Abstract
Human WRN and BLM genes are members of the conserved RECQ helicase family. Mutations in these genes are associated with Werner and Bloom syndromes. WRN and BLM proteins are implicated in DNA replication, recombination, repair, telomere maintenance, and transcription. Using microfluidics-assisted display of DNA for replication track analysis (ma-RTA), we show that WRN and BLM contribute additively to normal replication fork progression, and non-additively, in a RAD51-dependent pathway, to resumption of replication after arrest by hydroxyurea (HU), a replication-stalling drug. WRN but not BLM is required to support fork progression after HU. Resumption of replication by forks may be necessary but is not sufficient for timely completion of the cell cycle after HU arrest, as depletion of WRN or BLM compromises fork recovery to a similar degree, but only BLM depletion leads to extensive delay of cell division after HU, as well as more pronounced chromatin bridging. Finally, we show that recovery from HU includes apparent removal of some of the DNA that was synthesized immediately after release from HU, a novel phenomenon that we refer to as nascent strand processing, NSP.
Keywords: DNA replication/human, hydroxyurea, RECQ helicase, Werner syndrome, Bloom syndrome, mitosis
1. Introduction
RECQ helicases are a family of proteins conserved from bacteria to humans. Out of five human RECQ helicase genes, three are associated with heritable disorders. Mutations in the BLM [1], and WRN [2] genes cause, respectively, Bloom syndrome (BS) and Werner syndrome (WS), and mutations inRECQL4 [3] are seen in Rothmund-Thompson, RAPADILINO, and Baller-Gerold (BGS) syndromes.
Clinical manifestations of Werner syndrome mimic premature aging, while Bloom syndrome is associated with developmental abnormalities [4]. Bloom and Werner syndromes are cancer-prone diseases, albeit the spectra of cancers they predispose to are different. Cells mutated in BLM or WRN genes show phenotypes associated with genomic instability and perturbed replication: slower S phase, increased fraction of cells at the G2/M boundary of the cell cycle, and expression of some fragile sites (for review, see [5–8]). In vitro, several biochemical features are unique to BLM or WRN, warranting a systematic analysis of the redundancy and cooperation between these two RECQs within a cell. Studies in DT40 cells demonstrated synthetic hypersensitivity of WRN/BLM knock-out cells to a number of genotoxic drugs, including camptothecin [9], as well as unique genetic interactions between these RECQs and other genes [10], pointing towards WRN and BLM’s complementary roles within pathways of DNA metabolism, and inviting a more mechanistic inquiry.
The insight into roles of WRN and BLM in DNA replication is complicated by the facts that both RECQs are multifunctional proteins [4,11], and that replication fork metabolism is likely conducted through several interconnected pathways [8,12]. Briefly, when fork progression is interrupted by lesions in the template or by replisome poisoning, extra activities are turned on as part of the S phase checkpoint, and stabilize the replisome-DNA structure against collapse [12,13]. It is thought that collapsed replication forks are susceptible to double strand breaks (DSBs). These DSBs may be an intermediate in an active fork rescue pathway, or merely a breakdown product which necessitates repair (see refs. above). The exact balance between fork stabilization and fork collapse/rescue may depend on the cell type and the nature of interruption facing a fork.
Early studies have suggested that both WRN and BLM can be involved in elongation of DNA replication (reviewed in [8]). The use of DNA fiber technology allowed further insight into roles of RECQ helicases at a replication fork, demonstrating that WRN [14] and BLM [15] may be required for normal fork progression. In addition, complementing BS patient-derived human fibroblasts with BLM improves resumption of replication fork progression after an arrest with hydroxyurea (HU), a ribonucleotide reductase inhibitor [16]. Defects of fork recovery, albeit variable, were also demonstrated in WRN-depleted HeLa cells, in WS fibroblasts [17,18], and in WRN-depleted fibroblasts [19]. Both RECQs are targeted by the checkpoint kinase ATR [18,20–22] and affect checkpoint performance [16,23,24].
In order to delineate redundant vs. cooperative functions of WRN and BLM, we have established isogenic human fibroblasts depleted of WRN, BLM, or both RECQs [25]. Here, we undertake a detailed analysis of replication fork phenotypes in these cells, and describe both unique and shared functions of WRN and BLM at a replication fork, as well as uncover a novel process of metabolizing nascent strands during recovery from HU.
2. Materials and Methods
2.1. Cells and culture
SV40-transformed GM639 fibroblast cell line was obtained from the Coriell Institute Cell Repositories (Camden NJ). GM639cc1 is a pNeoA derivative of GM639 [19,25,26]. Unless stated otherwise, all experiments were performed using this cell line. The large T antigen is at least partially inactivated in this cell line since it does not support replication of SV40 origin-containing plasmids (JS, unpub.).
The primary human dermal fibroblasts were described [27]. All cell lines were grown in Dulbecco Modified Minimal Essential Medium (DMEM) supplemented with L-glutamine, sodium pyruvate, 10% fetal bovine serum (Hyclone, Ogden, UT) and antibiotics in a humidified 5% CO2, 37°C incubator.
2.2. Drugs and Dyes
Stock solutions of 5-bromodeoxyuridine (BrdU; 10 mM in water), 5-iododeoxyuridine (IdU, 2mM in PBS), 5-chlorodeoxyuridine (CldU, 10mM in water), 5-ethynyldeoxyuridine (EdU, 10mM in DMSO), hydroxyurea (HU, 1M in PBS), and cytochalasin-B (600μg/ml in DMSO) were stored at −20° C. All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) with the exception of EdU (Invitrogen). CldU, BrdU, and IdU were used at concentrations of 50μM and EdU was used at 10μM.
2.3. RNAi-mediated depletion of WRN, BLM, and RAD51
Short hairpin (sh) RNA constructs for depletion of WRN and BLM are described [19,25]. pLKO.1-based shRNA constructs against human RAD51 were purchased from Open Biosystems (Cat No. RHS4533-NM_002875). Depletions were carried out as described [19,25].
2.4. Western blotting
Western blotting of WRN was done as described [19,25] with the rabbit α-WRN (Novus Cat. No. NB100472A) or mouse α-WRN 195C (provided by Dr. Opresko). Rabbit α-BLM antibody against BLM C-terminal peptide (KPINRPFLKPSYAFS was described [25]. α-RAD51 antibodies were rabbit polyclonal (Cat. No. PC130, Calbiochem, La Jolla, CA), and mouse monoclonal, (Cat. No.05–530, Millipore, Temecula CA). Mouse α-CHK1 antibody was from Santa Cruz (Cat. No. sc-8408). Phosphorylation of CHK1 and CHK2 was analyzed with a Phospho-Chk1/2 Antibody Sampler Kit (Cell Signaling, Cat. No. 9931). All proteins were visualized by ECL (Amersham) and quantified using Storm Phosphorimager and ImageQuant software (Molecular Dynamics). For presentation, images were saved in TIFF format, adjusted for brightness/contrast and cropped using Adobe Photoshop, and assembled into figures in CorelDraw. Brightness/contrast adjustments were made to entire images.
2.5. Staining for BrdU incorporation and FACS
Staining for BrdU was done as described [19]. FACS data analysis and presentation were done with Summit software (Dako, Carpinteria, CA), and cell cycle phase quantitations were done with FACS express software (Phoenix Flow Systems, San Diego, CA).
