Abstract
Bacterial pathogens need to acquire nutrients from the host, but for many nutrients their importance during infection remain poorly understood. We have investigated the importance of methionine acquisition and synthesis for Streptococcus pneumoniae growth and virulence using strains with gene deletions affecting a putative methionine ABC transporter lipoprotein (Sp_0149, metQ) and/or methionine biosynthesis enzymes (Sp_0585 - Sp_0586, metE and metF). Immunoblot analysis confirmed MetQ was a lipoprotein and present in all S. pneumoniae strains investigated. However, vaccination with MetQ did not prevent fatal S. pneumoniae infection in mice despite stimulating a strong specific IgG response. Tryptophan fluorescence spectroscopy and isothermal titration calorimetry demonstrated that MetQ has both a high affinity and specificity for L-methionine with a KD of ∼25 nM, and a ΔmetQ strain had reduced uptake of C14-methionine. Growth of the ΔmetQ/ΔmetEF strain was greatly impaired in chemically defined medium containing low concentrations of methionine and in blood but was partially restored by addition of high concentrations of exogenous methionine. Mixed infection models showed no attenuation of the ΔmetQ, ΔmetEF and ΔmetQ/ΔmetEF strains in their ability to colonise the mouse nasopharnyx. In a mouse model of systemic infection although significant infection was established in all mice, there were reduced spleen bacterial CFU after infection with the ΔmetQ/ΔmetEF strain compared to the wild-type strain. These data demonstrate that Sp_0149 encodes a high affinity methionine ABC transporter lipoprotein and that Sp_0585 – Sp_0586 are likely to be required for methionine synthesis. Although Sp_0149 and Sp_0585-Sp_0586 make a contribution towards full virulence, neither was essential for S. pneumoniae survival during infection.
Introduction
The acquisition of essential nutrients from the host is a prerequisite for bacterial pathogens to be able to replicate and so to cause successful infection. Screens for virulence genes as well as targeted investigation of specific nutrient transporters has confirmed the importance of nutrient acquisition for the pathogenesis of infections for many microbial pathogens [1], [2], [3], [4], [5], [6], [7]. One group of nutrients required for bacterial growth are the amino acids, but there are only limited data on their importance for bacterial pathogenesis.
Methionine is one of the least abundant amino acids in physiological fluids (4 µg ml−1) [8], and yet is essential for protein synthesis and is a constituent of S-adenosylmethionine, the major biological methyl donor required for the biosynthesis of phospholipids and nucleic acids [9]. Two bacterial methionine transport systems have been described: the Methionine ABC (ATP binding cassette) Uptake Transporter (MUT) family [10], [11] and a secondary transporter termed BcaP [12]. In Escherichia coli, the MUT system is encoded by the metD locus and consists of the MetQ substrate binding protein (SBP), MetL transmembrane permease and the MetN cytoplasmic ATP-hydrolyzing protein (ATPase) [11]. E. coli metD mutants are unable to transport D-methionine or utilize this compound as a source of methionine [11]. Similar ABC transporters are the primary methionine transporters for Bacillus subtilis (metNPQ) [10], Streptococcus agalactiae (metQ1NP) [13] and Streptococcus mutans (atmBDE) [14]. The second methionine transport system, BcaP was described in Lactococcus lactis as a branched chain amino acids transporter, but is also involved in the transport of methionine [12]. Microorganisms and plants can also synthesize methionine by converting homoserine to homocysteine through addition of a sulphur group from either cysteine (requiring MetABC), sulfide (requiring MetA and CysD) or methionine using the SAM recycling pathway (MetK, Pfs, and LuxS) [15], [16]. Homocysteine is then methylated by methionine synthase (MetE) in conjunction with a methylenetetrahydrofolate reductase (MetF), with the methyl group supplied by 5-methyl tetrahydrofolate, to form methionine [15], [17]. Existing data show that methionine biosynthetic genes are required for the full virulence of Brucella melitensis [18], Haemophilus parasuis [19] and Salmonella enterica [20] and that mutation of the S. agalactiae methionine regulator MtaR attenuates virulence [8], suggesting methionine synthesis is essential for survival of many bacteria during invasive infection.
Streptococcus pneumoniae is a common nasopharyngeal commensal that is also an important pathogen frequently causing pneumonia, otitis media, septicaemia and meningitis. A recent investigation of the role of nine different ABC transporters for S. pneumoniae virulence identified an ABC transporter encoded by Sp_0148-52 that seemed to be important during pneumonia and septicaemia [7]. BLAST searches suggested this locus contained genes whose products have a high degree of identity to MetQNP and AtmBDE and therefore could be a S. pneumoniae methionine uptake ABC transporter. In this manuscript we describe in detail the Sp_0148-52 locus and the role of methionine during S. pneumoniae growth and virulence. Recombinant Sp_0149 was used to characterise the potential substrates of this ABC transporter, and deletion mutant strains of Sp_0149 and metEF were used to investigate role of methionine acquisition and synthesis during S. pneumoniae growth and virulence. In addition, as several S. pneumoniae lipoproteins have been shown to be effective vaccine candidates in animal models [21], [22], we also investigated the potential of recombinant Sp_0149 as a novel vaccine candidate.
Results
Results of BLAST alignments for Sp_0148-52
The Sp_0148-52 genetic locus was identified during a screen of ABC transporters for those involved in virulence. In this screen a mutant containing an insertion within Sp_0149 was attenuated in virulence in mouse models of pneumonia and sepsis [7]. The Sp_0148-52 region in the TIGR4 S. pneumoniae strain genome contains five genes and is 3669 bp in length (Fig. 1 A) [23]. Gene loci with identical organisation and high levels of identity at the amino acid level for the corresponding genes are present in S. agalactiae (SAN_1757-52) [13] and S. mutans (Smu.1942-38) [14] and contain the metQNP and atmBDE genes respectively that encode probable methionine ABC transporters. Both Sp_0148 and Sp_0149 possess potential lipoprotein signal sequences, LAACS (residues 21–25) and LAACG (residues 20–24) respectively, and are annotated in the TIGR4 genome as lipoproteins. Sp_0148 and Sp_0149 only share 21% identity and 34% similarity at the amino acid level, so are unlikely to have similar functions or have arisen by gene duplication. Sp_0148 has been annotated as an amino acid binding lipoprotein and is highly conserved amongst sequenced S. pneumoniae strains (98% identity at the amino acid level) and with proteins encoded by other streptococci (>49% identity). Sp_0149 is conserved in all the available S. pneumoniae genomes (>97% identity at the amino acid level) and has 62% identity at the amino acid level to the lipoprotein component of the S. mutans methionine transporter atmBCDE [14], suggesting it may have a similar function. BLAST searches demonstrate 73% identity at the amino acid level between the predicted proteins encoded by Sp_0151, atmD and metN (all predicted ATPases) and 70% for the predicted proteins encoded by Sp_0152, atmE and metP (all predicted permeases). Hence, Sp_0149-52 could encode a functional S. pneumoniae methionine ABC transporter, and therefore we have named Sp_0149 metQ, Sp_0151 metN, and Sp_0152 metP according to the nomenclature for the genes encoding a putative S. agalactiae methionine ABC transporter [13].
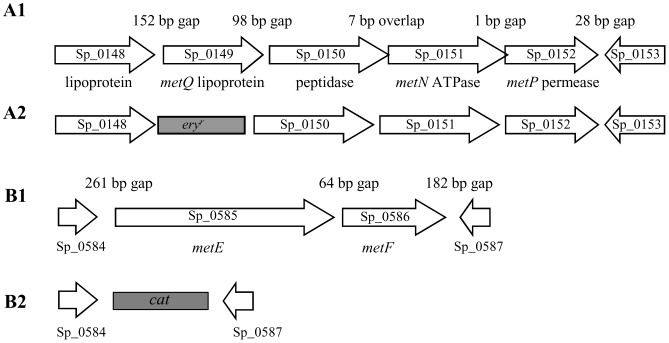
Figure 1. Schematic of the Sp_0148-53 and Sp_0584-0587 loci.
(A1) Structure of the Sp_0148-53 locus. Each arrow represents one gene, with the Sp number stated within the arrow and gene annotation and probable function written below. The number of bp between the stop codon of the upstream gene and the ATG of the downstream gene is given above the arrows in the corresponding gap. (A2) Representation of the structure of the Sp_0148-53 locus in the ΔmetQ strain; the erythromycin resistance cassette (eryr) is shaded grey. (B1) Structure of the Sp_0584-0587 locus, with the Sp number stated and gene annotation when available written below. The number of bp between the stop codon of the upstream gene and the ATG of the downstream gene is given above the arrows in the corresponding gap. (B2) Structure of the Sp_0584-0587 locus in the ΔmetEF mutant strain, showing replacement of metEFgenes with an in-frame copy of cat (shaded grey).
To investigate whether S. pneumoniae metQNP encode a methionine ABC transporter and the phenotype attributable to loss of its function, a metQ deletion mutant strain of 0100993 S. pneumoniae was constructed using a construct made by overlap extension PCR (Fig. 1A). Complete deletion of Sp_0149-0152 was not done as this would include deletion of Sp_0150 as well as metQNP. BLAST results suggested Sp_0150 encodes a protein belonging to the M20/M40/M45 family of peptidases. Genes encoding similar proteins are found within the S. mutans (atmC) and S. agalactiae (pdsM) methionine ABC transporter loci, but the function of the corresponding proteins is unknown. In addition the Sp_0148 lipoprotein may also utilise the MetN and MetP proteins to form an ABC transporter, making it difficult to attribute any phenotypes of an Sp_0149-0152 deletion mutant specifically to a single function. The correct identity of the mutant was confirmed using PCR and primers annealing to Sp_0148 or Sp_0150 and the erythromycin resistance gene eryr (data not shown). Attempts at complementing the metQ deletion mutant strain using reintegration of an intact copy of the full length gene were unsuccessful. However, qPCR demonstrated that expression of the downstream genes in the operon was not reduced by the ΔmetQ mutation, with fold differences (SD) in transcript levels from the ΔmetQ strain compared to the wild-type strain of 2.2 (2.3), 1.3 (1.2), and 1.1 (0.2) for Sp_0150, Sp_0151, and Sp_0152 respectively.
