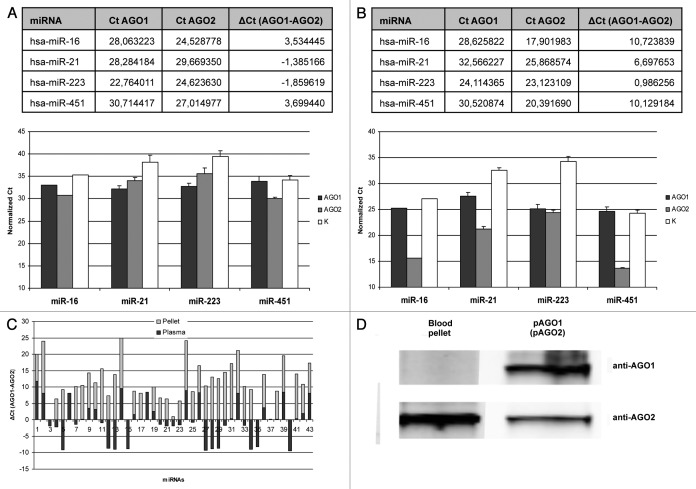
Unlike in cultured cells, human blood plasma AGO1 and AGO2 associated miRNAs profiles were very distinct (Fig. 2A; Fig. S4). Among 43 miRNAs detected on the arrays 19 (44%) were enriched in anti-AGO1 immunoprecipitates [ΔCt (AGO1-AGO2) ≤ -1], 16 miRNAs (37%) showed bias toward AGO2 [ΔCt (AGO1-AGO2) ≥ 1] and 8 miRNAs were detected in approximately equal amounts in both immunoprecpitates [-1 ≤ ΔCt (AGO1-AGO2) ≤ 1]. In another biological replicate (blood plasma from different individual) among 30 detected miRNAs 12 miRNAs (40%) and 13 miRNAs (43%) were AGO1 and AGO2 biased respectively (Fig. S4A). The correlation in miRNA content between samples from two different individuals was average (R2 = 0.61 for AGO1 immunoprecipitates and R2 = 0.75 for AGO2 immunoprecipitates) (Fig. S4C and D). Moreover, some miRNA species were uniquely present in the plasmas of particular individual (Fig. S4C and D). The results were further verified using individual TaqMan qPCR miRNA assays for miR-16, miR-21, miR-223 and miR-451 (Fig. 3A). These miRNA species were chosen due to their relatively low Ct values/high abundance in blood plasma what makes them stable detectible using individual miRNA assays. Additionally, these miRNAs were highly correlated between two different blood plasmas. In consistence with arrays data, significantly higher amounts of miR-16 and miR-451 were detected in AGO2 immunoprecipitates while miR-223 and miR-21 content was higher in AGO1 pellets as analyzed in three biological replicates (Fig. 3A). In general, lack of correlation between AGO1 and AGO2 bound miRNAs in blood plasma can be attributed to the fact that the different cell types and tissues/organs which contribute to circulating miRNA in plasma express AGO proteins in different proportions (and at the same time exhibit tissue specific miRNA profiles).
Figure 2. TaqMan miRNA Low Density Arrays performed on total RNA extracted from anti-AGO1 and anti-AGO2 immunoprecipitates of blood plasma (A) and whole blood pellet (B). Blood plasma (80 μL) was combined with 20 μg the antibody. Blood pellet lysate (5 μL) combined with 3 μg of the antibody. MiRNAs which were undetected on the array (Ct > 32) were assigned Ct values of 40. Note, (1) some miRNAs were presented exclusively in the coIP pellets of particular AGO protein; (2) unlike in blood plasma, in blood pellet overwhelming majority of miRNAs were AGO2 associated.