2.6. Microchannel fabrication, DNA fiber stretching and replication track analysis
These procedures were done as described [19,28]. The mouse antibody against total DNA was from Chemicon (Cat.No. MAB3034). Microscopy of stretched DNAs was performed on the Zeiss Axiovert microscope with a 63x objective. Lengths of tracks were measured in raw merged images (jpegs) using Zeiss AxioVision software. Details of statistical analysis are described in Figure Legends.
2.7. Nucleoplasmic bridge measurements
Cells were pulse-labeled with 10μM EdU for 1 hr and then arrested with 2mM HU for 6 hrs. After release from HU, cytochalasin-B was added at 2μg/ml. Cells were harvested by trypsinization in 20 hrs and cytospun onto poly-L-lysin-coated slides (Sigma). Cells were fixed for 10 min in 4% paraformaldehide, 0.2% Triton X-100, 20mM Pipes pH6.8, 1mM MgCl2, 10mM EGTA, washed with PBS, and stained for EdU incorporation (by Click-It reaction with AlexaFluor 594 azide according to the manufacturer’s recommendations (Invitrogen), as well as for total DNA using Hoechst33342. Slides were mounted in Vectashield (Vector labs) and examined under 40x magnification using Zeiss Axiovert microscope and AxioVision software. Images of binucleated cells were collected. Scoring was done according to [29]. 60–200 each of EdU+ and EdU− binucleated cells in each sample were inspected for nucleoplasmic bridges.
2.8. Microscopy image presentation
Visual scoring or measurement of features in microscopy images was done in sets of multicolor jpeg files in AxioVision. For presentation, images were adjusted for brightness/contrast and cropped in Adobe Photoshop, and assembled into figures in CorelDraw. Adjustments were always done to entire images. In some cases, brightness/contrast of individual color channels was adjusted separately.
3. Results
3.1. WRN and BLM contribute additively to fork progression rates during an unperturbed S phase
We depleted WRN and/or BLM with lentiviral shRNAs, as before ([19,25] and Figs. S1A, S3C, 4A, S4B), achieving at least 80–85% depletion of the target protein(s). Growth rate was lower in WRN-depleted and, more dramatically, in BLM-depleted cell populations, which reflected the size of replicating fraction. To account for it, every assay used in this study discriminated between replicating and non-replicating fractions, or focused exclusively on replicating fraction.
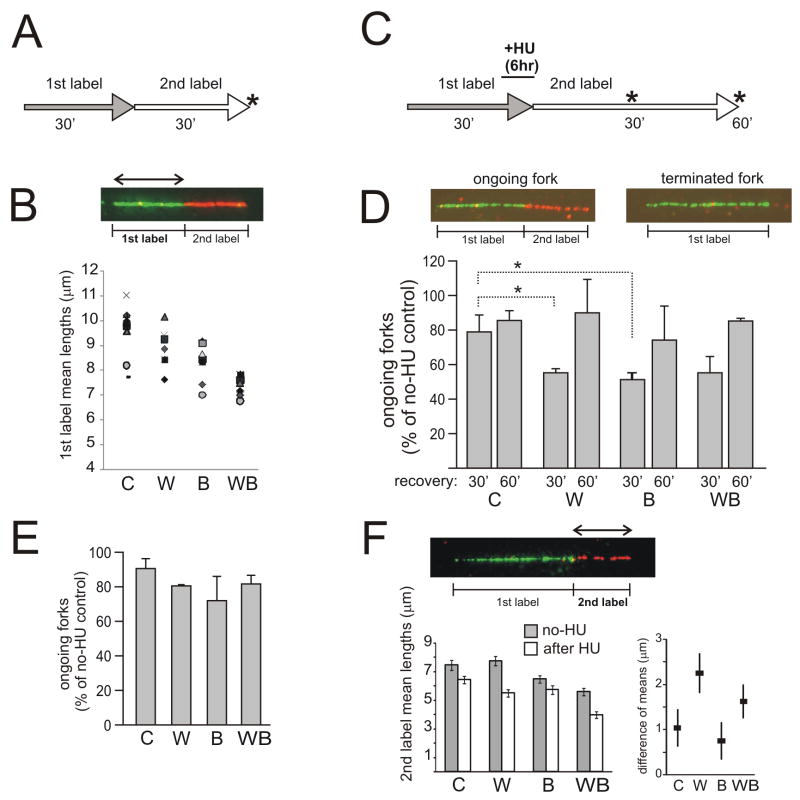
We labeled cells with two nucleotide analogs (CldU and IdU) for 30 min each and used immunofluorescence to visualize tracks of replication in DNA stretched using microfluidics [28]. We measured lengths of 1st and 2nd label segments in two-segment tracks that incorporated both labels in tandem and thus correspond to ongoing forks (Fig. 1A, B) Analysis of multiple independent experiments revealed statistically significant genotype-specific differences in track lengths (Supplemental Tables S1 and 2). Fig. 1B shows summary data for 1st label segments, as these may give a more accurate representation of fork progression rates than 2nd label segments, since they are less likely to be limited by replicon size [28]. 2nd label segment data are summarized in Tables S1 and 2. 1st segments of tracks (as well as whole tracks) were shorter in BLM-depleted cells compared to WRN-depleted cells. WRN/BLM-depleted cells had the shortest tracks, significantly different from WRN- or BLM-depleted cells. This result demonstrates that additive phenotypes can be observed using co-depletion in lieu of genetic manipulation.
Figure 1.
WRN and BLM exert additive effects on replication fork progression in a normal S phase, and non-additive effects on fork reactivation after HU. A) Labeling scheme. Asterisks mark sample collection time points. B) Example of a two-color, ongoing fork in which 1st label segments were measured. Lengths (in μm) of 1st and 2nd label segments in these ongoing forks were measured in up to 9 independent experiments. Mean values of 1st label segments derived in each experiment are plotted as a function of shRNA type. Different markers stand for individual experiments. See Supplemental Tables S1 and S2 for additional information. Designations here and elsewhere are: C, cells mock-depleted with no-shRNA lentivirus pLKO.1; W, WRN-depleted cells; B, BLM-depleted cells; WB, WRN/BLM-depleted cells. C) Labeling scheme for HU arrest/recovery experiments. 2mM HU was added at the end of a 30min 1st labeling interval. In 6 hrs, HU and the 1st label were removed and 2nd label was added. D) Ongoing forks (1st label–2nd label) and terminated or inactivated forks (1st label only) were counted to determine prevalence of ongoing forks. Track counts per experiment, for each cell type, totaled 361 on average. Fork reactivation after HU was expressed as prevalence of ongoing forks seen after HU, normalized to the prevalence of ongoing forks in untreated cells. These values were obtained from two independent experiments and averaged. Error bars are standard deviations. One-tailed Pvalues were determined in t-tests in pair-wise comparisons between control and depleted cells. PC/W=0.0417 (marked as *), PC/B=0.034, PC/WB=0.065. E) Fork reactivation in cells that have been labeled as in (C), but exposed to 0.5mM HU for 6 hrs and allowed to recover from HU for 30min. F) Mean lengths of 2nd label segments were measured in ongoing forks w/o HU and in forks reactivating after a 6-hr treatment with 0.5mM HU. The values were obtained in two independent experiments and pooled (the average number of measurements, N, per pooled sample was 363). Error bars are 95% confidence intervals of the means. The left panel shows mean lengths w/o HU and after 0.5mM HU, and the right panel shows differences between these means (μno-HU − μHU, black squares), and 95% confidence intervals for each difference (error bars). See Figure S1C and Table S3 for supporting information.