Localisation studies of Sp_0149 lipoprotein
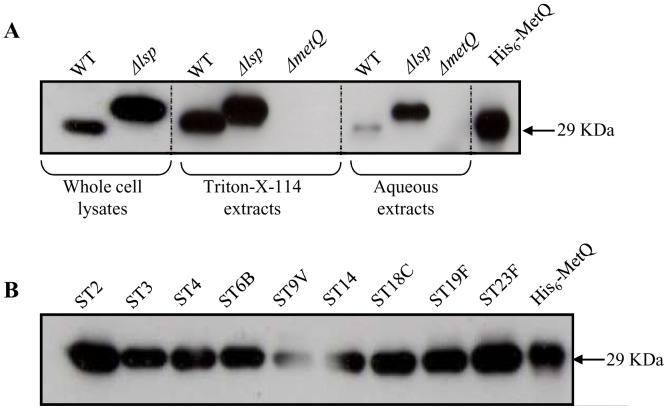
To confirm whether the lipoprotein MetQ is localised to the S. pneumoniae cell membrane, polyclonal anti-MetQ antibodies were obtained by intraperitoneal inoculation of recombinant MetQ into mice. Serum obtained after 4 weeks from inoculated mice recognised recombinant MetQ protein (Fig. 2A and B), and immunoblots of whole cell wild-type S. pneumoniae lysates identified a single band of similar size to the predicted size of MetQ excluding the lipoprotein signal peptide sequence. The same band was present when Triton X-114 extracts of membrane bound proteins were probed with polyclonal anti-MetQ antibodies but was only present as a weak band in the aqueous fraction (Fig. 2A). Previous studies have demonstrated that cleavage of the N terminal lipoprotein signal peptide sequence of lipoproteins requires lipoprotein signal peptidase (Lsp), and that lipoproteins in the S. pneumoniae Δlsp mutant are therefore slightly larger than in the wild type strain [24]. Immunoblotting of whole cell lysates and aqueous fractions of the Δlsp S. pneumoniae strain with anti-MetQ serum demonstrated that, as predicted, the band representing MetQ was slightly larger than in the wild-type strain (Fig. 2A). The band representing MetQ in the aqueous extracts from the Δlsp strain was also more prominent that in the wild-type strain, perhaps suggesting reduced retention of MetQ in the cell membrane in the absence of the Lsp enzyme. Overall, the immunoblot results demonstrate that MetQ is mainly associated with the membrane fraction and is processed by Lsp, confirming that MetQ is a lipoprotein.
Figure 2. Immunoblots of S. pneumoniae using polyclonal anti-MetQ antibodies.
(A) Probing of whole cell lysates, lipid and aqueous phases of Triton X-114 extracts of wild-type, Δlsp, and ΔmetQ 0100993 strains, and the recombinant MetQ protein (missing the N terminal signal sequence) with anti-MetQ antibodies. Equal numbers of bacteria were used for each strain, and the approximate sizes of the bands are displayed on the right. (B) Immunoblots of representative strains for common S. pneumoniae serotypes probed with anti-MetQ antibodies, showing the presence of a similar band in all strains.
MetQ binds L-methionine with high affinity and is required for methionine uptake by S. pneumoniae
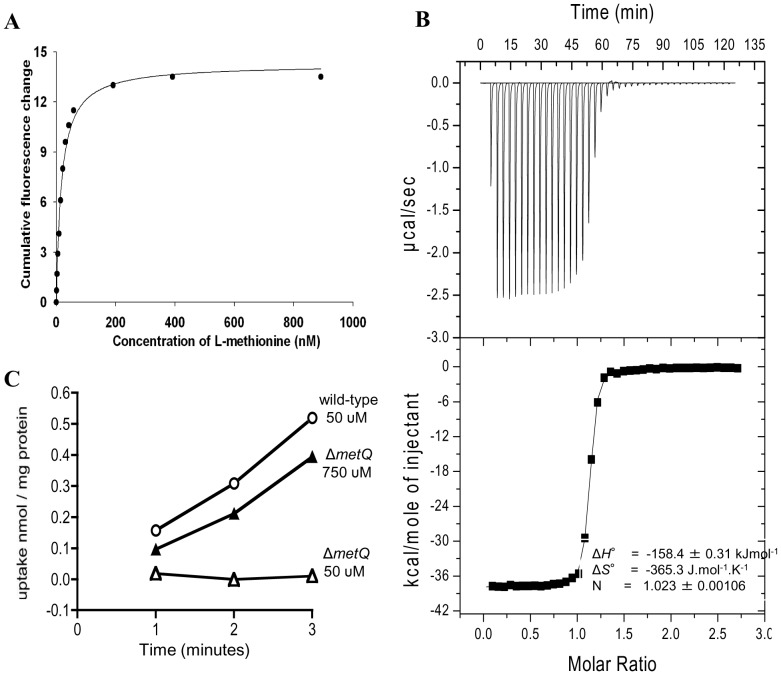
Potential substrates for the MetQNP ABC transporter were investigated using fluorescence spectroscopy of recombinant MetQ (Supplementary Figure 1 ), a method widely used to investigate ligand specificity for SBPs due to the large conformational changes that occurs upon ligand binding [25], [26]. We observed an approximately 30% enhancement of the intrinsic fluorescence of the protein in the presence of L-methionine (Supplementary Fig. 2) and we determined a K D of MetQ for L-methionine to be determined as 26.58±1.15 nM (Fig. 3A). We confirmed this tight binding using isothermal titration calorimetry, which yielded almost identical binding parameters of 26.55±1.28 nM (Fig. 3B), and also demonstrates 1∶1 binding of L-methionine to MetQ.
Figure 3. Analysis of L-methionine binding to MetQ.
(A) Using tryptophan fluorescence spectroscopy: MetQ (0.05 µM) in 50 mM potassium phosphate pH 8 was excited at 280 nm at 37°C. Example titration of the MetQ signal change at 342 nm with increasing concentrations of L-methionine. (B) Binding isotherms for the interaction of MetQ with L-methionine. The top panel display heat changes upon injection of ligand and the low panel the integrated hears of injection (▪) and the best fit (solid line) to a single site binding model. (C) Radioactive uptake of 14C-methionine expressed as nmol/mg protein over time in minutes by the wild-type (circles, 50 µM of methionine added) and ΔmetQ (open triangles 50 µM of methionine added, filled triangles 750 µM of methionine added) S. pneumoniae strains.
To investigate the specificity of MetQ for L-methionine, we examined ligand binding using a range of related ligands. Binding of D-methionine was around 1000 fold weaker with a K D of 27.4±0.96 µM, while binding of DL-homocysteine was only around 250-fold weaker with a K D of 7.0±0.26 µM (Supplementary Fig. 3). We also detected weak binding of α-methyl-DL-methionine with a K D of 30.1±0.86 µM (Supplementary Fig. 3), but could not detect any binding for either L-cysteine of glycine (Supplementary Fig. 2). These data demonstrate that MetQ has a high affinity and specificity for L-methionine, and confirm that methionine is the probable major ligand for the S. pneumoniae MetQNP ABC transporter. To confirm that MetQ is required for methionine uptake by S. pneumoniae, uptake assays of 14C-labelled methionine were performed in chemically defined medium with the wild-type and ΔmetQ strains [27]. As the mucoid phenotype of the serotype 3 strain prevents the efficient formation of bacterial pellets necessary for these assays, the ΔmetQ mutation was transferred into a serotype 2 S. pneumoniae strain (D39). In C+Y medium without added methionine, the ΔmetQ mutant strain was unable take up C14-methionine whereas the wild-type strain demonstrated significant uptake (Fig. 3C). In the presence of 750 µM methionine there was uptake of C14-methionine by the ΔmetQ mutant strain suggesting the presence of a lower affinity methionine transport mechanism ( Fig. 3C ). These results confirm that metQNP is a methionine ABC transporter that is required for methionine uptake by S. pneumoniae when methionine availability is limited.