Figure 3. (A) miRNA species in the blood plasma which showed significant bias toward either AGO1 (miR-21, miR-223) or AGO2 (miR-16, miR-451) proteins on TaqMan Low Density miRNA Array (above) and their verification with individual miRNA qPCR assays (below) in three independent experiments; (B) The same miRNA species demonstrated drastically different AGO1/AGO2 distribution in whole blood pellet: TLDA (above), individual qPCR assays (below). Data presented as Ct values normalized on cel-miR-39 and miR-16. Each bar represents mean + SD of three independent experiments (samples from different individuals). (C) The ΔCt (AGO1-AGO2) graph of miRNAs detected in both blood plasma and blood pellet. Note, for most of the miRNA there is no correlation of ΔCt (AGO1-AGO2) between plasma and pellet; (D) western blot analysis of AGO1 and AGO2 proteins in anti-AGO1 and anti-AGO2 coIP samples respectively. Note, unlike AGO2 protein, the amount of AGO1 in the immuprecipitate was below the detection limit.
It is worth noticing that blood samples were immediately processed and a second high-speed centrifugation step was applied in order to receive cell and cell-debris free plasma. This has been shown to be critical when analyzing circulating miRNA since blood cells miRNAs can significantly contaminate plasma if not processed properly.24-27
AGO1 and AGO2 miRNA profiles in blood pellet
Blood cells constitute about 45% of total blood volume and thus could be major contributors to extracellular plasma miRNA. To test this hypothesis we investigated if AGO1 and AGO2 associated miRNAs in plasma parallels the corresponding profiles in whole blood pellet. MiRNA from blood pellets (the red blood pellet + buffy coat remained after centrifuging the whole blood at 1.300 g) was co-immunoprecipitated using either anti-AGO1 or anti-AGO2 antibody. The data obtained in two independent replicates indicated that, as in blood plasmas, in blood pellets the AGO1 vs. AGO2 binding miRNA profiles did show poor correlation for most miRNAs (Fig. 2B; Fig. S5). However, in blood pellet a very strong, almost exclusive association of miRNAs with AGO2 was observed. Out 129 miRNA species only 27 were detected in AGO1 immunoprecipitates while 129 miRNAs were present in AGO2 pellets (for another biological replicate−41 and 126 respectively out of 128 detected miRNAs). Furthermore, AGO2 bound miRNAs had drastically lower Cp values as compared with miRNAs in AGO1 precipitates. Only 1 (0,8%) miRNA was enriched in anti-AGO1 immunoprecipitates [ΔCt (AGO1-AGO2) ≤ -1], 127 (98%) miRNAs showed bias toward AGO2 [ΔCt (AGO1-AGO2) ≥ 1] and 1 miRNA was detected in approximately equal amounts in both immunoprecpitates [-1 ≤ ΔCt (AGO1-AGO2) ≤ 1]. In another biological replicate (blood pellet from different individual) 2 (1,6%) and 116 (91%) miRNAs were AGO1 and AGO2 biased respectively while 10 miRNAs showed approximately equal AGO1/AGO2 distribution (Fig. S5).
The correlation coefficients (R2) of miRNAs for two independent blood pellets were 0.71 and 0.88 for AGO1 and AGO2 immunoprecipitates respectively and several miRNAs were uniquely present in the blood pellet of a particular individual (Fig. S5C and D).
The observed difference in miRNA amount between the AGO1 and AGO2 immunoprecipitates is also reflected by dramatically higher concentration of AGO2 protein in the blood pellet as compared with AGO1. In contrast to AGO2 protein we failed to detect any amounts of AGO1 protein in whole blood pellet on western blots (Fig. 3D). Interestingly, a few miRNAs including miR-223 had similar Cp values in AGO1 and AGO2 immunoprecipitates suggesting that those miRNAs are predominantly AGO1 bound (provided that the amount of AGO1 protein is dramatically lower in the blood pellet as compared with AGO2). The difference in AGO1 vs. AGO2 miRNA content in blood pellets was further verified using four individual TaqMan qPCR miRNA assays for miR-16, miR-21, miR-223 and miR-451 in three biological replicates (Fig. 3B). Importantly, the Ct values of miRNAs immunoprecipitated with anti-AGO2 antibody from 80 μL of blood plasma were ~10–15 Ct values higher than for miRNAs immunoprecipitated from as little as 5 μL of blood pellet. Such huge difference in miRNAs content in the blood pellet vs. plasma should be taken into account when isolating blood plasma for miRNA analysis. Obviously, a disruption of even a small number of blood cells (e.g., hemodialysis) can contaminate blood plasma with irrelevant miRNAs. This hypothesis is also confirmed by Kirschner et al., who demonstrated that the plasma levels of miR-16 and miR-451 (miRNAs which are present in red blood cells) showed huge variation in samples with the signs of hemolysis. In the absence of hemolysis the levels of both miR-16 and miR-451 were sufficiently constant to serve as normalizers.27 Our observation that human erythrocytes (~99% of cells in the blood pellet) contain large quantity of miRNA was also demonstrated before,26,28 however, the fact that most of it is AGO2 associated was unexpected.