3.2. Comparable, non-additive contributions of WRN and BLM to fork response to arrest by HU
Previous work suggested that BLM- or WRN-deficient cells have a decreased ability to restart and/or elongate replication forks stalled by HU [16–19]. We labeled WRN-, BLM-, and WRN/BLM- depleted cells and controls with the 1st label for 30 min prior to and then during a 6 hr arrest by 2mM HU, followed by the 2nd label after HU (Fig. 1C, S1B). Reactivation of forks after HU should result in tracks labeled in tandem with two labels. The prevalence of these two-segment tracks was quantified as a fraction of all tracks that contained the first label.
Without HU, prevalence of two-segment tracks was similar in all cell lines and measured around 70% (Fig. S1B). In HU-treated samples two-segment tracks were less prevalent, reflecting inactivation of forks by HU. Fig. 1D demonstrates this by comparing percent of ongoing forks relative to no-HU samples. As seen in Fig. 1D, WRN- or BLM-depleted cells displayed a very similar reduction in the fraction of forks that were able to resume replication within the first 30 min after release. WRN/BLM-depleted cells behaved as single-depleted cells. Both WRN- and BLM depleted cells were able to reactivate additional forks if recovery was measured at 60 min after release.
3.3. A distinct effect of WRN-depletion on fork progression during recovery
The lengths of tracks incorporated immediately after HU can be shorter than lengths of tracks incorporated over the same period of time if no HU was present [19,30]. This effect is more pronounced in WRN-depleted cells [19]. We also observed a similar phenotype in BLM-depleted cells (not shown). However, short tracks synthesized within the first 30–60 minutes after HU may reflect a delay in fork reactivation, rather than a specific post-HU elongation defect. To distinguish between these possibilities, we sought conditions of HU treatment that minimize fork inactivation.
We found that incubating control and RECQ-depleted cells with 0.5mM instead of 2mM HU permitted measurable though slow fork progression (on average 0.1Kb/min), and did not appear to substantially inactivate forks (Fig. 1E). We next measured the lengths of post-HU segments in these ongoing forks and compared them to no-HU controls. Whenever WRN was depleted, either alone or along with BLM, the lengths of the post-HU segments were comparatively more shortened than in controls or in BLM-depleted cells (Figs. 1F, S1C, Table S3). This agrees with the notion that coordinating fork progression during recovery may be a specific and separate function that involves WRN.
3.4. Nascent strand processing is observed during fork reactivation
While performing track length measurements in reactivated forks after HU, we noticed small but consistent fluctuations in lengths of 1st label segments. Depending on the time point of recovery, we could detect either lengthening or shortening of these segments. We reasoned this may indicate either that different populations of stalled forks are activated at different times during recovery, or that an additional event happens to the forks that have resumed replication.
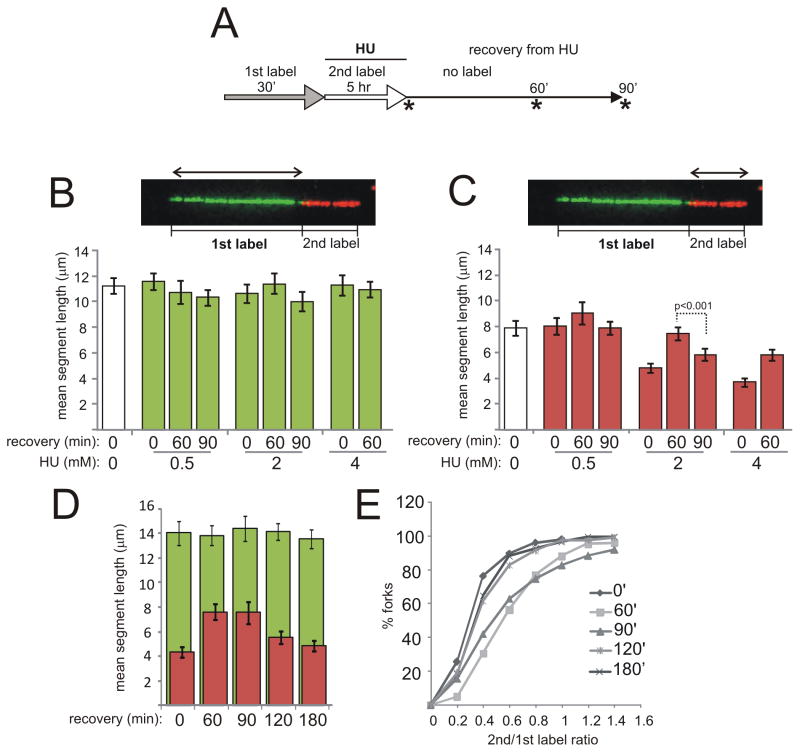
In order to distinguish between the above possibilities and verify that the observed phenomenon is not peculiar to transformed cells, we used a different labeling scheme and primary human fibroblasts (Fig. 2). We incubated cells with the 1st label (IdU) for 30min, then replaced it with the 2nd label (CldU) together with HU. After 5hrs, cells were released into label-free media. Samples were taken at 0, 60, and 90 min after release (Fig. 2A). Tracks of forks ongoing before and during HU (i.e. containing 1st and 2nd label segments) were analyzed.
Figure 2.
Nascent strand processing is observed in replication forks of primary human fibroblasts recovering from HU. The Figure presents data from two independent, representative experiments (panels B, C and panels D, E). The average N per sample was 134. A) Labeling scheme. Samples were collected at time points marked by asterisks. The types of track segment length measurements collected in this experiment are shown in images in (B) and (C). Mean lengths of 1st label (B) and 2nd label (C) segments in reactivated forks were determined for each HU treatment and recovery regimen. White bars in each graph are reference values for ongoing forks labeled without HU and for 30 min each with 1st and 2nd label. Error bars are 95% confidence intervals of the means, and a representative P value shown above bars was determined in Mann-Whitney U test. See Figure S2A and B for supporting information. D) An independent experiment in primary fibroblasts showing mean 2nd label segment gain and loss after release from 2mM of HU as well as respective 1st label segment means. E) The same data as in (D) were plotted as ratios of 2nd to 1st label segment lengths for each two-segment fork. Shown is a cumulative distribution, i.e. a Y axis shows a fraction of values in a dataset that are equal to or less than a given value on an X axis.
First, we found that in the presence of HU, on average 4–8 μm (16–24 Kb) of DNA was synthesized in 5 hrs, depending on HU concentration (Fig. 2C). Second, during the first 60 min of recovery, from 1–3μm (4–12 Kb) more of labeled DNA was added, presumably from a residual intracellular pool of the 2nd label. Higher concentrations of HU were associated with post-HU addition of longer tracks of labeled DNA. This may be expected, assuming that if less DNA is synthesized during HU arrest, then the residual intracellular pool of labeled nucleotide is higher.
Importantly, at later times during recovery (90 min or later) some of the additional length gain that 2nd label segments had experienced was apparently reversed. This was evident in a reduction of average lengths of 2nd label segments (Fig. 2C, D, Fig. S2A), as well as in a decrease in 2nd to1st segment ratios for each fork (Fig. 2E).
No comparable change occurred in the corresponding 1st label segments synthesized before HU (Fig. 2B, D). The fraction of two-segment tracks among all tracks containing the 1st label did not increase between 60 and 90 min of recovery, but in fact decreased (Fig. S2B), suggesting that reduction in overall lengths of 2nd label segments was not due to emergence of a new population of reactivated forks. Instead, the data are more consistent with removal of some of the incorporated 2nd label in the already reactivated forks, resulting, in a subset of cases, in tracks that even appear not to contain any 2nd label. We will refer to this novel phenomenon as nascent strand processing, or NSP, with the caveat that we make no inference to its mechanism.