Investigation of the lipoprotein MetQ as a vaccine candidate
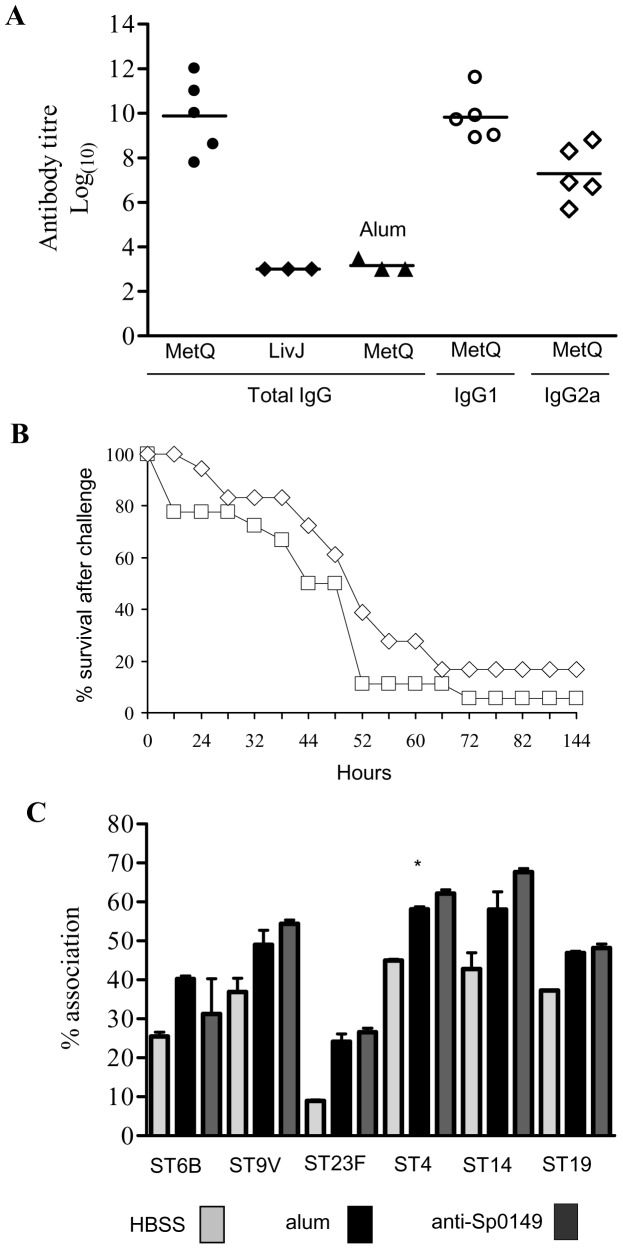
Lipoprotein are attached to the external surface of bacterial membranes and have been shown to be effective vaccines that protect against S. pneumoniae infection in mouse models [21], [22]. Hence, MetQ was investigated as a potential novel vaccine candidate. PCR (data not shown) and immunoblots demonstrated that MetQ was present in all of a range of S. pneumoniae strains from common MLST sequence types for clinically important capsular serotypes (Fig. 2B). Mice were immunised by three intraperitoneal injections of 10 ug recombinant MetQ with alum as the adjuvant separated by 7 or 8 days, which resulted in significant serum titres of IgG (mainly IgG1) against MetQ that did not cross-react with the control lipoprotein LivJ (2) (Fig. 4A). However, when challenged by intraperitoneal inoculation with a potentially lethal dose of the serotype 2 S. pneumoniae strain D39 there were no differences in the speed of development of fatal infection and survival between vaccinated and control (given PBS and the adjuvant alone) mice (Fig. 4B). To investigate whether this lack of vaccine efficacy despite good anti-MetQ titres was due to lack of MetQ expression by S. pneumoniae during infection, semi-quantitative reverse transcriptase PCR (RT-PCR) for metQ transcripts was performed using S. pneumoniae RNA extracted from the blood of mice with severe S. pneumoniae septicaemia. As a positive control RT-PCR was also performed for psaA, which encodes another lipoprotein that is expressed during infection. RT-PCR readily identified a product of the right size for metQ, but at a lower level of expression compared to psaA, suggesting that relative lack of expression of metQ could be one reason why vaccination with the MetQ was not protective (Fig. 5B). These data were supported by transcriptome data calculating the relative abundance of metQ, metE, metF, PsaA and another lipoprotein PiuA at three anatomical sites during infection (Fig. 5C). The results show that transcript abundance for metQ, metE and metF were similar for bacteria found in the nasopharynx, lungs and blood. Importantly, the amount of metQ transcripts was almost two log10 lower and one log10 lower than the amount of psaA and piuA transcripts respectively, both of which encode effective lipoprotein vaccine candidates [28], [29], [30], [31]. These results support the hypothesis that lack of MetQ adundance during infection could explain the failure of vaccination with MetQ to provide protection against systemic disease. Furthermore, when assessed using an established flow cytometry assay polyclonal anti-MetQ sera did not significantly improve phagocytosis of other S. pneumoniae strains by a neutrophil cell line (Fig. 4C).
Figure 4. Investigation of MetQ as a vaccine candidate.
(A) Mouse sera antibody titres to recombinant His-tagged MetQ after intraperitoneal immunisation with His6-MetQ. Target protein (His6-MetQ or the negative control SBP His6-LivJ) and IgG class are labelled along the X axis, and results were obtained using serum from His6-MetQ vaccinated mice with the exception of the filled triangles which represent results for serum from control mice (labelled alum). Filled symbols represent titres for individual mice for the total IgG titre, open circles for IgG1, and open diamonds for IgG2A titres. Bars represent median values. (B) Development of fatal disease in mice vaccinated with His6-MetQ (square symbols) and control (given alum and PBS alone, diamonds) mice after IP inoculation of 104 CFU of D39 (n = 20). There were no statistical differences in survival (log rank test). (C) Phagocytosis of representative S. pneumoniae vaccine serotypes. Key: bacteria opsonised with: grey bars, HBSS; solid bars, serum from alum immunised mice; dark grey bars, polyclonal anti-MetQ sera from MetQ vaccinated mice.
Figure 5. Gene expression of Sp_0149 during infection.
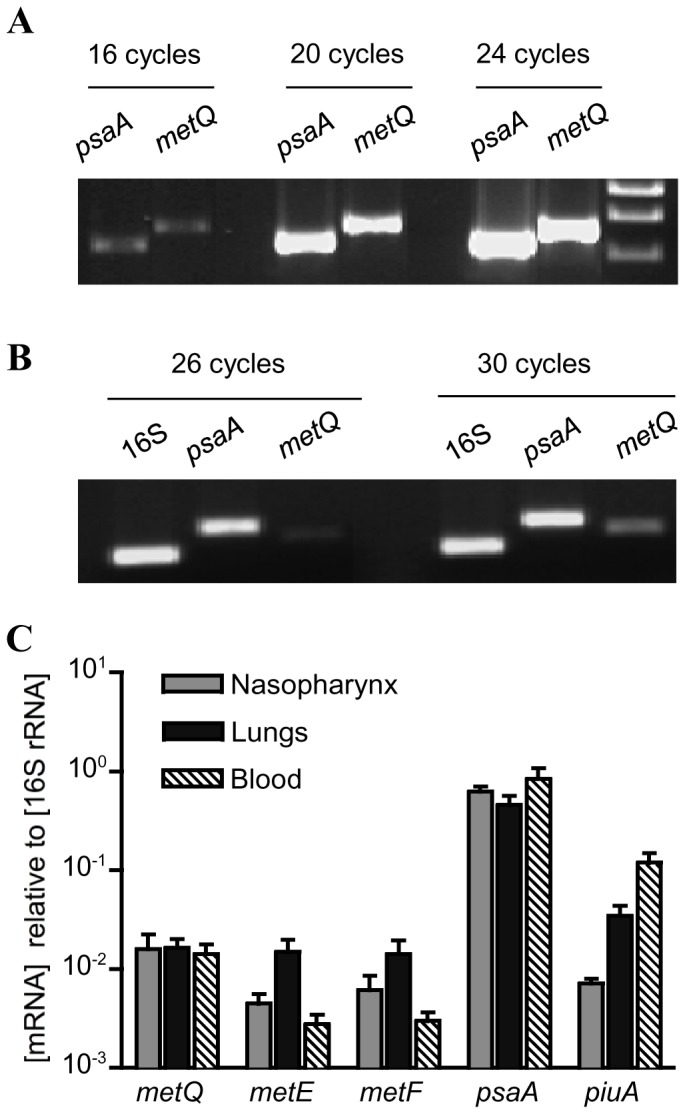
(A) Ethidium stained agarose gel showing equal amplification efficiency for the primer pairs used for PCR of psaA and metQ when using S. pneumoniae 0100993 genomic DNA as the template. (B) Ethidium stained agarose gel of cDNA products generated by RT-PCR after 26 and 30 cycles using S. pneumoniae 0100993 total RNA extracted from the blood of infected mice and primers for 16S rRNA (internal control), psaA (positive control) and metQ. (C) Relative mRNA concentrations of selected genes of S. pneumoniae WCH43 (serotype 4) in various in vivo niches at 72 h post-intranasal infection of mice. Transcript abundance for each gene was obtained by microarray analysis, and normalized against those obtained for the 16S rRNA control. Quantitative fold differences for each transcript were determined as a ratio of its abundance to that of the 16S rRNA control. Data are means ± SEM for each gene transcript from three replicate challenge experiments.
In vitro phenotypes of the ΔmetQ, ΔmetEF and ΔmetEF/ΔmetQ strains
As synthesis of methionine may reduce the effects of loss of transport of methionine due to the metQ mutation on S. pneumoniae phenotype, additional mutant strains were constructed containing deletions of the S. pneumoniae metE and metF [32]. BLAST searches of S. pneumoniae TIGR4 genome identified the adjacent (and likely cotranscribed) genes Sp_0585 and Sp_0586 as homologues of the S. mutans metE and metF genes (47% and 38% identity at the amino acid level respectively) [14] (Fig. 1B). The genome structure suggested metE and metF were likely to be contranscribed independent of the adjacent genes, due to the large gap between Sp_0584 and Sp_0585, and because Sp_0587 is transcribed in the opposite direction to Sp_0586 (Fig. 1B). Deletion mutants replacing metE and metF with a chloramphenicol resistance marker were made in the wild-type (ΔmetEF) or in combination with metQ deletion (ΔmetEF/ΔmetQ) S. pneumoniae 0100993 strains (Fig. 1B). The correct identity of the mutants was confirmed using PCR and primers annealing to Sp_0584 or Sp_0587 and the chloramphenicol resistance gene cat (data not shown).
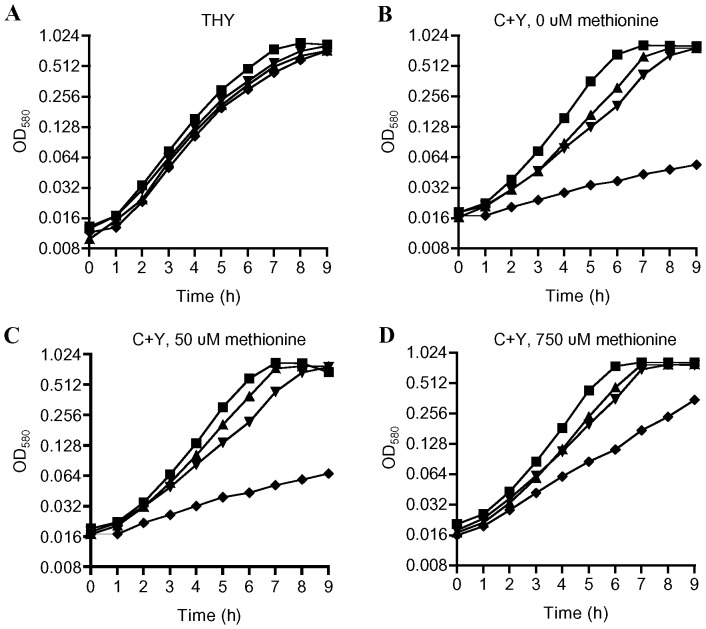
To investigate the functional importance of methionine synthesis and uptake for S. pneumoniae, growth of the wild-type, ΔmetQ, ΔmetEF and ΔmetEF/ΔmetQ strains were compared in the complete medium THY and in the semi-defined medium C+Y with restricted methionine content. In THY there were no significant differences in growth between the wild-type and ΔmetQ, ΔmetEF and ΔmetEF/ΔmetQ strains (Fig. 6A). In C+Y without added methionine (Fig. 6B), both ΔmetQ and ΔmetEF mutant strains had slightly impaired growth compared to the wild-type strain, but still reached similar growth levels to the wild-type strain after 9 hours. However, in C+Y with either no added methionine or supplemented with only 50 µM of methionine the double mutant ΔmetEF/ΔmetQ had markedly impaired growth due to reduced generation times (Fig. 6B and C). These results provide further evidence that MetQ is required for methionine uptake and indicate that MetEF are necessary for methionine synthesis. The impaired growth of the ΔmetEF/ΔmetQ strain in C+Y medium was partially restored by addition of 750 µM of methionine (Fig. 6D), suggesting there are additional weaker affinity secondary transport systems that allow the ΔmetEF/ΔmetQ strain to obtain methionine in the presence of high concentrations of substrate.
