Abstract
Analysis of the kinetics of hybridization in liquid of labeled herpes simplex virus (HSV) 1 and 2 DNAs with excess unlabeled RNA extracted at 2 (early) and 8 (late) h postinfection revealed the following. (i) The RNA transcripts present in the HSV-1-infected cells at 2 and 8 h postinfection are complementary to 44 and 48% of HSV-1 DNA. The RNA transcripts present in the HSV-2-infected cells at 2 and 8 h postinfection are complementary to 21 and 50% of HSV-2 DNA. (ii) The transcripts present in 2-h HSV-1- or HSV-2-infected cells treated with cycloheximide are complementary to 44 and 45% of the respective DNAs. (iii) The RNA transcripts present in the HSV-1-infected cells at 2 h postinfection and in HSV-2-infected cells at 8 h postinfection form 2 classes, abundant and scarce, differing in molar concentrations. The RNA transcripts present in the HSV-2-infected cells at 2 h postinfection form only one abundance class. (iv) The transcripts present in the HSV-1-infected cells at 8 h postinfection are complementary to 24% of HSV-2 DNA and therefore 50% of the transcribed HSV-1 sequences are shared by the two viruses. Of the RNA sequences complementary to HSV-2 DNA, 13% arise from HSV-1 templates specifying abundant RNA and 11% arise from HSV-1 templates specifying scarce RNA. Thus, the DNA sequences shared in common by HSV-1 and HSV-2 DNAs constitute 71% of the HSV-1 templates specifying abundant RNA and 39% of sequences specifying scarce RNA.