If no HU had been added, no change in 2nd label segment lengths in ongoing forks occurred during the first hour after labeling (Fig. S2C). Also, neither post-arrest addition of new DNA, nor NSP occurred if aphidicolin was substituted for HU (Figure S2D).
3.5. WRN and BLM differentially affect post-HU addition of new DNA
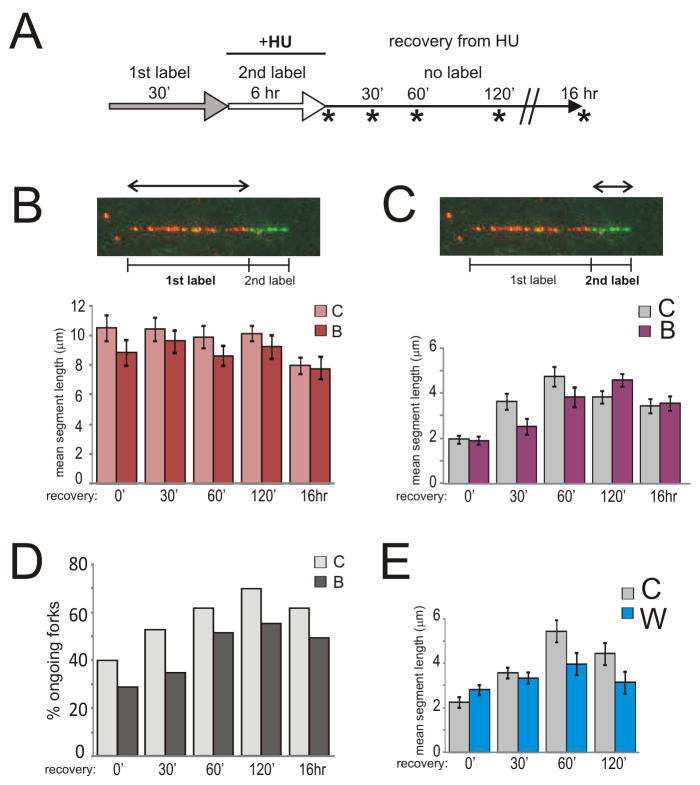
WRN- or BLM-depleted SV40 transformed fibroblasts were labeled with the 1st label (CldU) before HU and with the 2nd label (IdU) during HU incubation, then released into label-free media and harvested 0, 30, 60, 120 min and 16 hrs after release (Fig. 3A).
Figure 3.
WRN and BLM helicases differentially affect post-HU DNA synthesis. A) Labeling scheme. Mean lengths of 1st label (B) and 2nd label (C) segments in reactivated forks were determined for the specified time points during recovery from 2mM HU/6hr arrest in controls and BLM-depleted cells. D) Percent of reactivated forks (i.e. forks containing consecutive 1st and 2nd label segments) among all forks containing the 1st label measured in samples shown in (B) and (C). Panels B, C, and D describe a representative experiment. The average N per sample was 125 ongoing fork tracks for (B) and (C), and 306 total tracks for (D). E) Mean lengths of 2nd label segments in reactivated forks were determined for the specified time points during recovery from 2mM HU/6hr arrest in controls and WRN-depleted cells. Shown are results of a representative experiment. The average N per sample was 103. Error bars in panels B, C, E are 95% confidence intervals of the means.
Only 2nd label segments changed lengths within 2 hrs after HU (Fig. 3B, C, Fig. S2F), although we could detect minor shortening of 1st label segments in all cells at 16 hr post HU. In BLM-depleted cells, 2nd label segments gained and lost as much length as in controls, but they did it on a delayed schedule (Fig. 3C, peak lengths at 120 min for BLM-depleted cells vs. 60 min for control). The relative abundance of two-segment tracks, i.e. DNA labeled both pre- and post-HU, mostly paralleled gain and loss of 2nd label segment length (Fig. 3D).
WRN-depleted cells behaved differently than BLM-depleted cells. In WRN-depleted cells lengths of 2nd label segments did not increase as much as in controls (Fig. 3E). However, both the small gain and loss of length in 2nd label segments appeared to occur at the same time as in control. Fig. S2F shows an independent comparison of WRN-depleted and BLM-depleted cells in one experiment, illustrating the difference between the effects exerted by these two RECQ helicases on post-HU DNA synthesis.
3.6. WRN and BLM are involved in fork reactivation via a pathway that may include RAD51
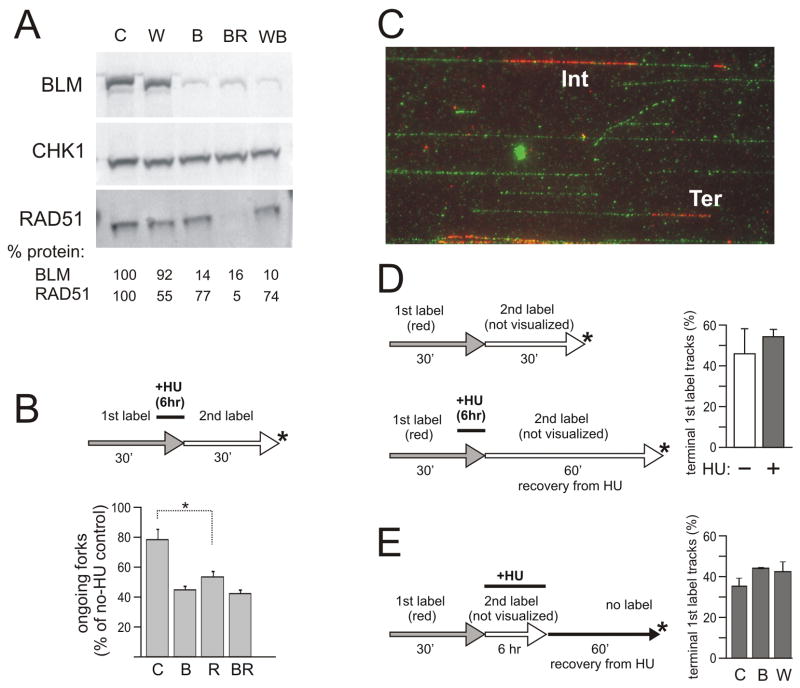
Numerous studies described physical and/or functional interactions between BLM or WRN, and RAD51 [31–35], as well as altered RAD51 function in BLM or WRN mutant cells in response to HU [17,18,35–37]. RAD51 directly participates in at least a subset of pathways of fork reactivation, where it restores a fork by enabling invasion of a DNA duplex by a single-stranded 3′ DNA tail [38,39]. To ask whether activities of WRN or BLM in replication fork resumption depend on RAD51, we depleted RAD51 in SV40-transformed fibroblasts using shRNAs cloned in the same lentiviral vector backbone as WRN or BLM shRNAs. Two out of five shRNAs depleted 80–90% of the protein (Fig. S3A), and these had the most negative impact on cell growth and cell cycle progression (Fig. S3B), causing a delay of the G2/M transition, as expected [40]. We were also able to co-deplete WRN or BLM together with RAD51, using the same approach as previously with WRN/BLM co-depletions (Fig. 4A). Co-depletion of WRN and RAD51 appeared to reduce the attainable level of WRN depletion (Figure S3C).