Differential AGO1 and AGO2 miRNA profiles in blood plasma and blood pellet
Another important observation was that many miRNA species which were highly presented in blood pellet was not detected in the blood plasma and vice versa (Figs. S5 and S6). Among those miRNAs which have been detected in both blood plasma and blood pellet the correlation of their ΔCt (AGO1-AGO2) values was absent for the majority of miRNAs (Fig. 3C). Only 14 out of 43 miRNAs showed similar ΔCt (AGO1-AGO2) in both plasma and pellet. We verified the content of miR-146a, one of the miRNAs which demonstrated reverse correlation of ΔCt (AGO1-AGO2) in blood plasma (ΔCt = -1.34) and in the pellet (ΔCt = 10.24). Consistently, in blood plasmas from different individuals miR-146a was enriched in AGO1 IP samples while in the blood pellets it was predominately AGO2 bound (Fig. S6). Therefore, some miRNAs in the plasma do not derive from blood cells under normal conditions (no disruption of blood cells). It is likely that different organs and cell types (especially those having high contact with the blood) contribute to the extracellular miRNA content. For instance, miR-122 constitutes about 75% of all miRNAs in human liver and its amount in the blood plasma is elevated after liver injury.15,29 We consistently detected miR-122 in AGO1 precipitates obtained from blood plasmas with Cp values of ~28 while in the blood pellets miR-122 was not detected (Figs. S4 and S5).
Discussion
The regularity of the association of human AGO proteins with different miRNAs is poorly understood. Intracellular binding of several investigated miRNA species was found to be unbiased toward exogenously overexpressed Argonautes in early reports.10,11 However, sensitive qPCR methods for miRNA detection and quantification were not available at that time. Deep sequencing of total RNA extracted from endogenous human AGO1, AGO2 and AGO3 immunoprecipitats revealed that miRNAs associate with approximately equivalent efficacy with three Argonauts, however, with few exceptions.12,13 In accordance with the13 study we found miR-222 to be associated predominantly with AGO1. We measured the relative amount of miRNAs in anti-AGO1 and anti-AGO2 immunoprecipitates using qPCR based techniques in MCF7 and MCF10a cells. We confirmed that on average miRNAs associate with AGO1 and AGO2 proteins with equal efficacy. However, our observation that many miRNA species were significantly biased toward particular AGO proteins were unexpected. We further noticed that AGO1-biased miRNAs were predominantly (but not all) 3p (derived from antisense strand of corresponding pre-miRNA), while AGO2-biased miRNAs were mostly 5p (derived from sense strand of pre-miRNA). Our data also indicate that majority of miRNAs associate with AGO1 and AGO2 proteins indiscriminately of their sequence. Importantly, only AGO2 possess target RNA cleavage activity while AGO1, AGO3 and AGO4 are only capable to guide translational repression. Interestingly, the degree of complementarity between the miRNA/miRNA* strands defines how miRNAs are sorted into AGO1 and AGO2 proteins in D. melanogaster.30,31 However, in our experiments both AGO1 and AGO2 biased miRNAs shared similar patterns of sequence complementarily (as judged by miRNA/miRNA* predicted structures available at www.mirbase.org).