Figure 6. Growth of the wild-type, ΔmetQ, ΔmetEF and ΔmetEF/ΔmetQ S. pneumoniae strains in nutrient rich and semi defined media.
Growth measured using OD580 of the wild-type (squares), ΔmetQ (inverted triangles), ΔmetEF (triangles) and ΔmetEF/ΔmetQ (diamonds) S. pneumoniae strains grown in THY (A), C+Y with no added methionine (B), C+Y with 50 µM methionine (C), or C+Y with 750 µM methionine (D). Each data point is the mean log2 OD580 for three samples.
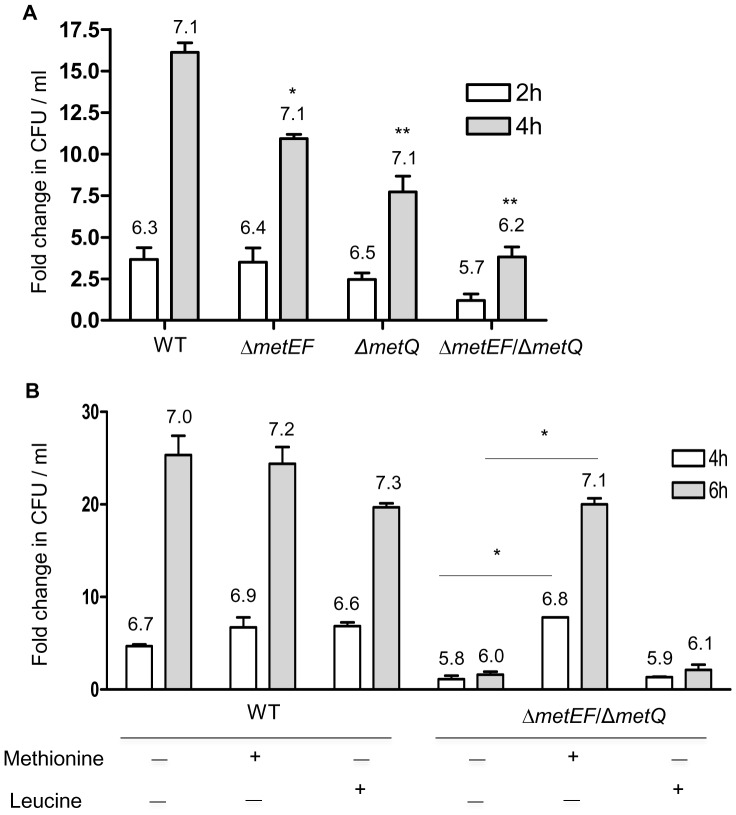
To assess the requirement for methionine for S. pneumoniae growth in physiologically relevant conditions, replication of these strains was assessed in freshly obtained human blood. The ΔmetQ and ΔmetEF strains were both significantly impaired in growth in blood (inoculum size 1.5×106 CFU, 10.9 and 7.7 fold increase in CFU, respectively) compared to the wild type strain (inoculum size 0.5×106 CFU, 16.2 fold increase, P<0.001, ANOVA) (Fig. 7A), although all three strains reached similar total CFU per ml. Moreover, the ΔmetEF/ΔmetQ strain had markedly reduced growth in blood with only 3.8 fold increase in CFU after 4 hours and reaching a total CFU of nearly 1 log10 fewer than the wild-type strain (Fig. 7A). Addition of 1 mM methionine but not leucine (a branched-chain amino acid not linked to the methionine biosynthetic pathways) largely reversed the growth defect of the double mutant in blood, without affecting growth of the wild-type strain (Fig. 7B). These results demonstrate that absence of methionine uptake and synthesis strongly impairs S. pneumoniae growth in physiological conditions found during infection.
Figure 7. Growth of the wild-type, ΔmetQ, ΔmetEF and ΔmetEF/ΔmetQ S. pneumoniae strains in human blood or in human blood supplemented with amino acids.
Data is presented as the mean (SD) fold change in the bacterial CFU per ml at 2 h (clear columns) and at 4 h (grey columns) in human blood (A) or at 4 h (clear columns) and 6 h (grey columns) in human blood supplemented with amino acids (B). P values for the differences between the wild-type and the mutant strains at 4 h (A) and between the ΔmetEF/ΔmetQ strain with and without added methionine at 4 h or 6 h (B) were obtained using ANOVA and post hoc tests, *P<0.05, ** p<0.01. Log10 bacterial CFU per ml results for each condition are given in figures above the corresponding bar in the graphs.
In vivo phenotypes of the ΔmetQ, ΔmetEF and ΔmetEF/ΔmetQ strains
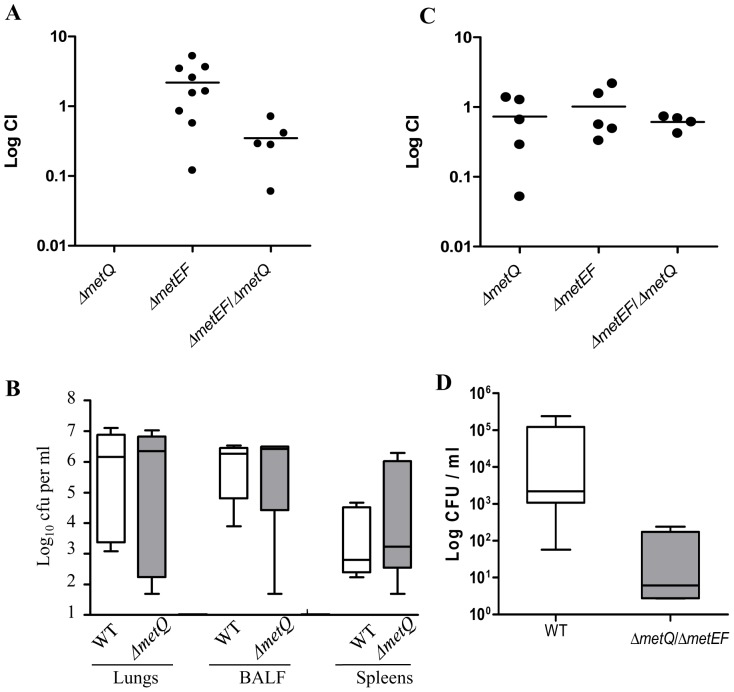
To investigate the effects of deletion of metQ and/or metEF on the ability of S. pneumoniae to colonise the nasopharynx, competitive indices (CIs) for mixed infection with the wild-type and the ΔmetQ, or ΔmetEF or ΔmetEF/ΔmetQ strains were determined in a mouse model of colonisation. None of the mutant strains were reduced in virulence in the colonisation model when CIs were calculated for nasal washes obtained 5 days after inoculation (Fig. 8A). Previously we have shown that a metQ disruption strain was attenuated in virulence in mouse models of sepsis and pneumonia when investigated using CIs [7]. To further assess whether the ΔmetQ mutation affected the development of infection, groups of 5 CD1 mice each were intranasally inoculated with 107 CFU of the wild-type or ΔmetQ strain, and bacterial CFU from the BALF, lungs and spleens determined after 48 hours (Fig. 8B). There were no significant differences in the bacterial loads between the wild-type and ΔmetQ strains in the BALF, lungs and spleens. Hence although previously we have shown disruption of metQ strain reduced virulence in a competitive model of lung infection [7], the ΔmetQ strain was still able to cause significant infection when given as a pure inoculum. The severe growth defect of the double mutant ΔmetEF/ΔmetQ in human blood prompted us to investigate its growth in a mouse sepsis model. After intraperitoneal inoculation the number of ΔmetEF/ΔmetQ strain CFU recovered from the blood was approximately two log10 fewer than CFU recovered for the wild-type strain (P = 0.032, Mann Whitney U test) (Fig. 8C). These data suggested that the inability of the ΔmetEF/ΔmetQ strain to either transport or synthesise methionine has resulted in decreased but not completely abrogated virulence of this strain in a mouse sepsis model.
Figure 8. Virulence of the ΔmetQ, ΔmetEF and ΔmetEF/ΔmetQ strains in mouse models of infection.
(A) CIs for mixed infections with the ΔmetQ, ΔmetEF and ΔmetEF/ΔmetQ and wild-type strains for bacteria recovered from the nasal washes 5 days after colonisation. Each point represents results for a single mouse, and bars median CIs for the group. (B) Log10 ml−1 bacterial CFU recovered from lungs, BALF and spleens of mice 24 hours after IN inoculation with 1×107 CFU of either the wild-type or ΔmetQ strain. There were no statistical differences between the strains for any organ site. (C) Log10 ml−1 bacterial CFU recovered from blood 24 hours after IP inoculation with 3×103 CFU of either the wild-type or ΔmetEF/ΔmetQ strain. P = 0.0317, Mann Whitney U test.