Figure 4.
A, B) BLM and RAD51 exert comparable, non-additive effects on fork reactivation after HU. A) A Western blot showing examples of shRNA-mediated depletion of BLM, and/or RAD51. CHK1 was used as a loading control [19,25]. Values below images represent BLM or RAD51 levels normalized to CHK1 levels. Note that depletion of BLM appears to mildly affect RAD51 levels. B) Ongoing forks seen after 30 min recovery from 2mM HU/6hrs in control, BLM-, RAD51- and BLM/RAD51-depleted cells (values expressed as percent of untreated control). Two measurements per cell type obtained in one experiment were averaged. The average N per sample was 308. Error bars are standard deviations. The one-tailed P value PC/R=0.01 was calculated in a t-test. C–E) Comparable Replication Associated breakage is detected in WRN, BLM-depleted cells and controls. C) An example of stretched DNA stained with anti-dT antibody (green) and anti-CldU antibody (red). IN, a replication track is internal to a DNA molecule. TER, a replication track is located at a terminus of a DNA molecule. D, E) Labeling schemes with sample collection points and the graphs showing average percent of terminally located tracks among all 1st label, red tracks in the collected samples from 2 independent experiments. Average N per sample was 147 (panel D) and 160 (panel E). Error bars are standard deviations. HU was added at 2mM.
RAD51 depletion reduced the efficiency of fork reactivation after 6 hrs of HU (as expected, [38]), while BLM/RAD51-depleted cells had only a slightly lower efficiency of fork reactivation than either RAD51- or BLM-depleted cells (Fig. 4B). Thus, combining RAD51 and BLM deficiencies did not have a synthetic negative effect on overall efficiency of fork reactivation. A similar result was obtained when we co-depleted WRN and RAD51 (Fig. S3D), though these data may be considered less definitive given the lower depletion level of WRN attainable in WRN/RAD51-depleted cells.
We also measured post-HU addition of new DNA and NSP in RAD51-depleted cells (Figure S3E). Similar to WRN-depleted cells, RAD51-depleted cells added shorter segments of new DNA than controls to preexisting forks during a post-arrest “spurt” of DNA synthesis. This phenotype precludes conclusive determination whether NSP occurred or not.
At least in some cell types, RAD51 contributes to fork reactivation only after relatively short HU arrests when forks have not yet collapsed to DSBs [38]. Under these conditions RAD51 is recruited to chromatin but does not form foci, which is in contrast to long HU arrests where both RAD51 foci and DSBs are readily detectable. We looked for RAD51 foci formation under our HU treatment conditions and specifically in cells that have been replicating DNA prior to HU addition (this is the population in which ma-RTA measures fork reactivation). Cells were labeled with EdU just prior to incubation with HU (Fig. S4). We found RAD51 foci in 50–70% of EdU+ cells, whether wild type, or RECQ-depleted. In EdU- cells, percent of RAD51 foci-positive cells was much lower and, as expected, was RECQ-dependent (control, 9.4±3.6%, WRN-depleted, 17.3±8.4%, BLM-depleted, 27.6±8%). Prevalence of RAD51 foci in EdU+ cells likely corresponded to S phase per se rather than being a response to EdU, since nuclear density and localization of RAD51 foci did not correlate with that of EdU foci (not shown). Importantly, there was no increase in RAD51 foci-positive EdU+ cells during the first hour of recovery from HU arrest in either cell line (instead, there was a slight decline in percent of EdU+ cells containing RAD51 foci, Fig. S4). This observation is consistent with the idea that our HU treatment regimen does not cause significant fork collapse.
3.7. No selective susceptibility of replication intermediates to breakage in BLM-depleted versus WRN-depleted cells
We previously showed that in our model system, depleting BLM but not WRN resulted in reduced cell survival after a 24 hr arrest with 0.5mM HU [25]. However, our results thus far suggest no correlation between fork reactivation efficiency and increased HU sensitivity of BLM- versus WRN-depleted cells. On the other hand, both WRN or BLM-deficient cells have been shown to develop more DSBs than controls after prolonged, 12–24 hr [17,35], and in some cell lines even relatively short, ~6 hr [18] HU arrests, which is suggestive of fork breakage. While our RAD51 foci data may suggest that there is no wide-spread fork breakage in any of our cell lines, it is still possible that differential HU sensitivity of BLM-depleted cells in due to a minority of forks that do not resume replication after HU and instead develop DSBs or breakage-sensitive intermediates.
We used a modification of ma-RTA to measure if sites of HU-stalled replication are susceptible to breakage. DNA from some of the experimental sets described above was stretched and stained with antibodies to CldU (red) and dT (green). CldU (1st label) tracks marked locations of DNA segments that were replicated before and/or during HU arrest (Fig. 4C, D). Total DNA staining by anti dT antibody let us quantify percentage of Replication-Associated breaks, i.e. the fraction of CldU tracks located at the ends of DNA molecules rather than within them. It should be noted that this approach does not discriminate between breaks that occurred in vivo and those occurring in vitro during sample processing. Instead, it merely evaluates a relative susceptibility of DNA to breakage.
We quantified RA breaks in samples from control cells arrested with HU for 6 hrs and allowed to recover for 60 or 120 min, and compared them to HU-untreated cells (Fig. 4D). These time points were chosen in order to let all forks that did reactivate, clear the vicinity of CldU tracks. This measure ensured that breakage susceptibility of the ends of CldU tracks could be associated only with truly stalled or collapsed forks, and/or defects left behind forks. We found that our HU treatment led to only a slight elevation in RA breaks above the level seen in no-HU controls (Fig. 4D). For a reference, a 20 hr arrest with HU leads to a 50% increase in terminal tracks (to 75%) over a no-HU control (51%) in normal human fibroblasts (JS unpub.).
We next measured RA breaks in control, BLM-, or WRN-depleted cells that were recovering from HU for 120 min (Fig. 4E). The percentage of RA breaks was only slightly higher in WRN- and BLM-depleted samples than in controls, and no significant difference was observed between WRN- and BLM-depleted cells. We also used neutral comet assay as a readout for DSBs. Cells were labeled with BrdU prior to HU arrest, and we measured comet tail parameters of BrdU-positive cells recovering from HU for 60 min (Fig. S4E). We detected no increase and in fact a slight HU-dependent decrease in % DNA in the tail (Fig. S4E) or tail moment (not shown) across cell lines, suggesting that breakage susceptibility is not a major player during recovery from 6 hr HU arrests in our model system.
3.8. BLM-depleted cells have a slightly longer active period for the S phase checkpoint
Another explanation for the increased HU sensitivity of BLM-depleted cells is that these cells have an exaggerated replication stress or DNA damage checkpoint response despite the fact that replication forks do reactivate. We tested this by analyzing phosphorylation state of the CHK1 and CHK2 kinases in RECQ-depleted cells over a time course of recovery from HU (Fig. S5A).
We saw robust phosphorylation of CHK1 on Serines 317 and 345 in HU (Fig. S5B). Phosphorylation of CHK1 on S317 is required for recovery of replication and viability after HU arrest, while phosphorylation on S345 may have an extra role during normal mitosis [41]. After HU, clearance of S317P species was only minimally delayed in BLM-depleted cells compared to controls or WRN-depleted cells (Fig. S5C, D), and by 12 hrs after HU, CHK1 phosphorylation of S317 was back to baseline in all cell lines (Fig. S5E). Ser345-phosphorylated forms of CHK1 appeared and disappeared with similar kinetics in all cell lines for the first 10 hrs after HU (Fig. S5F) and did not reappear at later time points (up to 23 hrs, not shown). We did not detect phosphorylation of CHK2 on Thr68 above baseline in any of the samples (not shown).