Differential expression levels of four human AGO proteins in different cell lines may point to further complexity of regulation of RNA silencing in a cell-type specific manner.13 Thus, the analysis of the protein and mRNA levels of endogenous Argonauts in several cell lines have indicated similar content of AGO1 and AGO2 but drastically lower level of AGO3 and AGO4. Furthermore, the efficacy of AGO3 and AGO4 overexpression using recombinant constructs in cell lines were dramatically lower than of AGO1 and AGO2 on both mRNA and protein level.10,32 It is therefore conceivable that AGO3 and AGO4 mediated miRNA pathway do not play significant role in somatic cells.
Unlike in mammalian cells, the difference in miRNA sorting into distinct AGO proteins has been consistently shown in fruit fly and C. elegans. In D. melanogaster, most mature miRNA sequences associate with AGO1 while most antisense miRNA* sequences either degraded or associate with AGO2.33,34 In C. elegans, distinct miRNA associate with different AGO-like proteins.35,36 Our observation that in human cells AGO1 and AGO2 biased miRNAs derived mostly from 3p (antisense) and 5p (sense) strands of pre-miRNA respectively may indicate on the presence of selective sorting mechanism for some miRNAs in mammals which is yet to be studied in more details.
Extracellular circulating miRNA had been recently found in human body fluids and hold great promise as a new class of diseases biomarkers due to their high stability in nuclease rich environment.14,22 It has been recently shown that most of the circulating miRNA in blood plasma is microvesicles free and bound to Argonaut proteins.21,22 However, little is known about the origin of circulating miRNAs and what factors may influence levels of circulating miRNA biomarkers. Previous studies also demonstrated that at least some miRNAs are likely to originate from different blood cells.26,27 Since blood cells including erythrocytes constitute about 45% of total blood volume we hypothesized that they can be the main contributors to miRNA in plasma. However, unlike in blood plasma, the miRNA content in AGO1 immunoprecipitates obtained from whole blood cells fraction was dramatically lower than in AGO2 immunoprecipitates. This can be explained by the fact that AGO2 protein is present at significantly higher levels in the blood pellet as compared with AGO1. Furthermore, most AGO2 associated miRNAs in blood pellet and plasma did not correlate on a whole-profile level. Our notion that blood cells (presumably erythrocytes only) are packed with AGO2-associated miRNA strongly support a recent report indicating that procedures disrupting blood cells (e.g., hemolysis) alters plasma miRNA level significantly and masks diseases derived miRNAs.26,27 We suggest that immunoenrichment using anti-AGO1 antibody may help to reduce the masking of the “true” disease-derived miRNA.
Leukocytes and platelets together constitute less than 1% of total blood volume and it remains to be tested whether they can significantly impact the profile of extracellular plasma miRNA. However, blood plasma has more extensive contacts with other cell types of the body (e.g., liver, spleen, and kidney) than it has with leukocytes and platelets. Therefore, it is unlikely that white blood cells and platelets are the major contributors to extracellular miRNA content in normal conditions. Furthermore, since the expression of four AGO proteins is tissue specific37-39 differential profiles of AGO1 and AGO2 associated miRNAs in blood plasma, observed in our experiments, support a multi-organ origin of circulating miRNA.
The observation that particular miRNA can be simultaneously enriched or depleted across two AGO proteins suggest that blood plasmas have not only their miRNA signatures, but also miRNA-AGO signatures. For instance, during cytotoxicity in particular tissue or organ having high expression of particular AGO protein, an elevated level of miRNAs associated with the corresponding AGO can be anticipated. Whether individual miRNA-AGO profiles can be of any use for molecular diagnosis of diseases (besides total miRNA profiles) remains to be elucidated. Emerging data consistently suggest that other two human Argonautes–AGO3 and AGO4 are expressed at dramatically lower levels in most organs and tissues.37-39 Thus, it is conceivable that blood plasma miRNA are predominantly AGO1 and AGO2 associated. However, germ-line cells express relatively high quantities both AGO3 and AGO4 mRNA.38 Therefore, elevation of AGO3 and AGO4 level in the blood plasma (or other body fluids) may indicate cytotoxicity or cancer in these organs.