Full text
PDF
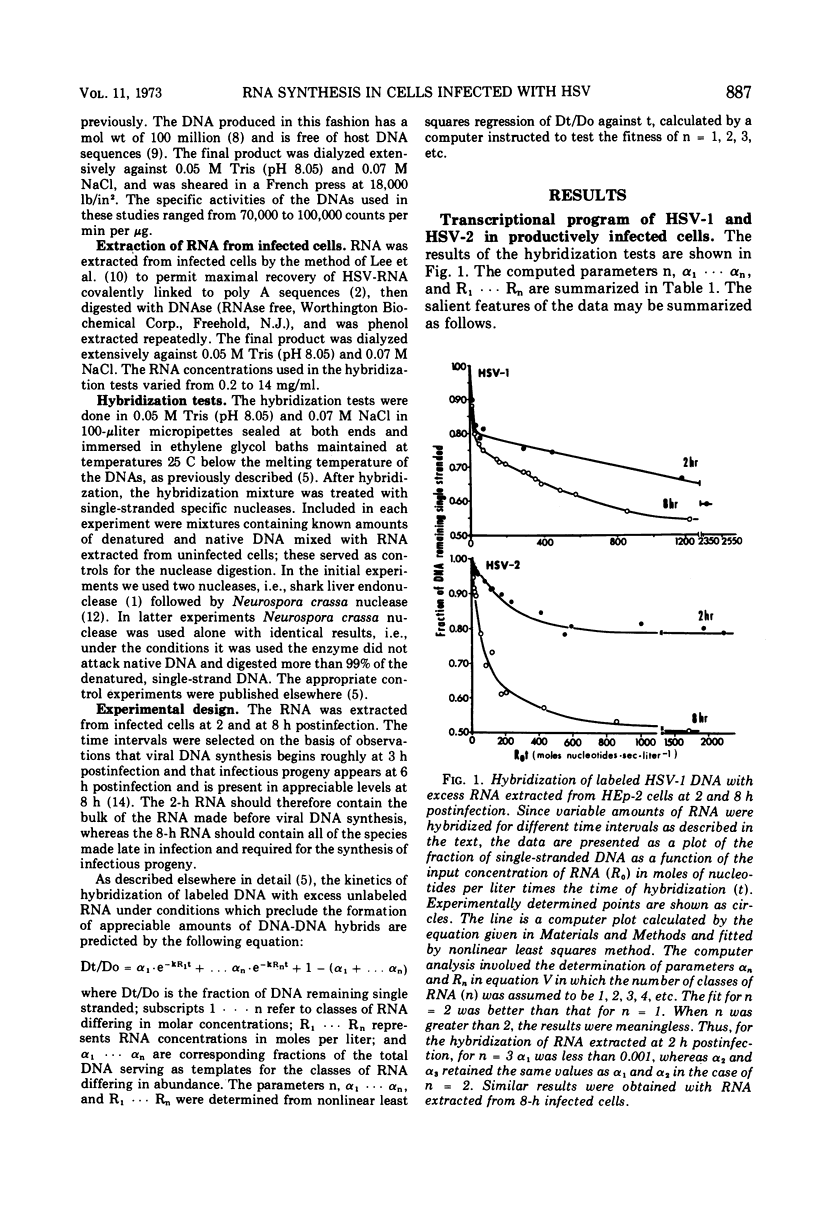
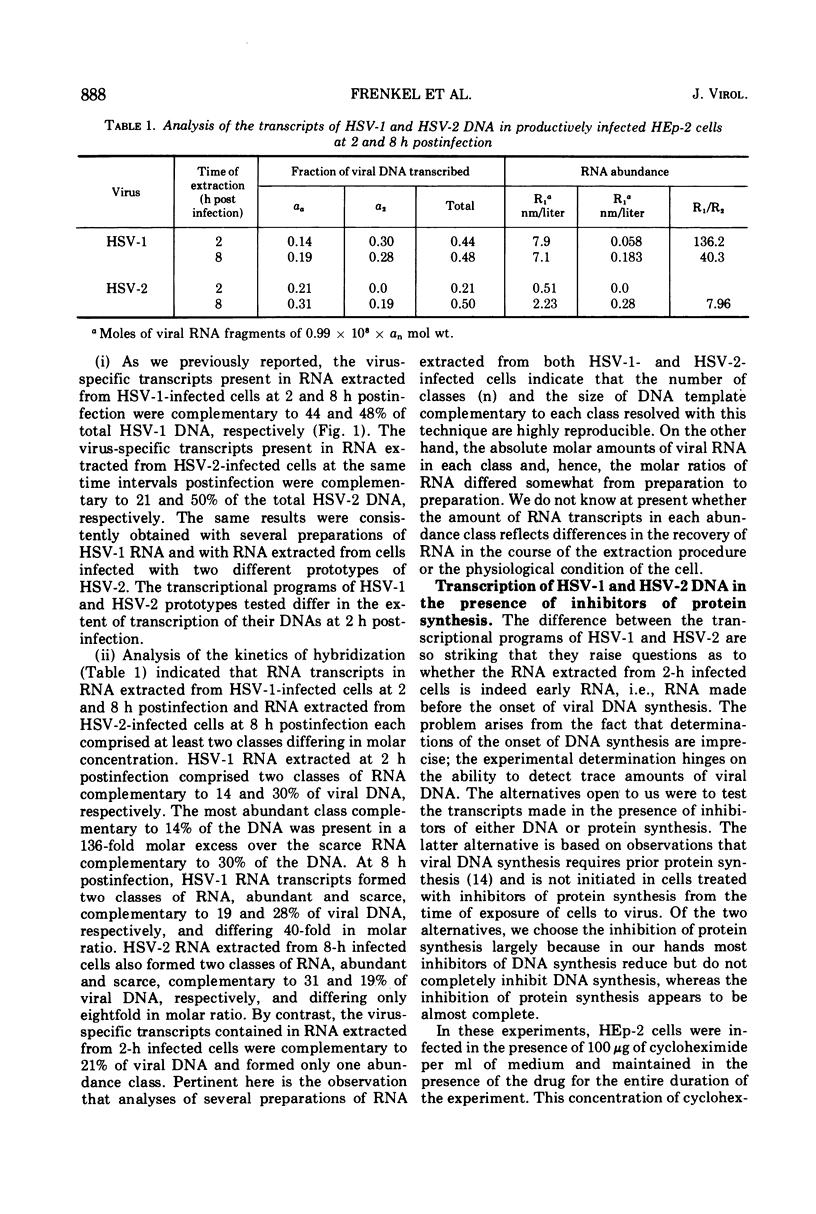
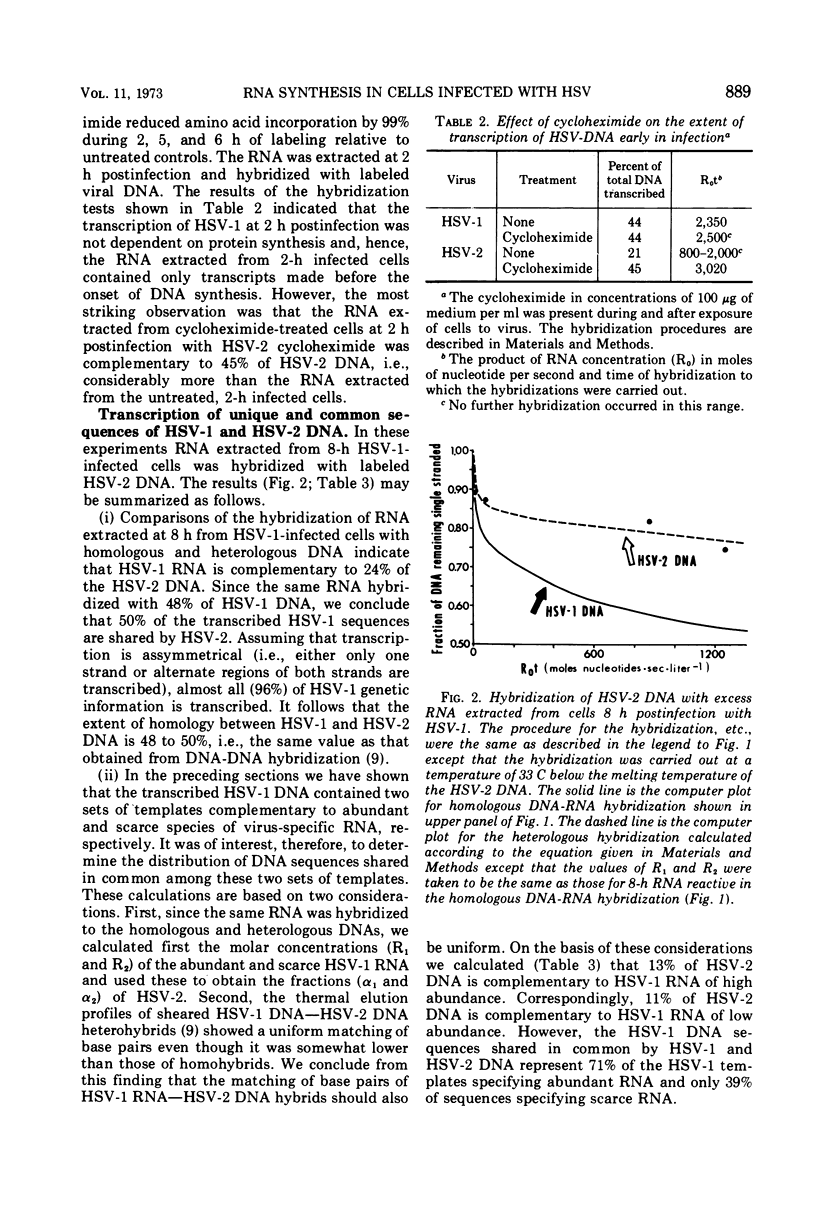
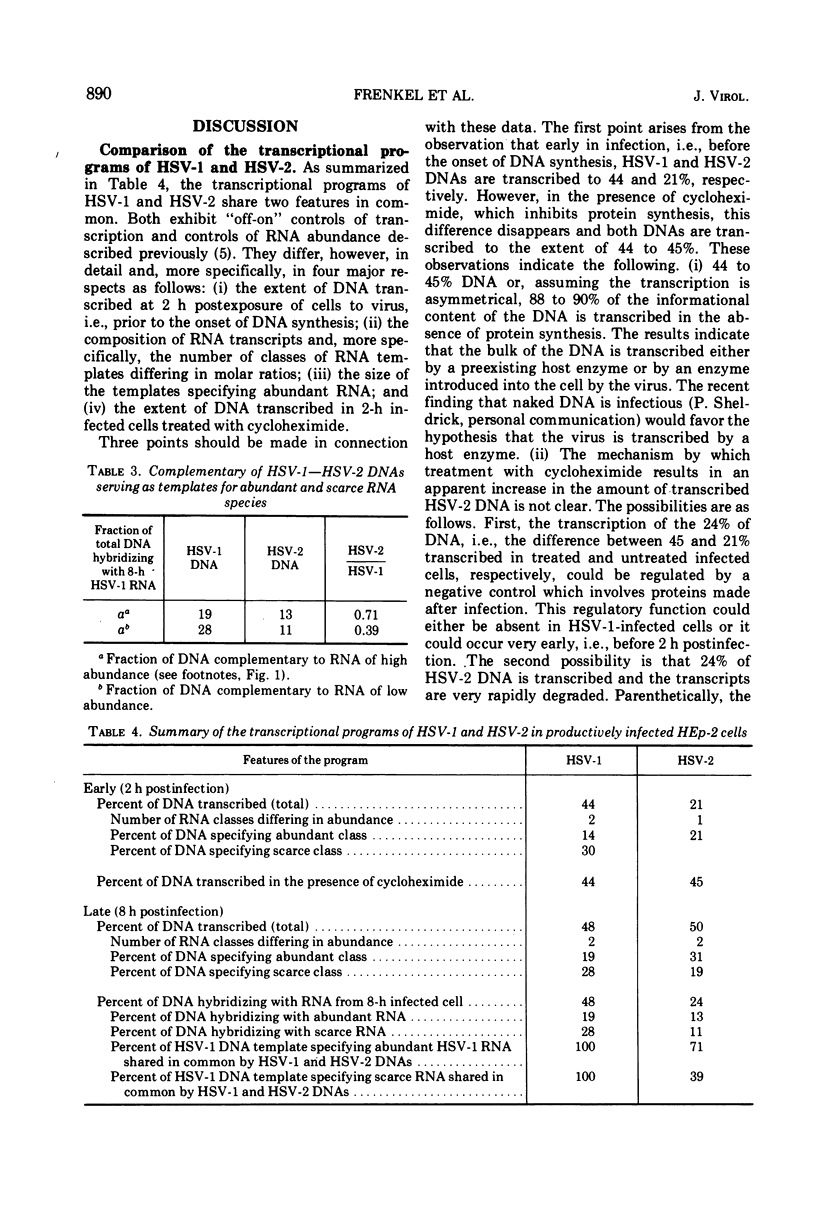


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ASHE H., SEAMAN E., VANVUNAKIS H., LEVINE L. CHARACTERIZATION OF A DEOXYRIBONUCLEASE OF MUSTELUS CANIS LIVER. Biochim Biophys Acta. 1965 May 18;99:298–306. doi: 10.1016/s0926-6593(65)80126-6. [DOI] [PubMed] [Google Scholar]
- Bachenheimer S. L., Roizman B. Ribonucleic acid synthesis in cells infected with herpes simplex virus. VI. Polyadenylic acid sequences in viral messenger ribonucleic acid. J Virol. 1972 Oct;10(4):875–879. doi: 10.1128/jvi.10.4.875-879.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejercito P. M., Kieff E. D., Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968 May;2(3):357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- Frenkel N., Roizman B., Cassai E., Nahmias A. A DNA fragment of Herpes simplex 2 and its transcription in human cervical cancer tissue. Proc Natl Acad Sci U S A. 1972 Dec;69(12):3784–3789. doi: 10.1073/pnas.69.12.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel N., Roizman B. Herpes vimplex virus: genome size and redundancy studied by renaturation kinetics. J Virol. 