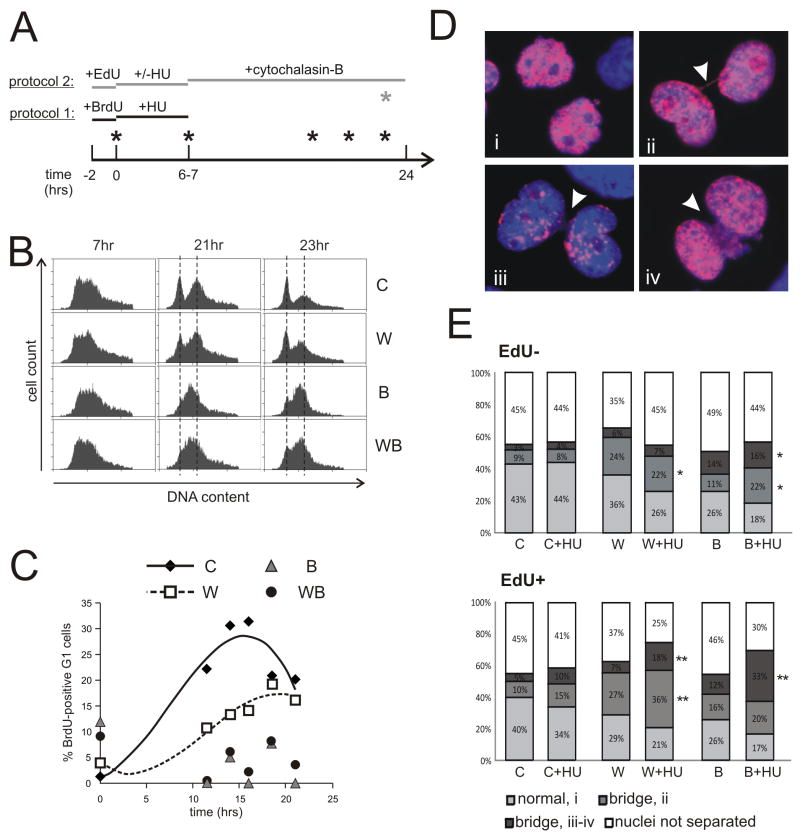
3.9. After a transient exposure to HU, BLM- or BLM/WRN-depleted cells experience a longer G2/M delay than WRN-depleted cells and a more extensive chromatin bridging
We next asked whether the window of activation/deactivation of CHK1 correlated with an altered cell cycle progression after HU in any of the cell lines. We used flow cytometry to compare kinetics of completion of the cell cycle by BrdU-labeled S phase cells exposed to HU (Fig. 5A, protocol 1). In the absence of HU, BLM and WRN/BLM depleted cells were only slightly slower than both control and WRN-depleted cells (not shown). After HU, WRN-depleted cells traversed to G1 somewhat slower than control (Fig. 5B, C). However, BLM-depleted cells were profoundly slower than both control and WRN-depleted cells, and appeared to persist in the late S-G2/M compartment of the cell cycle for at least 13–16 hrs after HU. WRN/BLM depleted cells behaved similarly to BLM-depleted cells. Importantly, BLM-depleted cells with late S/G2 DNA content persisted even after their CHK1 phosphorylation had returned to baseline, as only background levels of BrdU-positive G1 cells were found in these populations between 12 and 20 hrs of the time course. This cell cycle delay was the largest difference observed between BLM- and WRN-depleted cells after HU, and it extended well past the window of time taken by fork recovery, nascent strand processing, and CHK1 deactivation.
Figure 5.
RECQ helicase depletion causes variable degree of late S and/or G2/M delay after HU. A) Experimental designs. Protocol 1: cells were labeled with BrdU for 2hrs and then 2mM HU was added for 6–7 hrs. After removal of HU, samples were taken for up to 24 hrs to determine cell cycle distributions of BrdU-positive cells by FACS. Protocol 2: cells were labeled with EdU for 2hrs and then 2mM HU was added for 6 hrs. After removal of HU, cytochalasin-B was added for 20 hours, after which cells were harvested and analyzed. B) Examples of cell cycle profiles of BrdU-positive cells at time points 7, 21 and 23 hrs. C) Data from an independent experiment performed as in (A) and (B) were quantified and plotted as percent of BrdU-positive G1 cells versus time, using trendlines to connect time points. The decrease in percentage of G1 cells seen at later time points in control cells reflects their entry into the next S phase. BLM- and WRN/BLM-depleted cells exhibited only baseline quantities of BrdU-positive G1 cells. 0 time point corresponds to the addition of HU. D) Examples of binucleated cells with no (i) nucleoplasmic bridging, (ii) thin thread bridging, and thick cord-like (iii) or webbing-like (iv) bridging. Blue: Hoechst 33342, red: Alexa 594-EdU. E, F) Average distribution of normal and bridged binucleated EdU− (E) or EdU+ (F) cells in HU-treated and untreated control or RECQ-depleted cells. Results are means of three independent experiments. Statistical significance was determined in t-test in pair-wise comparisons between controls and WRN- or BLM-depleted cells under the same treatment conditions and for the same class of bridges. Two-tailed P values are denoted as **, P<0.005, *, P<0.05, and are next to the respective bridge classes. P values are also given in the Results section.
One mitotic function of BLM may be to dissolve chromatin bridges [42] that may originate from sister chromatid linkage, among other causes [43,44]. Anaphase bridges are elevated in HeLa cells both in HU and BLM-dependent manner [37]. We looked for chromatin bridges in WRN- or BLM-depleted cells after HU (Figure 5A protocol 2). Cells were pulse-labeled with EdU prior to HU for quick visualization of S phase cells, and cytochalasin-B was added after HU to prevent loss of bridges due to cytokinesis. EdU incorporation did not affect chromatin separation (compare EdU− and EdU+ cells without HU in Figure 5E)
We found several types of binucleated cells (Figure 5D). Among cells with clearly separated nuclei, we detected normal separation (Figure 5D, i), and nucleoplasmic bridges. Among the latter, we observed thin, single-thread bridges (ii), as well as more extensive bridging seen as thick chords or “webbing” of nucleoplasmic material between the nuclei (examples iii and iv). As expected, EdU− cells were virtually unaffected by HU, since these cells were not in S phase when HU was added, and only a small fraction of them may have entered S phase during incubation with HU. However, these cells showed higher levels of chromatin bridging associated with RECQ depletion. For example, WRN-depleted cells had an increased level of ii-type bridges compared to control (PC/W=0.0046), and BLM-depleted cells had a higher level of iii-iv-type bridges (PC/B=0.037).
Among EdU+ cells, the patterns were more complex. HU treatment caused less than twofold increases in ii-type bridging in control and WRN-depleted cells relative to their respective no-HU baselines (PW/W-HU =0.045). Interestingly, this was not observed in BLM-depleted cells (PB/B-HU =0.11), suggesting that WRN may participate in a BLM-independent pathway of chromatin resolution, for example, affecting the resolution step of homologous recombination [45]. On the other hand, iii-iv type bridging underwent >2-fold, HU-dependent increases in each cell type. In BLM-depleted cells in particular, iii-iv-type bridges were almost 3 times more prevalent after HU than without HU (PB/B-HU =0.014). In these cells, iii-iv-type bridges were seen almost in 50% of well-separated nuclei after HU. By comparison, in WRN-depleted cells, these bridges were only seen in a quarter of all well-separated nuclei. While overall bridging after HU was at the same level in WRN-depleted or BLM-depleted cells when all types of bridges were considered, it was evident that BLM-depleted cells had a larger proportion of extensive, iii-iv-type bridges than other two cell types (for example, PW-HU/B-HU=0.018).