Materials and Methods
Cell culture and transfection
All cell lines were obtained from the American Type Culture Collection (ATCC). Human MCF7 breast adenocarcinoma cells were grown in αMEM supplemented with 10% FBS, 1% non-essential amino acid and penicillin/streptomycin at 37°C in 5% CO2. Human MCF10a cells were grown in DMEM/F12 supplemented with 5% Horse Serum, 20 ng/mL EGF, 0.5 mg/mL Hydrocortisone, 10 μg/mL Insulin, 100 ng/mL Cholera Toxin and penicillin/streptomycin. For the overexpression of AGO proteins HEK 293FT cells (maintained in DMEM, 10% FBS, 1% non-essential amino acid and penicillin/streptomycin) were transiently transfected with pIRESneo-FLAF-HA-Ago1 or pIRESneo-FLAF-HA-Ago2 using Turbofect transfection reagent (Fermentas) according to the manufacturer’s protocol. Next day after transfection cells were lysed in either Laemmli loading buffer or Qiazol reagent and used for western blot analysis. For the immunoprecipitation experiments ~5 × 106 cells were lysed in 1 mL of Lysis/Wash buffer containing 1% NP.
Collection of human blood, plasma and blood cell pellet
EDTA-Blood was collected from healthy donors and processed for plasma isolation immediately after collection. The total blood was centrifuged at 1.300 g for 20 min at 10°C and the plasma was immediately replaced into the new tube. To remove any cells and cell-debris blood plasma was further centrifuged at 16.000 g for 30 min before co-immunoprecipitation and RNA isolation. For a single immunoprecipitation reaction 80 μL of blood plasma were used. The blood pellets remaining after the 1.300 g centrifugation step, consisted of the red blood cells fraction and the intermediate buffy coat (leucocytes and platelets), were thoroughly mixed to achieve the homogenous distribution of different blood cells in the pellet. Blood pellets were lysed in 9V of Lysis/Wash buffer containing 1% NP, cleared by the centrifugation at 16.000 g for 10 min and 50 μL of the lysate (corresponds to 5 μL of total pellet) were used per immunoprecipitation reaction. This study has been approved by the ethics committee, Heidelberg, Germany.
Immunoprecipitation
Immunoprecipitation of miRNA was performed using monoclonal anti-AGO1 (clone 4B8, Sigma) and anti-AGO2 (clone 11A9, Sigma) produced in rat using Pierce Classic IP kit (Thermo Scientific). Antibody to α-tubulin (Sigma) was used in control samples. To form AGO/anti-AGO complexes, 300 μL (600 μg) of MCF7 cells lysates or 50 μl of blood cells lysates were combined with 3 μg of antibody and incubated overnight at +4°C. Blood plasma (80 μL per reaction) was combined with 20 μg of antibody and incubated overnight at +4°C. To capture the immune complexes, 20 μL Pierce Protein A/G Agarose suspended in 300 μL of IP Lysis/Wash buffer was added to the antibody/lysate samples and incubated for 2 h on a shaker at RT. The A/G Agarose resin was washed two times with 1 mL of IP Lysis/Wash buffer, lysed in 700 μL of Qiazol and processed with the miRNeasy kit (Qiagen) as described below.
Isolation of MicroRNAs
Isolation of miRNA from protein A/G Agarose was performed using miRNeasy kit (Qiagen). Briefly, 700 mL of Qiazol reagent were added to immunoprecipitates, mixed by vigorous shaking for 10 sec and incubated for 10 min at RT to ensure complete dissociation of nucleoprotein complexes. Then, (1) 5 pg of synthetic miRNA-39 from C. elegans (cel-miRNA-39) was added as a spike-in control for miRNA isolation efficiency; (2) 1.2 μL of Glycogen (10 mg/mL) were added to enhance the efficiency of RNA column binding. After addition of 140 μL of chloroform, the mixture was vigorously shaken for 45 sec and allowed to stand for 5 min at RT. Following the centrifugation at 16.000 g for 20 min total RNA was precipitated from the upper (aqueous) phase by the addition of 1.5 V of 100% ethanol. Purification of extracted total RNA was performed with miRNeasy columns (Qiagen) according to the manufacturer's instructions. RNA was eluted in 50 μL of RNase-free water.