1971 Oct;8(4):591–593. doi: 10.1128/jvi.8.4.591-593.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel N., Roizman B. Ribonucleic acid synthesis in cells infected with herpes simplex virus: controls of transcription and of RNA abundance. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2654–2658. doi: 10.1073/pnas.69.9.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel N., Roizman B. Separation of the herpesvirus deoxyribonucleic acid duplex into unique fragments and intact strand on sedimentation in alkaline gradients. J Virol. 1972 Oct;10(4):565–572. doi: 10.1128/jvi.10.4.565-572.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieff E. D., Bachenheimer S. L., Roizman B. Size, composition, and structure of the deoxyribonucleic acid of herpes simplex virus subtypes 1 and 2. J Virol. 1971 Aug;8(2):125–132. doi: 10.1128/jvi.8.2.125-132.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieff E., Hoyer B., Bachenheimer S., Roizman B. Genetic relatedness of type 1 and type 2 herpes simplex viruses. J Virol. 1972 May;9(5):738–745. doi: 10.1128/jvi.9.5.738-745.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. Y., Mendecki J., Brawerman G. A polynucleotide segment rich in adenylic acid in the rapidly-labeled polyribosomal RNA component of mouse sarcoma 180 ascites cells. Proc Natl Acad Sci U S A. 1971 Jun;68(6):1331–1335. doi: 10.1073/pnas.68.