4. Discussion
4.1. WRN and BLM contribute additively to fork progression during unstressed replication
BLM-deficient cells exhibit slowed fork progression [15], while WRN-deficient cells have an increased level of asymmetrically diverging forks in early S phase, suggesting fork inactivation [14]. Our study confirmed that BLM, and to a lesser degree, WRN, are needed for normal fork progression, and we showed for the first time that when both RECQs were depleted, fork progression was slower than in single-depleted cells. Thus, BLM and WRN can partially substitute for each other or perform parallel functions, each contributing to fork progression. WRN and BLM could assist in processing Okazaki fragments [7], or unwind secondary structures [46]. Reduced fork progression in BLM deficient cells has also been connected to pyrimidine pool imbalance [47].
4.2. Roles of WRN and BLM during recovery from HU arrest
HU-sensitivity of BLM or WRN-deficient cells has proved to vary depending on cell type, depletion vs. knock-out, drug concentration, and duration of the arrest [17,21,25,48,49]. Innate cell type or cell line variations in fork resistance to collapse and in the extent of fork progression inhibition by a given HU dose will likely emerge as factors contributing to this variability. That notwithstanding, one well-developed line of evidence suggests that WRN is recruited to stalled forks where it interacts with the 9-1-1 complex to prevent DSB formation and recruitment of RAD51 [17,18,22]. On the other hand, BLM can both stimulate and counteract RAD51 activities in vitro, and sumoylated BLM may be recruiting RAD51 to collapsed forks in vivo [33–35].
We were interested to explore the less well studied situation where forks have been stalled but have not yet collapsed into DSBs, and where most of them are reactivated within 60 min. We thus adhered to relatively short arrests with 2mM HU. Under these conditions, depleting WRN or BLM similarly and non-additively delays reactivation of a fraction of forks, yet BLM-depleted cells subsequently experience a more prolonged delay of cell division than WRN-depleted cells. We found no selective increase in DNA breakage susceptibility of replication forks after HU in BLM-depleted S phase cells, no evidence of increased RAD51 foci formation, and only a very minor delay in deactivation of the replication stress checkpoint. However, we saw an increase specifically in “webbing”-like chromatin bridging in BLM-depleted cells that undergo their first mitosis after HU.
As expected [38], RAD51 was important for fork recovery under our conditions, and the data were consistent with RAD51, BLM, and WRN acting within the same pathway. We also found evidence that fork reactivation and the speed with which a reactivated fork progresses for the first 30 min, may represent separate phenomena, with WRN being one of the factors involved in fork progression after HU.
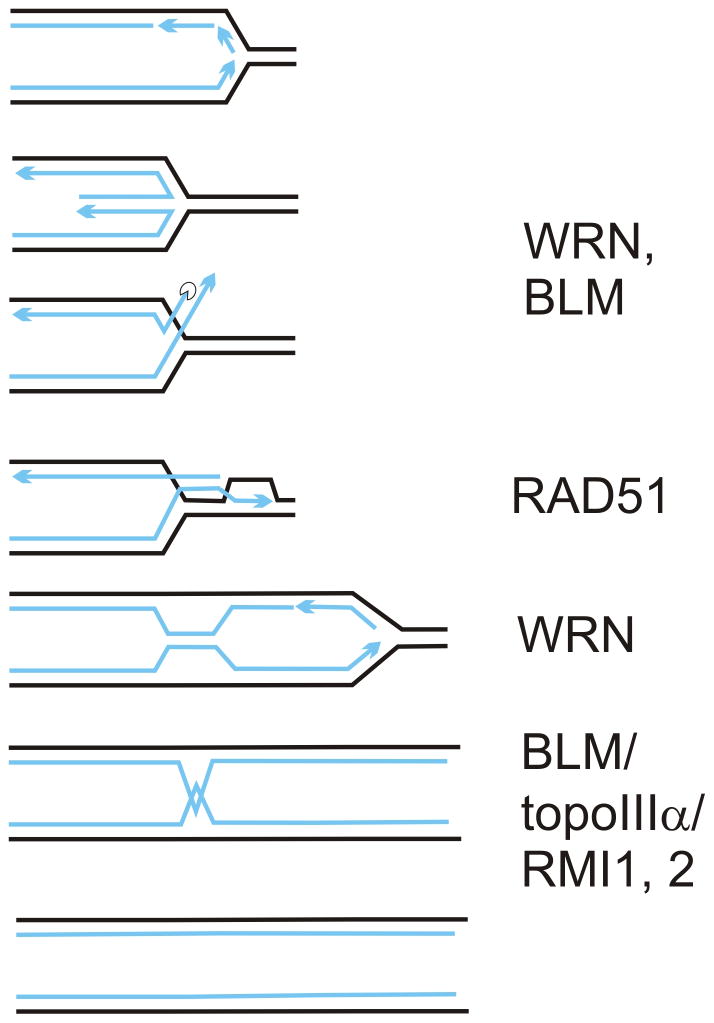
To explain these results, we propose that in HU forks are remodeled in terms of their constituent regulatory proteins and DNA polymerases, and perhaps progress in a regression/reversion cycle. One efficient pathway of exiting this cycle when HU is removed may be a RAD51-mediated reconstitution of an active fork via a D-loop ([8,12] and refs. therein, Fig. 6). In vitro, WRN and BLM display activities that can place them at virtually any point in these processes. Both RECQs regress fork-like substrates and reverse regression [50–53], WRN degrades a recessed 3′ end in a fork [52] and displaces RPA [54], and BLM displaces RAD51 from DNA and also stimulates strand exchange by RAD51 and D-loop extension [33,34]. In vivo, all these activities may be channeled in a particular direction by protein interactions and regulatory modifications of WRN and BLM to generate a substrate for RAD51 [55,56], and optimize D-loop formation and extension. WRN and BLM physically and/or functionally interact with RAD51 in vivo and/or in vitro (refs. above, also, [32,35,36]), and in our model they may, though are not absolutely required to, affect recruitment of RAD51 to forks.
Figure 6.
A model of one of the pathways of replication fork recovery from HU-induced arrest. An arrested fork regresses, forming a daughter/daughter strand duplex. This duplex is resected and/or unwound to expose a 3′ single-stranded tail. WRN and BLM can perform unwinding or assist resection. The tail invades the parental duplex in a RAD51-dependent manner, forming a D-loop. Extension of a D-loop may be stimulated by WRN. Note that if resection/unwinding of the daughter/daughter duplex is incomplete, remaining linkage of paired strands can persist after the D-loop is converted into a reactivated fork, and even after S phase is completed. At mitosis, this hemicatenation between sister chromatids can be dissolved by BLM (in complex with topoIIIα and RMIs).