miRNA profiling
The content of miRNA in anti-AGO1 and anti-AGO2 immunoprecipitates was profiled using stem-loop RT-PCR based TaqMan Human MicroRNA Arrays (Applied Biosystems). The arrays represent 667 mature miRNAs and miRNAs* present in the Sanger miRBase v12 in a two-card set of arrays (Array A and B). RT-PCR reactions were performed according to the manufacturer's instructions. Briefly, total RNA (3 μL per reaction) was reverse transcribed using the TaqMan miRNA Reverse Transcription Kit (Applied Biosystems) in combination with the stem-loop Megaplex primers pool sets A and B in total volume of 7.5 μL. The reverse transcription cycling conditions were: 40 cycles of 16°C for 2 min, 42°C for 1 min and 50°C for 1 sec. Megaplex RT products were further pre-amplified using TaqMan PreAmp Master Mix and Megaplex PreAmp primers (Applied Biosystems). Briefly, 2.5 μL of the Megaplex RT products were mixed with 2.5 μl of Megaplex PreAmp Primers (pool A or B) and 12.5 μl TaqMan PreAmp Master Mix in a 25 μl PCR reaction. The pre-amplification cycling conditions were 95°C for 10 min, 55°C for 2 min and 75°C for 2 min followed by 12 cycles of 95°C for 15 sec and 60°C for 4 min. The pre-amplified cDNA was diluted with ddH2O to 100 μl and 10 μl of diluted cDNA was used in each plate for real-time PCR reactions. RNA isolated from blood pellet immunoprecipitates was not pre-amplified. The expression profiles of miRNA were acquired using TaqMan Low-Density Arrays. Briefly, quantitative real-time PCR was performed on Applied BioSystems 7900HT thermocycler using the manufacturer’s recommended cycling conditions. Cycle threshold (Ct) values were calculated with the SDS software v2.3 using automatic baseline settings with an assigned minimum threshold of 0.2. Those miRNAs that showed close Ct values (ΔCt < 4, on the negative control samples arrays were removed from the analysis. TaqMan Array experiments on MCF7 cells lysates were performed using two biological replicates (lysates from two independent passages) and selected miRNA species (miR-16, miR-21, miR-24, miR-27a, miR-222, miR-331) were verified using individual TaqMan miRNA Assays (Applied Biosystems) in three independent biological replicates (lysates from three independent passages). Raw Ct values from two replicates were quantile normalized. TaqMan Array experiments on blood plasmas and blood pellets were performed using two independent samples (different individuals) and shown separately in Supplemental Material. Selected miRNAs (miR-16, miR-21, miR-223, miR-451) in immunoprecipitates from blood pellets and blood plasmas were verified on three independent replicates using individual TaqMan miRNA Assays (Applied Biosystems).
Quantitative real-time PCR
A fixed volume of 5 μl of total RNA from 50 μL eluates obtained after RNA isolation was used as an input into a reverse transcription (RT) reaction. The levels of mature hsa-miR-16, hsa-miR-21, hsa-miR-24, hsa-miR-27a, hsa-miR-222, hsa-miR-223, hsa-miR-331, hsa-miR-451 and cel-miR-39 were measured using individual TaqMan microRNA Assays (Applied Biosystems) according to the manufacturer’s instructions with minor modifications. All Ct values were normalized on cel-miR-39 signals to account for difference in isolation efficacy. Reverse transcriptase reactions contained 5 μL of purified total RNA, 50/N nM stem–loop RT primers (where N is a number of analyzed miRNAs), 1 × RT buffer, 0.25 mM each of dNTPs, 3.33 U/µl MultiScribe reverse transcriptase and 0.25 U/µl RNase inhibitor. RT reaction had final volume of 15 µL were incubated for 30 min at 16°C, 30 min at 42°C, 5 min at 85°C and then held at 4°C. RT product was further diluted six times with RNase-free water. Real-time PCR was performed using Taqman Universal PCR Master Mix. A 10 µL PCR reaction included 2 µl of diluted RT product, 1x TaqMan Universal PCR Master Mix (Applied Biosystems) and 1x of the corresponding miRNA assay primers. The reactions were incubated in 384-well plates at 95°C for 10 min, followed by 50 cycles of 95°C for 15 ses and 60°C for 1 min. All reactions were run in duplicate. Real-time PCR was performed using the LightCycler 480 Real-Time PCR System (Roche). Data was analyzed with the LightCycler 480 software (Roche), determining the threshold cycle (Ct) by the second derivative max method.