Discussion
In this study, we provide evidence that the S. pneumoniae Sp_0148-52 locus encodes a methionine uptake ABC transporter and that Sp_0585 and Sp_0586 encode MetE and MetF required for methionine biosynthesis. BLAST searches demonstrated that the Sp_0148-52 and Sp_0585/Sp_0586 encode proteins with high degrees of similarity to S. mutans AtmBDE and MetEF respectively, which have been shown to be required for methionine uptake and synthesis [13], [14]. Deletion of the gene encoding the lipoprotein component Sp_0149 (which we have called metQ) resulted in a strain that had reduced growth in methionine restricted media and no detectable uptake of radioactive methionine. Tryptophan fluorescence spectroscopy confirmed MetQ bound to L-methionine with high affinity as well as D-methionine and homocysteine, an amino acid that is related to methionine. Deletion of Sp_0585/Sp_0586 increased the growth defect of the metQ deletion strain in methionine restricted media and in blood, supporting a role for the products of these genes for methionine synthesis. Genetic complementation would be helpful to link the metEF and metQ mutations to these phenotypes but was not possible to achieve, as is often the case with S. pneumoniae. However the successful biochemical complementation of the in vitro phenotypes by exogenous methionine and the lack of impaired transcription of the rest of the operon in the metQ mutant strongly support that the observed phenotypes were caused by the mutations. Overall the data indicate that S. pneumoniae methionine acquisition and synthesis is largely dependent on the Sp_0148-52 locus and Sp_0585 and Sp_0586 loci respectively. Unlike the equivalent S. mutans mutants [14], the S. pneumoniae metQ and metEF mutant strains were still able to grow in the absence of supplementation with methionine. This observation and the persisting growth of the S. pneumoniae ΔmetQ/metEF double mutant in media containing high concentrations of methionine suggest that there are additional mechanisms for S. pneumoniae methionine uptake, possibly similar to BcaP of L. lactis [12]. One candidate is the Sp_0481-4 locus which contains an upstream consensus sequence for the streptococcal methionine regulator MtaR/MetR (data not shown).
As well as encoding the MetQNP methionine ABC transporter, the Sp_0148-52 locus also encodes another SBP (Sp_0148) unrelated to MetQ and a peptidase (Sp_0150). The functions of these proteins and why their genes are associated with the metQNP genes are unknown. The peptidase could be required for the breakdown of extracellular peptides to provide substrates for MetQNP, but this hypothesis will require further investigation. Previously it has been suggested that two separate zinc uptake lipoproteins AdcA and AscAII share the same permease and ATPase components [33]. Similarly, the Sp_0148 SBP could associate with MetN and MetP to make an ABC transporter with separate affinities to the MetQ lipoprotein, perhaps another amino acid as Sp_0148 has some similarities to proteins annotated as glutamine SBPs.
Existing vaccines against S. pneumoniae are unable to provide protection against all strains, prompting the investigation of surface exposed antigens such as SBPs that are generally highly conserved between different S. pneumoniae strains as novel vaccine candidates. Existing data show that several ABC transporters SBPs are effective vaccines in animal models of S. pneumoniae [28], [30], [34], [35]. However, vaccination with MetQ did not prevent fatal systemic infection despite inducing a reasonable antibody response and the surface location of MetQ. RT-PCR and transcriptome data suggested that expression of metQ during actual infection was considerably lower than that of the SBP vaccine candidates psaA and piuA [36]; this low level of expression could readily contribute to the lack of protective efficacy of high titres to the S. pneumoniae MetQ. In addition, unlike IgG raised against other SBPs [31], [34], [35], anti-MetQ antibodies did not promote neutrophil phagocytosis of S. pneumoniae, suggesting these antibodies are functionally ineffective at promoting immunity.
Several S. pneumoniae ABC transporters have been shown to be required for full virulence, including those involved in the acquisition of metals (manganese, zinc, and iron), phosphate, polyamines, and branched chain amino acids [5], [6], [7], [29], [34], [35], [37]. As methionine is essential for bacterial growth and has restricted availability in mammalian systems, with a concentration of 4 µg ml−1 in physiological fluid, the MetQNP ABC transporter might also be expected to be important for S. pneumoniae virulence. Indeed, our previous data using mixed infection experiments with the wild-type strain and a metQ mutant created by insertional mutagenesis showed the metQ mutant strain was significantly less competitive in both pneumonia and sepsis infection models [7]. However, the competitive nature of the mixed infection model makes it a very stringent test of virulence for mutant strains, and in this manuscript we have shown that when given a pure inoculum of the deletion ΔmetQ or wild-type strains the progression of infection in mouse model of pneumonia was very similar. These results are similar to those we have described for the S. pneumoniae branched chain amino acid ABC transporter, which also has reduced virulence when competing against the wild-type strain but could still establish fatal infection [7]. The double mutant ΔmetEF/metQ strain was attenuated in virulence in a sepsis model with lower bacterial CFU than the wild-type strain, suggesting that methionine synthesis helped to maintain virulence of the ΔmetQ strain. However, somewhat surprisingly the mutant Δ metEF/metQ strain was still able to establish systemic infection in mice, despite its poor growth in blood in vitro and a circulating methionine level in mammalian blood of 7 to 43 µM, a level that is associated with a strong growth defect for the this strain in medium. Similarly, there was no evidence that deletion of either metQ or metEF individually or in combination affected colonisation when assessed using highly sensitive competitive infection assays. These results may indicate that local methionine availability at the site of infection is relatively high, perhaps due to cell lysis, and adequate for S. pneumoniae growth even in a strain containing deletions affecting both high affinity methionine uptake and methionine biosynthesis. The effect on virulence of some S. pneumoniae mutations vary with strain background, and it is possible that in other strains metQ and metEF deletion will impair virulence to a greater extent than seen in the serotype 3 strain used for these studies.
To conclude, we have assessed the role of the MetQNP ABC transporter and MetEF methionine synthesis enzymes for S. pneumoniae growth and virulence. The results demonstrate that MetQNP is required for methionine uptake, and that dual mutation of metQ and the metEF locus has a profound effect on growth of S. pneumoniae in methionine restricted conditions. Methionine uptake and synthesis assisted, but was not essential, for the development of S. pneumoniae infections. These data provide more information on the micronutrient requirements of S. pneumoniae during the development of infection.
Materials and Methods
Ethics statement
All animal experiments were approved by the UCL Biological Services Ethical Committee and the UK Home Office (Project Licence PPL70/6510). Experiments were performed according to UK national guidelines for animal use and care, under UK Home Office licence.
Bacterial strains, media and growth conditions
The E. coli strain M15 (Qiagen) were used for the cloning of Sp_0149 lipoprotein gene. Capsular serotypes 2 D39 strain and capsular serotype 3 S. pneumoniae 0100993 strain were used to construct S. pneumoniae mutant strains for the in vitro and in vivo phenotype analysis [1]. A range of S. pneumoniae strains from common multi-locus sequence typing (MLST) sequence types (STs) for clinically important capsular serotypes were kind gifts from Professor Spratt, Imperial College [38]. E. coli was cultured at 37°C using Luria Bertani (LB) broth or agar plates and S. pneumoniae strains were cultured in the presence of 5% CO2 at 37°C on Columbia agar (Oxoid) supplemented with 5% horse blood (TCS Biosciences), or in Todd-Hewitt broth supplemented with 0.5% yeast-extract (Oxoid) or C+Y media [39]. Plasmids and mutant strains were selected by using appropriate antibiotics (carbenicillin 100 µg ml−1 and kanamycin 25 µg ml−1 for E. coli, 0.2 µg ml−1 erythromycin for S. pneumoniae). Stocks of S. pneumoniae were stored as single use 0.5 ml aliquots of THY broth culture (O.D580 0.3–0.4) at −70°C in 10% glycerol. Growth of S. pneumoniae strains in broth was monitored by measuring optical density at 580 nm (OD580). Replication of S. pneumoniae strains in freshly obtained human blood was determined by inoculating with 0.5 to 2.0×106 CFU ml−1 (depending on strain and experiment). After 2, 4, 6 h of growth at 37°C under 5% CO2, serial dilutions were plated on to blood agar plates to enumerate the CFU. Fold change was calculated by dividing the results by the inoculum CFU.
Nucleic acid manipulations, reverse transcriptase PCR and transcriptome analysis
S. pneumoniae genomic DNA was extracted from bacteria grown in THY using a modified Wizard genomic DNA kit (Promega) and RNA using SV total RNA extraction kit (Promega) as previously described [6]. Total RNA from S. pneumoniae found in mouse blood was extracted as described by Ogunniyi et al [40]. S. pneumoniae were harvested from human and mouse blood by brief centrifugation at 825 g at 4°C for 5 minutes. The resulting supernatant was centrifuged further at 15500 g at 4°C for 5 minutes, and the bacterial pellet resuspended in 300 µl of pre-warmed (65°C) acid-phenol (Ambion) and incubated for 5 minutes at 65°C followed by further addition of 300 µl of pre-warmed NAES buffer and incubation at 65°C for 5 minutes with intermittent mixing. The mixture was cooled on ice for 1 minute and centrifuged at 15500 g for 1 minute, and the aqueous phase was re-extracted twice with acid-phenol and NAES buffer followed by twice with 300 µl of chloroform. To this mixture, sodium acetate was added at a final concentration of 300 mM followed by addition of 2 volumes of ethanol and RNA was precipitated at −20°C overnight, centrifuged at 6000 rpm for 5 minutes and washed in 70% ethanol before resuspension in 50 µl of nuclease-free water. To the resulting RNA, recombinant RNasin ribonuclease inhibitor (Promega N251A) was added to a final concentration of 1 U µl−1 and then treated with 0·5 U µl−1 RQ1 RNase-free DNase (Promega M610A) at 37°C for 40 minutes. Aliquots of this RNA were stored at −70°C until use. An aliquot was used to check the purity by RT-PCR with and without reverse transcriptase using gene specific primers. qPCR was performed as previously described [24] using cDNA amplified from 0.4 µg of total RNA using the Applied Biosystems Geneamp RNA PCR core kit (Applied Biosystems, UK), and target-specific primers used for Sp_0150, Sp_0151 and Sp_0152 expression ( Table 1 ). For each gene, crossing point (Cp) values were determined from the linear region of the amplification plot and normalized by subtraction of the Cp value for 16S RNA generating a ΔCp value. Relative change was determined by subtraction of the ΔCp value for the wild-type strain from the ΔCp value for the mutant strain (ΔΔCp value), and fold change calculated using the formula 2−ΔΔCp. Transcriptome data for RNA extracted from S. pneumoniae serotype 4 strain WCH43 recovered from mouse nasopharynx, lungs and blood was obtained as described [41] , using whole-genome S. pneumoniae PCR arrays obtained from the Bacterial Microarray Group at St George's Hospital Medical School, London, England ( http://bugs.sghms.ac.uk/ ). Transcript abundance for each gene was obtained, and quantitative fold differences determined as a ratio of a specific gene abundance to that of the 16S rRNA control.