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig H. O., Biswal N., Benyesh-Melnick M. Studies on the relatedness of herpesviruses through DNA-DNA hybridization. Virology. 1972 Jul;49(1):95–101. doi: 10.1016/s0042-6822(72)80010-2. [DOI] [PubMed] [Google Scholar]
- ROIZMAN B., ROANE P. R., Jr THE MULTIPLICATION OF HERPES SIMPLEX VIRUS. II. THE RELATION BETWEEN PROTEIN SYNTHESIS AND THE DUPLICATION OF VIRAL DNA IN INFECTED HEP-2 CELLS. Virology. 1964 Feb;22:262–269. doi: 10.1016/0042-6822(64)90011-x. [DOI] [PubMed] [Google Scholar]
- Rabin E. Z., Preiss B., Fraser M. J. A nuclease from Neurospora crassa conidia specific for single-stranded nucleic acids. Prep Biochem. 1971;1(4):283–307. doi: 10.1080/00327487108081946. [DOI] [PubMed] [Google Scholar]
- Rakusanova T., Ben-Porat T., Himeno M., Kaplan A. S. Early functions of the genome of herpesvirus. I. Characterization of the RNA synthesized in cycloheximide-treated, infected cells. Virology. 1971 Dec;46(3):877–889. doi: 10.1016/0042-6822(71)90088-2. [DOI] [PubMed] [Google Scholar]
- Roizman B., Spear P. G. Preparation of herpes simplex virus of high titer. J Virol. 1968 Jan;2(1):83–84. doi: 10.1128/jvi.2.1.83-84.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B. The herpesviruses--a biochemical definition of the group. Curr Top Microbiol Immunol. 1969;49:3–79. [PubMed] [Google Scholar]