Western blot analysis
Pellets from obtained after immunoprecipitation with anti-AGO1 or anti-AGO2 antibody or HEK 293FT cells transfected with pIRESneoAGO1 or pIRESneoAGO2 were lysed in Laemmli loading buffer (250 mM Tris–HCl pH 6.4; 2% SDS; 100 nM b-MeEtOH), separated SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked in Odyssey blocking buffer (LI-COR) for 1h at RT. Incubation with rabbit polyclonal anti-FLAG (Sigma), rat anti-AGO1 (clone 4B8, Sigma), rat anti-AGO2 (clone 11A9, Sigma) or mouse anti-α-tubulin (Sigma) was performed at 4°C overnight in a 1:1000 dilution. After incubation with the primary antibody, membranes were washed twice in 1 × PBST for 10 min and incubated for 1h at RT with infrared fluorescent goat anti-rabbit or goat anti-mouse IgG diluted in blocking solution. Membranes were washed four times for 10 min in PBST and signals were then acquired with Odyssey infrared imaging system (LI-COR).
Conclusion
In this work we confirmed that in human cells the efficiency of miRNA binding toward AGO1 and AGO2 is similar for most miRNAs. However, many miRNA species demonstrated clear bias toward one of the Argonautes. Furthermore, we observed a dramatic difference in AGO1 and AGO2 associated miRNA profiles in blood plasma. The lack of correlation between AGO1 and AGO2 miRNA content in the plasma can be explained by the fact that many tissues contribute to the extracellular miRNA content. Unlike in blood plasma, in blood cells almost all miRNAs were AGO2 bound suggesting that plasma miRNA is unlikely to derive mostly from blood cells. The cells fraction of the blood is heterogeneous and dominated by red blood cells. Moreover different cell types have different lifetime in the blood. This can explain the very low correlation between AGO1 and AGO2 bound miRNAs in the whole blood. We hypothesize that individual AGO profiles of extracellular miRNA can be used as biomarkers for diseases and/or organs damage.
Supplementary Material
Acknowledgments
This work was supported by the Dietmar-Hopp Foundation, the Helmholtz Society and the German Cancer Research Center. Authors’ contributions: AT designed and performed experimental part and wrote this manuscript; BB conceived of the study, participated in its design, data analysis, coordination and revision of the manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/21083
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 3.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–8. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 5.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 6.Mallory AC, Elmayan T, Vaucheret H. MicroRNA maturation and action--the expanding roles of ARGONAUTEs. Curr Opin Plant Biol. 2008;11:560–6. doi: 10.1016/j.pbi.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 8.Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Höck J, Meister G. The Argonaute protein family. Genome Biol. 2008;9:210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–97. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–41. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 12.Azuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, et al. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci U S A. 2008;105:7964–9. doi: 10.1073/pnas.0800334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Suzuki H, Hayashizaki Y, et al. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol. 2011;8:158–77. doi: 10.4161/rna.8.1.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A. 2009;106:4402–7. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118–26. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 17.Zhao H, Shen J, Medico L, Wang D, Ambrosone CB, Liu S. A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer. PLoS One. 2010;5:e13735. doi: 10.1371/journal.pone.0013735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roth P, Wischhusen J, Happold C, Chandran PA, Hofer S, Eisele G, et al. A specific miRNA signature in the peripheral blood of glioblastoma patients. J Neurochem. 2011;118:449–57. doi: 10.1111/j.1471-4159.2011.07307.x. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Jia Y, Zheng R, Guo Y, Wang Y, Guo H, et al. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin Chem. 2010;56:1830–8. doi: 10.1373/clinchem.2010.147850. [DOI] [PubMed] [Google Scholar]