Table 1. Strains, plasmids and primers used in this study. Restriction enzyme sites in primers 5′ linkers are underlined.
| Name | Description (source/reference) |
| Plasmids | |
| pID701 | Shuttle vector for IDM transformation of S. pneumoniae: Cmr (1) |
| pPC111 | pID701 containing an internal portion of Sp_0149 in the XbaI site: Cmr (this study) |
| ΔmetQ | pGEM-Teasy containing a deletion of Sp_0149: Ampr (this study) |
| pPC138 | pQE30UA carrying full length Sp_0149: Kmr, Ampr (this study) |
| Strains | |
| 0100993 | S. pneumoniae capsular serotype 3 clinical isolate (2) |
| D39 | S. pneumoniae capsular serotype 2 |
| TIGR4 | S. pneumoniae capsular serotype 4 |
| JSB6B | S. pneumoniae capsular serotype 6B (42) |
| JSB9V | S. pneumoniae capsular serotype 9V (42) |
| JSB14 | S. pneumoniae capsular serotype 14 (42) |
| JSB18C | S. pneumoniae capsular serotype 18C |
| JSB19F | S. pneumoniae capsular serotype 19F |
| JSB23F | S. pneumoniae capsular serotype 23F (42) |
| ΔmetQ | 0100993 containing the Sp_0149 deletion construct: Eryr (this study) |
| ΔmetQD39 | D39 containing the Sp_0149 deletion made with ΔmetQ genomic DNA: Eryr (this study) |
| ΔmetEF | 0100993 containing the Sp_0585and Sp_0586 deletion construct: Cmr (this study) |
| ΔmetQ/ΔmetEF | 0100993 containing the metQ and metEF deletion constructs: Eryr and Cmr (this study) |
| Expression and qPCR Primers | |
| 16s.1 | GGT GAG TAA CGC GTA GGT AA |
| 16s.2 | ACG ATC CGA AAA CCT TCT TC |
| PsaA.1 | CGT TCC GAT TGG GCA AGA C |
| PsaA.2 | GCA CTT GGA ACA CCA TAG |
| Sp0149.1 | GGC TCT TGC AGC TTG CGG |
| Sp0149.2 | GGC TTT CGT TTG TAG CGT C |
| Sp0150.F | GAGCTATACAGCGCCCTTTG |
| Sp0150.R | CGCTGGCACAGTGTCATAGT |
| Sp0151.F | ACCGTGTTGCAGTTATGCAG |
| Sp0151.R | CCATGGCTTCGTCAATACCT |
| Sp0152.F | GGAGCTGGTGGTATCGGTAA |
| Sp.0152.R | TCTCCCAAGAATTGGATTGC |
| Mutation primers | |
| Sp0148F | CAC CAA TTG CCC AAA ATC C |
| Ery-Sp0148R | TATT TTAT ATT TTT GTT CAT GAT TCT TTC TCC TTA AAA ATA |
| Ery-Sp0150 | ATT ATT TAA CGG GAG GAA ATA A TAA GAA ACA GGG AGG TGG GAG |
| Sp0150R | CCA AGG CAT TTT TGG TCC C |
| EryF | ATGAACAAAAATATAAAATA |
| EryR | TTATTTCCTCCCGTTAAATAAT |
| Sp0583F | GCGTGGGACAGTCCGACAATG |
| Cm-Sp0584R | CATTATCCATTAAAAATCAAAGATGTGTCCTCCAAAATTTGTTGTTG |
| Cm-Sp0587F | CTAATGACTGGCTTTTATAATAAAAGCAAACCATTCTTCTCAGG |
| Sp0587R | CTCACGACCCGCAAAAGTC |
| Cm.1 | TTATAAAAGCCAGTCATTAG |
| Cm.2 | TTTGATTTTTAATGGATAATG |
Restriction digests, ligation of DNA fragments, fractionation of DNA fragments by electrophoresis, and transformation of E. coli (by heat shock) were performed according to established protocols [42]. DNA fragments were purified from electrophoresis gels using the QIAquick gel extraction kit. RT-PCR was performed using the Access RT-PCR system (Promega) and gene specific primers (Table 1). The NCBI website (http://www.ncbi.nlm.nih.gov/blast) was used to perform BLAST searches and alignments of the available complete and incomplete bacterial nucleotide and protein databases. Primers for both PCR and RT-PCR were designed using sequences displayed by the ARTEMIS software.
Construction of ΔmetQ, ΔmetEF and ΔmetEF/ΔmetQ S. pneumoniae mutant strains
Strains, plasmids and primers used for this study are described in Table 1. Target genes were amplified by PCR from S. pneumoniae 0100993 genomic DNA using primers designed from the TIGR4 genome sequence (http://www.tigr.org) [23]. The ΔmetQ strain was constructed by overlap extension PCR [43] using a transformation fragment in which the Sp_0149/metQ gene had been replaced by the erythromycin resistance cassette ery. Two products corresponding to 630 bp 5′ (primers Sp0148F and Ery-Sp0148R) and 672 bp 3′ (primers Ery-Sp0150F and Sp0150R) to metQ were amplified from S. pneumoniae genomic DNA by PCR carrying 3′ and 5′ linkers complementary to the 5′ and 3′ portion of the ery gene respectively. ery was amplified from pACH74 using PCR and the primers EryF and EryR [24]. Similarly, For the in-frame deletion of metEF (Sp_0585-Sp_0586), a construct was created in which 921 bp of flanking DNA 5′ to the Sp_0585 ATG (primers Sp0583F and Cm-Sp0584R) and 725 bp of flanking DNA 3′ to the Sp_0586 ORF (primers Cm-Sp0587F and Sp0587R) were amplified by PCR from S. pneumoniae genomic DNA and fused with the chloramphenicol resistance marker (cat, amplified from pID701, a suicide vector containing cat gene, with primers CmF and CmR) by overlap extension PCR. The constructs were transformed into S. pneumoniae by homologous recombination and allelic replacement using competence stimulating peptide (CSP-1) (kind gift from D. Morrison) and selection with antibiotics according to established protocols [1], [5]. Deletion of metQ and/or metEF was confirmed by PCR and sequencing of the PCR products (performed by Lark Technologies Inc. UK or UCL sequencing services using the Big Dye™ Terminator technique and gene specific PCR primers).
Cloning, expression and purification of His6-MetQ
Recombinant MetQ protein was expressed in E. coli and purified using an N terminal His-tagged QIAexpressionist™ system. Primers Sp0149.1 and Sp0149.2 amplified a full-length metQ from the S. pneumoniae D39 strain (excluding the 5′ portion encoding the predicted N-terminal signal peptide), which was then ligated into the pQE30 expression vector to make the plasmid pPC138 and transformed into E. coli strain M15. For synthesis of recombinant proteins, the transformed cells were grown aerobically in 5 ml of Lennox broth (LB) for 8 h, and used to inoculate 50 ml LB for overnight aerobic growth at 37°C. This was then used to inoculate 625 ml of LB at 30°C. Cells were allowed to grow aerobically to an A 650 of 0.4–0.6 before production of recombinant protein was induced with 1 mM isopropyl 1-thio-β-D-galactopyranoside (IPTG), followed by four hours of incubation at 30°C. Cells were harvested by centrifugation at 4430× g for 15 min at 4°C and resuspended in buffer A (50 mM potassium phosphate, pH 7.8, 500 mM NaCl, 10 mM immidazole and 20% glycerol). The cells were lysed by sonication and cell debris was removed by centrifugation at 38000× g for 30 min at 4°C and the supernatant containing overexpressed protein was collected. Nickel affinity chromatography was used for the purification of the hexahistidine tagged protein using 5 ml His-trap column [GE Healthcare]. The soluble fraction containing the over produced protein was passed over column pre-equilibrated with buffer A. Weakly bound contaminants were removed by washing the column with 20 CV of buffer A containing 40 mM imidazole and the recombinant protein was eluted with elution buffer containing 500 mM imidazole. Purification products were analysed by SDS-PAGE gel and shown to contain 99% pure protein of the expected size for His6-MetQ. Purified protein was dialysed in 50 mM potassium phosphate pH 7.8 and concentrated using Vivaspin 5 kDa MWCO concentrators. Protein concentration was determined from the absorbance at 280 nm ( A 280) using a calculated molar extinction coefficients of 45780 cm−1 M−1 for MetQ. His6-Sp0749 (LivJ) was also purified using an N terminal His-tagged QIAexpressionist™ system as described in [7].
Preparation of ligand free His6-MetQ
Binding proteins often co-purify with their cognate ligands and thus must be made ligand free in order to check binding with alternate ligands in vitro. Co-purified ligand was removed from MetQ using guanidinium HCl (GdmHCl) as described previously [44] . The protein was partially unfolded while bound to the His-trap column, by including additional wash steps (in buffer A with 30 mm imidazole) of 40 CV with 2 m GdmHCl, 4 CV with 1.5 m GdmHCl, 4 CV with 1 m GdmHCl, 4 CV with 0.5 m GdmHCl, and finally 8 CV with 0 m GdmHCl. The protein was eluted with elution buffer as described previously.
Immunoblots and Triton-X-114 extract preparation
S. pneumoniae were grown in THY medium until the OD580 reached 0.6 and the cells were centrifuged, the pellet resuspended in sterile phosphate buffered saline (PBS) and sonicated. The cell lysate was centrifuged, and the supernatants used for further analysis by immunoblotting. Lipid associated proteins were extracted according to Khandavilli et al [24] using Triton X-114. Protein samples from Triton X-114 extracts were separated on SDS-PAGE 10% and 12% resolving gels respectively, blotted onto nitrocellulose membranes and probed with polyclonal mouse antisera raised against MetQ (1∶2500 dilution) according to standard protocols [45].