- 20.Brase JC, Johannes M, Schlomm T, Fälth M, Haese A, Steuber T, et al. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer. 2011;128:608–16. doi: 10.1002/ijc.25376. [DOI] [PubMed] [Google Scholar]
- 21.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–8. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–33. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beitzinger M, Meister G. Experimental identification of microRNA targets by immunoprecipitation of Argonaute protein complexes. Methods Mol Biol. 2011;732:153–67. doi: 10.1007/978-1-61779-083-6_12. [DOI] [PubMed] [Google Scholar]
- 24.McDonald JS, Milosevic D, Reddi HV, Grebe SK, Algeciras-Schimnich A. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem. 2011;57:833–40. doi: 10.1373/clinchem.2010.157198. [DOI] [PubMed] [Google Scholar]
- 25.Duttagupta R, Jiang R, Gollub J, Getts RC, Jones KW. Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLoS One. 2011;6:e20769. doi: 10.1371/journal.pone.0020769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 2012;5:492–7. doi: 10.1158/1940-6207.CAPR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirschner MB, Kao SC, Edelman JJ, Armstrong NJ, Vallely MP, van Zandwijk N, et al. Haemolysis during sample preparation alters microRNA content of plasma. PLoS One. 2011;6:e24145. doi: 10.1371/journal.pone.0024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen SY, Wang Y, Telen MJ, Chi JT. The genomic analysis of erythrocyte microRNA expression in sickle cell diseases. PLoS One. 2008;3:e2360. doi: 10.1371/journal.pone.0002360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–13. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 30.Förstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130:287–97. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valdmanis PN, Gu S, Schüermann N, Sethupathy P, Grimm D, Kay MA. Expression determinants of mammalian argonaute proteins in mediating gene silencing. Nucleic Acids Res. 2012;40:3704–13. doi: 10.1093/nar/gkr1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Czech B, Zhou R, Erlich Y, Brennecke J, Binari R, Villalta C, et al. Hierarchical rules for Argonaute loading in Drosophila. Mol Cell. 2009;36:445–56. doi: 10.1016/j.molcel.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okamura K, Liu N, Lai EC. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell. 2009;36:431–44. doi: 10.1016/j.molcel.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steiner FA, Hoogstrate SW, Okihara KL, Thijssen KL, Ketting RF, Plasterk RH, et al. Structural features of small RNA precursors determine Argonaute loading in Caenorhabditis elegans. Nat Struct Mol Biol. 2007;14:927–33. doi: 10.1038/nsmb1308. [DOI] [PubMed] [Google Scholar]
- 36.Jannot G, Boisvert ME, Banville IH, Simard MJ. Two molecular features contribute to the Argonaute specificity for the microRNA and RNAi pathways in C. elegans. RNA. 2008;14:829–35. doi: 10.1261/rna.901908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sasaki T, Shiohama A, Minoshima S, Shimizu N. Identification of eight members of the Argonaute family in the human genome small star, filled. Genomics. 2003;82:323–30. doi: 10.1016/S0888-7543(03)00129-0. [DOI] [PubMed] [Google Scholar]
- 38.González-González E, López-Casas PP, del Mazo J. The expression patterns of genes involved in the RNAi pathways are tissue-dependent and differ in the germ and somatic cells of mouse testis. Biochim Biophys Acta. 2008;1779:306–11. doi: 10.1016/j.bbagrm.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 39.García-López J, Del Mazo J. Expression dynamics of microRNA biogenesis during preimplantation mouse development. Biochim Biophys Acta. 2012;1819:847–54. doi: 10.1016/j.bbagrm.2012.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.