Enzyme Linked Immunosorbent assay (ELISA)
ELISA was performed according to Jomaa et al [31]. Microtitre ELISA plates were coated with 100 µl of antigen at a concentration of 5 µg ml−1 in TSA buffer and incubated overnight at 4°C. The plates were washed 5 times with ELISA wash buffer and soaked in 150 µl of 2% BSA-Tween for 2–4 hours at 37°C and washed again for 3–4 times with ELISA wash buffer. A 1/1000 dilution of mouse serum was made in BSA-Tween diluent buffer and added to the first well, and then 2 fold dilutions transferred to subsequent wells and incubated at 37°C for 4 hours. The wells were then washed 6 times with ELISA wash buffer and incubated overnight at 4°C in 100 µl of 1/15000 dilution of goat anti-mouse IgG conjugated to alkaline phosphate (Sigma) diluted in enzyme diluent. The ELISA plate was then washed 5 times in ELISA wash buffer and 100 µl of dinitrophenol (dNP) substrate solution added to each well and incubated for 1 hour at 37°C (dNP substrate solution was prepared by adding 1 tablet of dNP (Sigma) in 5 ml of water). The absorbance was read at 405 nm and antibody titre was calculated as the lowest dilution giving an OD equal to or greater than 0.3.
Opsonophagocytosis assays
To assess the effect of anti-metQ antibodies on the interaction of S. pneumoniae with phagocytes, we measured the proportion of a neutrophil cell line associated with fluorescent bacteria using a previously described flow cytometry opsonophagocytosis assay [31], [46]. The complement source used was commercially available baby rabbit serum (Sigma S7764; rabbit HLA-ABC). S. pneumoniae strains were fluorescently labelled by incubation with 5, 6-carboxyfluorescein-succinimidyl ester (FAM-SE; Molecular Probes, Eugene, Oreg.) solution (10 mg/ml in dimethyl sulfoxide; Sigma) in 0.1 M sodium bicarbonate buffer for 1 h at 37°C and then washed six times with Hanks balanced salt solution (HBSS) in 0.2% bovine serum albumin and stored in aliquots at −70°C in 10% glycerol (approximately 109 CFU/ml). The human cell line HL-60 (promyelocytic leukemia cells; CCL240; American Type Culture Collection, Manassas, Va.) was used to provide the effector cells after differentiation into granulocytes by using previously described protocols. Differentiated HL60 cells were harvested by centrifugation (160× g, 8 min, 4°C) and washed twice with HBSS and once with HBSS in the presence of Ca2+ and Mg2+. FAM-SE-labelled bacteria (106 CFU) were opsonized with 1/100, 1/40, and 1/10 dilutions of serum in a 96-well plate for 20 min at 37°C with horizontal shaking (170 rpm). Negative controls were included, using the same volume of HBSS. HL60 cells (105) were added to the opsonized bacteria in the microplate plate and incubated for 30 min at 37°C with shaking, after which the bacteria and cells were fixed using 3% paraformaldehyde and analyzed using flow cytometry. A minimum of 6,000 cells per sample were analyzed.
14C-methionine uptake assays
Radioactive uptake assays were performed by the rapid filtration method as previously described [27] with minor modifications. S. pneumoniae strains were grown in C+Y medium until the OD620 reached 0.2–0.4. These experiments were performed using a capsular serotype 2 S. pneumoniae strain (D39) as the mucoid colonies of the capsular serotype 3 strain prevented effective pelleting of the bacteria for these assays. Bacteria were harvested at 13000 g for 20 minutes and resuspended in 50 mM potassium phosphate buffer (pH 7.2) with 1 mM MgCl2 to an OD620 between 0.8–1.1. Uptake of methionine was determined in 1 ml assays containing 0.85 ml of bacterial cells and a final concentration of 50 or 750 µM methionine with 0.125 µCi 14C-methionine (GE Healthcare, United Kingdom). Samples (150 µl) containing bacteria, radioactive and non-radioactive substrates were removed at time intervals (0, 1, 2, and 3 minutes) and immediately filtered through glass fibre filters (Whatman GF/F), washed twice with 50 mM potassium phosphate buffer, and then washed filters placed in scintillation vials in 5 ml of Ready Safe scintillation cocktail (Beckman Coulter) and radioactivity determined using a Wallac 1214 RackBeta liquid scintillation counter. Bichinchoninic acid protein assay (Sigma Aldrich, UK) determined that an optical density (620 nm) of 1 was equivalent to 0.238 mg protein, and this figure was used to convert the radioactivity counts to nmol solute per mg protein.
Fluorescence Spectroscopy
Protein fluorescence experiment used a FluoroMax 4 fluorescence spectrometer (Horiba Jobin-Yvon) with connecting water bath at 37°C. Ligand free protein (MetQ) was used at a concentration of 0.05 µM in 50 mM potassium phosphate, pH 7.8. Protein was excited at 281 nm with slit widths of 5 nm and emission was monitored at 352 nm with slit widths of 5 nm. To examine alternative ligands for MetQ, potential ligands were added at concentrations up to 5 mM. To determine the KD for ligand binding, the protein was titrated with increasing concentrations of the ligand and the corresponding fluorescence change was monitored in time acquisition mode. The cumulative fluorescence change was plotted in SigmaPlot and the KD was calculated from the hyperbolic fit of the binding curve. L-methionine, D-methionine, DL-Homocysteine, α-Methyl-DL-methionine, L-Cysteine and Glycine were purchased from Sigma-Aldrich.
Isothermal Titration Calorimetry
Calorimetry experiment was performed using a VP-ITC instrument (MicroCal Inc., GE Heath Sciences) by taking 1.4 ml of ligand free MetQ in the calorimeter cell and 400 µl of L-methionine ligand in the syringe. The concentration of ligand in the syringe was typically 10 times that in the cell, whereas the cell concentration was chosen according to c value of 50, where c = [macromolecule]/(predicted) KD . Experiments were carried out in 50 mM potassium phosphate buffer, pH 7.8, at 37°C. Both cell and syringe solutions were degassed at 35°C for 10 min before use. The titrations were performed as follows. A single preliminary injection of 3 µl of ligand solution was followed by 40 injections (6 µl), delivered at an injection speed of 10 µl s−1. Injections were made at 3-min intervals with a stirring speed of 307 rpm. Raw titration data were integrated and fit to a one-site model of binding using MicroCal Origin version 7.0.
Animal models of infection models
Infection experiments were performed in age and sex matched groups of outbred CD1 mice (Charles River Breeders) between 4 to 8 weeks old. For mixed infections, equivalent numbers of bacteria from stocks of wild-type and mutant S. pneumoniae strains were mixed and diluted to the appropriate concentration. For the nasopharyneal colonisation model, 107 CFU of bacteria in 10 µl were administered by intranasal inoculation under halothane general anaesthesia, and nasal washes were obtained 5 days post colonisation. Serial dilutions of the samples were plated onto Columbia blood agar plates containing optochin (50 µg ml−1) and/or gentamycin (5 µg ml−1) to differentiate pneumococcus from other contaminating streptococci and to enumerate CFU. The CI was calculated using the formula: ratio of mutant to wild-type strain recovered from mice divided by the ratio of mutant to wild-type strain in the inoculum [47]. A CI of less than 1 indicates the mutant strain is attenuated in virulence compared to the wild-type strain, and the lower the CI the more attenuated the mutant strain. Experiments were performed using pure inocula of wild-type or mutant strains to calculate bacterial CFU in recovered target organs for each strain at specific time points [47]; for the systemic model of infection 1×103 CFU bacteria in 100 µl were inoculated by intraperitoneal (IP) injection and spleen homogenates obtained at 24 hours for plating [5], [6], [24]; and for the pneumonia model, 5×106 CFU bacteria in 40 µl were given by intranasal (IN) inoculation under halothane general anaesthesia, and lung homogenates obtained at 48 hours for plating [6], [24]. For the vaccination infections, mice were given by IP injection 100 µg of MetQ or just PBS mixed with 10% alum on three occasions separated by 7 to 8 days. Serum was obtained for immunoblots and antibody titres ELISAs from five mice four weeks after the last vaccination, and the remaining vaccinated mice challenged by IP inoculation with 1×104 S. pneumoniae D39 strain CFU. Disease progression was monitored using previously established criteria to identify mice likely to progress to fatal disease [5].
Statistical analysis
All the in vitro growth curves were performed in triplicates and represented as means and standard deviations. Results of growth curves, radioactive uptake and binding assays were analysed using two-tailed t-tests. Target organ CFU were compared between strains using the Mann Whitney U test, and the survival of infected mice using the log rank test.
Funding Statement
This work was undertaken at UCLH/UCL, which received a proportion of funding from the Department of Health's NIHR Biomedical Research Centre's funding scheme, and was supported by the UCL Charities funding and grants from the British Lung Foundation (P05/3) and the Wellcome Trust (grant reference 076442). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lau GW, Haataja S, Lonetto M, Kensit SE, Marra A, et al. (2001) A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol Microbiol 40: 555–571. [DOI] [PubMed] [Google Scholar]
- 2. Darwin AJ (2005) Genome-wide screens to identify genes of human pathogenic Yersinia species that are expressed during host infection. Curr Issues Mol Biol 7: 135–149. [PubMed] [Google Scholar]
- 3. Mei JM, Nourbakhsh F, Ford CW, Holden DW (1997) Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol Microbiol 26: 399–407. [DOI] [PubMed] [Google Scholar]
- 4. Himpsl SD, Lockatell CV, Hebel JR, Johnson DE, Mobley HL (2008) Identification of virulence determinants in uropathogenic Proteus mirabilis using signature-tagged mutagenesis. J Med Microbiol 57: 1068–1078. [DOI] [PubMed] [Google Scholar]
- 5. Brown JS, Gilliland SM, Holden DW (2001) A Streptococcus pneumoniae pathogenicity island encoding an ABC transporter involved in iron uptake and virulence. Mol Microbiol 40: 572–585. [DOI] [PubMed] [Google Scholar]
- 6. Brown JS, Gilliland SM, Ruiz-Albert J, Holden DW (2002) Characterization of pit, a Streptococcus pneumoniae iron uptake ABC transporter. Infect Immun 70: 4389–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Basavanna S, Khandavilli S, Yuste J, Cohen JM, Hosie AH, et al. (2009) Screening of Streptococcus pneumoniae ABC transporter mutants demonstrates that LivJHMGF, a branched-chain amino acid ABC transporter, is necessary for disease pathogenesis. Infect Immun 77: 3412–3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shelver D, Rajagopal L, Harris TO, Rubens CE (2003) MtaR, a regulator of methionine transport, is critical for survival of group B streptococcus in vivo . J Bacteriol 185: 6592–6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fontecave M, Atta M, Mulliez E (2004) S-adenosylmethionine: nothing goes to waste. Trends Biochem Sci 29: 243–249. [DOI] [PubMed] [Google Scholar]
- 10. Hullo MF, Auger S, Dassa E, Danchin A, Martin-Verstraete I (2004) The metNPQ operon of Bacillus subtilis encodes an ABC permease transporting methionine sulfoxide, D- and L-methionine. Res Microbiol 155: 80–86. [DOI] [PubMed] [Google Scholar]
- 11. Merlin C, Gardiner G, Durand S, Masters M (2002) The Escherichia coli metD locus encodes an ABC transporter which includes Abc (MetN), YaeE (MetI), and YaeC (MetQ). J Bacteriol 184: 5513–5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. den Hengst CD, Groeneveld M, Kuipers OP, Kok J (2006) Identification and functional characterization of the Lactococcus lactis CodY-regulated branched-chain amino acid permease BcaP (CtrA). J Bacteriol 188: 3280–3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bryan JD, Liles R, Cvek U, Trutschl M, Shelver D (2008) Global transcriptional profiling reveals Streptococcus agalactiae genes controlled by the MtaR transcription factor. BMC Genomics 9: 607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sperandio B, Gautier C, McGovern S, Ehrlich DS, Renault P, et al. (2007) Control of methionine synthesis and uptake by MetR and homocysteine in Streptococcus mutans . J Bacteriol 189: 7032–7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greene RC (1996) Biosynthesis of methionine. In: F.C.Neidhardt, et al.., editors. Escherichia coli and Salmonella: cellular and molecular biology: ASM Press, Washington, DC. pp. 542–560.
- 16. Kovaleva GY, Gelfand MS (2007) Transcriptional regulation of the methionine and cysteine transport and metabolism in streptococci. FEMS MicrobiolLett 276: 207–215. [DOI] [PubMed] [Google Scholar]
- 17. Ravanel S, Gakiere B, Job D, Douce R (1998) The specific features of methionine biosynthesis and metabolism in plants. Proc Natl Acad Sci USA 95: 7805–7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lestrate P, Delrue RM, Danese I, Didembourg C, Taminiau B, et al. (2000) Identification and characterization of in vivo attenuated mutants of Brucella melitensis . Mol Microbiol 38: 543–551. [DOI] [PubMed] [Google Scholar]
- 19. Hill CE, Metcalf DS, MacInnes JI (2003) A search for virulence genes of Haemophilus parasuis using differential display RT-PCR. Vet Microbiol 96: 189–202. [DOI] [PubMed] [Google Scholar]
- 20. Ejim LJ, D'Costa VM, Elowe NH, Loredo-Osti JC, Malo D, et al. (2004) Cystathionine beta-lyase is important for virulence of Salmonella enterica serovar Typhimurium . Infect Immun 72: 3310–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lei B, Liu M, Chesney GL, Musser JM (2004) Identification of new candidate vaccine antigens made by Streptococcus pyogenes: purification and characterization of 16 putative extracellular lipoproteins. J Infect Dis 189: 79–89. [DOI] [PubMed] [Google Scholar]
- 22. Garmory HS, Titball RW (2004) ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect Immun 72: 6757–6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, et al. (2001) Complete genome sequence of a virulent isolate of Streptococcus pneumoniae . Science 293: 498–506. [DOI] [PubMed] [Google Scholar]
- 24. Khandavilli S, Homer KA, Yuste J, Basavanna S, Mitchell T, et al. (2008) Maturation of Streptococcus pneumoniae lipoproteins by a type II signal peptidase is required for ABC transporter function and full virulence. Mol Microbiol 67: 541–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Severi E, Randle G, Kivlin P, Whitfield K, Young R, et al. (2005) Sialic acid transport in Haemophilus influenzae is essential for lipopolysaccharide sialylation and serum resistance and is dependent on a novel tripartite ATP-independent periplasmic transporter. Mol Microbiol 58: 1173–1185. [DOI] [PubMed] [Google Scholar]
- 26. Thomas GH, Southworth T, Leon-Kempis MR, Leech A, Kelly DJ (2006) Novel ligands for the extracellular solute receptors of two bacterial TRAP transporters. Microbiology 152: 187–198. [DOI] [PubMed] [Google Scholar]
- 27. Webb AJ, Homer KA, Hosie AH (2008) Two closely related ABC transporters in Streptococcus mutans are involved in disaccharide and/or oligosaccharide uptake. J Bacteriol 190: 168–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brown JS, Ogunniyi AD, Woodrow MC, Holden DW, Paton JC (2001) Immunization with components of two iron uptake ABC transporters protects mice against systemic Streptococcus pneumoniae infection. Infect Immun 69: 6702–6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Johnston JW, Myers LE, Ochs MM, Benjamin WH Jr, Briles DE, et al. (2004) Lipoprotein PsaA in virulence of Streptococcus pneumoniae: surface accessibility and role in protection from superoxide. Infect Immun 72: 5858–5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jomaa M, Terry S, Hale C, Jones C, Dougan G, et al. (2006) Immunization with the iron uptake ABC transporter proteins PiaA and PiuA prevents respiratory infection with Streptococcus pneumoniae . Vaccine 24: 5133–5139. [DOI] [PubMed] [Google Scholar]
- 31. Jomaa M, Yuste J, Paton JC, Jones C, Dougan G, et al. (2005) Antibodies to the iron uptake ABC transporter lipoproteins PiaA and PiuA promote opsonophagocytosis of Streptococcus pneumoniae . Infect Immun 73: 6852–6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chu J, Shoeman R, Hart J, Coleman T, Mazaitis A, et al. (1985) Cloning and expression of the metE gene in Escherichia coli . Archives of biochemistry and biophysics 239: 467–474. [DOI] [PubMed] [Google Scholar]
- 33. Bayle L, Chimalapati S, Schoehn G, Brown J, Vernet T, et al. (2011) Zinc uptake by Streptococcus pneumoniae depends on both AdcA and AdcAII and is essential for normal bacterial morphology and virulence. Mol Microbiol 82: 904–916. [DOI] [PubMed] [Google Scholar]
- 34. Shah P, Swiatlo E (2006) Immunization with polyamine transport protein PotD protects mice against systemic infection with Streptococcus pneumoniae . Infect Immun 74: 5888–5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shah P, Briles DE, King J, Hale Y, Swiatlo E (2009) Mucosal immunization with polyamine transport protein D (PotD) protects mice against nasopharyngeal colonization with Streptococcus pneumoniae . Exp Biol Med (Maywood) 234: 403–409. [DOI] [PubMed] [Google Scholar]
- 36. Arevalo MT, Xu Q, Paton JC, Hollingshead SK, Pichichero ME, et al. (2009) Mucosal vaccination with a multicomponent adenovirus-vectored vaccine protects against Streptococcus pneumoniae infection in the lung. FEMS Immunol Med Microbiol 55: 346–351. [DOI] [PubMed] [Google Scholar]
- 37. Novak R, Cauwels A, Charpentier E, Tuomanen E (1999) Identification of a Streptococcus pneumoniae gene locus encoding proteins of an ABC phosphate transporter and a two-component regulatory system. J Bacteriol 181: 1126–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yuste J, Khandavilli S, Ansari N, Muttardi K, Ismail L, et al. (2010) The effects of PspC on complement-mediated immunity to Streptococcus pneumoniae vary with strain background and capsular serotype. Infect Immu 78: 283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomasz A (1964) Studies on the competence (for genetic transformation) of Diplococcus pneumoniae in a synthetic medium, abstr G87. Abstr 64th Gen Meet Am Soc Microbiol American Society for Microbiology. Washington, DC.
- 40. Ogunniyi AD, Giammarinaro P, Paton JC (2002) The genes encoding virulence-associated proteins and the capsule of Streptococcus pneumoniae are upregulated and differentially expressed in vivo . Microbiol 148: 2045–2053. [DOI] [PubMed] [Google Scholar]
- 41. Ogunniyi AD, Mahdi LK, Trappetti C, Verhoeven N, Mermans D, et al. (2012) Identification of genes that contribute to the pathogenesis of invasive pneumococcal disease by in vivo transcriptomic analysis. Infect Immun 80: 3268–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook J EFF, and Maniatis T (1989) Molecular cloning: a laboratory cloning, 2nd ed. New York: Cold Spring Harbor Laboratory Press.
- 43. Shevchuk NA, Bryksin AV, Nusinovich YA, Cabello FC, Sutherland M, et al. (2004) Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res 32: e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Maqbool A, Levdikov VM, Blagova EV, Herve M, Horler RS, et al. (2011) Compensating stereochemical changes allow murein tripeptide to be accommodated in a conventional peptide-binding protein. J Biol Chem 286: 31512–31521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Saier MH Jr (2000) A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol Mol Biol Rev 64: 354–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yuste J, Ali S, Sriskandan S, Hyams C, Botto M, et al. (2006) Roles of the alternative complement pathway and C1q during innate immunity to Streptococcus pyogenes . J Immunol 176: 6112–6120. [DOI] [PubMed] [Google Scholar]
- 47. Beuzon CR, Holden DW (2001) Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo . Microbes Infect 3: 1345–1352. [DOI] [PubMed] [Google Scholar]