If the daughter/daughter duplex of a regressed fork is not completely unwound or resected, as may happen in the absence of WRN or BLM [57,58], the twists between daughter strands can persist as hemicatenation between sister chromatids (Fig. 6), and show as chromatin bridges at mitosis. BLM, in complex with topoIIIa and RMI proteins can dissolve such structures [59], and it localizes to ultrafine anaphase bridges [60]. Incomplete processing of daughter/daughter duplex combined with a failure to dissolve interchromatid linkage predicts an increase in chromatin bridges in BLM-depleted cells in the first M phase after HU, and can explain the prolonged cell cycle delay in BLM though not WRN-depleted cells after HU. Consistent with this, we observed an HU- and BLM-dependent rise in extensive, webbing-like nucleoplasmic bridging. This may suggest a more profound defect in decatenating of chromatin.
Reactivated forks appear to move slower immediately after HU. This may be due to dNTP pool imbalances as well as switching to low fidelity polymerases [61–63]. We show (Figs. 1, 3, S2, S3) that WRN or RAD51 depletion exacerbates slow fork progression after HU. One mechanistically attractive possibility is that without WRN or RAD51 a reactivated fork is extended by different polymerase(s) than in wild type, and/or these polymerases are more prone to pausing. Polymerase η was shown to extend a D-loop and it is recruited to reactivating forks with RAD51 [39,64,65], and WRN can facilitate activities of polymerase η in vitro[66]. However, it is unclear why the same slow extension phenotype is not observed in BLM-depleted cells, as BLM can facilitate D-loop extension in vitro [33]. Further complexity is introduced by the fact that at least one of our fork extension assays (Figs. 2–3) uses residual intracellular pool of labeled analog. In BLM-depleted cells endogenous dU pool size may be smaller [47], thus raising the relative concentration of the residual label.
4.3. Nascent strand processing during reactivation of forks after HU
We found that some of the label incorporated within the first 30–60 min after HU, appears to be lost within the next hour, a phenomenon we refer to as nascent strand processing, NSP. One possibility is that NSP is a response to DNA synthesis under conditions of unbalanced dNTP pools and/or NTP/dNTP ratios. Initial extension of reactivated forks by low fidelity polymerases may also invite NSP. Ratios of dNTP concentrations undergo changes during incubation with HU, and altering dNTP ratios can affect DNA polymerase misincorporation rate [67–69]. HU treatment causes misincorporation in vivo [70]. Also, post-HU DNA may contain NMPs [71]. Misincorporated dNMPs and NMPs may trigger DNA repair, resulting in concomitant loss of label from post-HU DNA. One prediction from this hypothesis is that NSP may be reduced in MMR-deficient cells, or in ribonucleotide excision deficient, RNAse H2-depleted cells [71].
In BLM-depleted cells, both post-HU spurt and NSP appear to occur, though are delayed, consistent with a delay in fork reactivation. In RAD51-depleted or WRN-depleted cells post-HU segment length gain is smaller, thus making it possible that NSP is proportionately reduced. However, it is also possible that post-HU DNA in these cells contains fewer mismatches, being replicated by high-fidelity polymerases, which reduces the need for NSP.
Cases of degradation of nascent DNA strands during replication stress have been recently described as persistence of MRE11, RAD51-dependent,≤300 nt gaps in MMS-damaged DNA replicating in Xenopus extracts [72], or as MRE11-dependent degradation of Kb-sized stretches of DNA at stalled forks during HU arrest in BRCA2-deficient but not in normal cells [73]. In contrast, we observe loss of Kb-sized nascent DNA in normal human cells during recovery from HU. Further studies will be needed to establish whether these processes are related and have a similar mechanistic significance.
Supplementary Material
Highlights.
WRN and BLM RECQ helicases contribute additively to support fork progression rate during normal replication in human fibroblasts
WRN and BLM make similar and non-redundant contributions to the proper recovery of replication forks from HU arrest, and are non-redundant with the RAD51-dependent pathway of fork recovery
BLM but not WRN-depleted cells experience a prolonged G2/M delay, which is not associated with activated CHK1
BLM but not WRN-depleted cells show elevated level of chromatin bridging in the first mitosis after HU. We propose a requirement for chromatid decatenation function after fork restart.
Human fibroblasts appear to replace or degrade some of the DNA synthesized by recovering forks immediately after release from HU. This novel phenomenon, NSP, or nascent strand processing, is unaffected in BLM absence, however, in the absence of WRN or RAD51, less DNA is made during recovery from HU, and NSP may be obviated or actively suppressed.
We propose that NSP may be a response to misincorporation of dNTPs and NTPs into nascent DNA as a consequence of HU-induced nucleotide pool imbalance.
Acknowledgments
We are grateful to Drs. Bonita Brewer, M.K. Raghuraman, and members of their lab for support and discussions, and to Drs. Wenyi Feng and Katharina Schlacher for communicating unpublished results and for critical discussions. Thanks are due to Dr. Babak Parviz and Ehsan Saeedi for manufacturing a microchannel template, to Dr. Albert Folch and his lab for the use of Plasma Preen, and to Alden Hackmann for help with figures. This work was supported by an NCI P01 grant CA77852 to R.M. Jr.
Footnotes
Conflict of Interest Statement
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 2.Yu CE, Oshima J, Wijsman EM, Nakura J, Miki T, Piussan C, Matthews S, Fu YH, Mulligan J, Martin GM, Schellenberg GD. Mutations in the consensus helicase domains of the Werner syndrome gene. Werner’s Syndrome Collaborative Group. Am J Hum Genet. 1997;60:330–341. [PMC free article] [PubMed] [Google Scholar]
- 3.Kitao S, Shimamoto A, Goto M, Miller RW, Smithson WA, Lindor NM, Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 4.Monnat RJ., Jr Human RECQ helicases: Roles in DNA metabolism, mutagenesis and cancer biology. Seminars in Cancer Biology. 2010;20:329–339. doi: 10.1016/j.semcancer.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rossi ML, Ghosh AK, Bohr VA. Roles of Werner syndrome protein in protection of genome integrity. DNA Repair. 2010;9:331–344. doi: 10.1016/j.dnarep.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 7.Bachrati CZ, Hickson ID. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma. 2008;117:219–233. doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
- 8.Sidorova JM. Roles of the Werner syndrome RecQ helicase in DNA replication. DNA Repair. 2008;7:1776–1786. doi: 10.1016/j.dnarep.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imamura O, Fujita K, Itoh C, Takeda S, Furuichi Y, Matsumoto T. Werner and Bloom helicases are involved in DNA repair in a complementary fashion. Oncogene. 2002;21:954–963. doi: 10.1038/sj.onc.1205143. [DOI] [PubMed] [Google Scholar]
- 10.Otsuki M, Seki M, Kawabe Y-i, Inoue E, Dong YP, Abe T, Kato G, Yoshimura A, Tada S, Enomoto T. WRN counteracts the NHEJ pathway upon camptothecin exposure. Biochemical and Biophysical Research Communications. 2007;355:477–482. doi: 10.1016/j.bbrc.2007.01.175. [DOI] [PubMed] [Google Scholar]
- 11.Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA Helicases in DNA Repair. Annual Review of Genetics. 2010;44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol. 2010;11:683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- 13.Nam EA, Cortez D. ATR signalling: more than meeting at the fork. Biochem J. 2011;436:527–536. doi: 10.1042/BJ20102162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez-Lopez A, Jackson D, Iborra F, Cox L. Asymmetry of DNA replication fork progression in Werner’s syndrome. Aging Cell. 2002;1:30–39. doi: 10.1046/j.1474-9728.2002.00002.x. [DOI] [PubMed] [Google Scholar]
- 15.Rao VA, Conti C, Guirouilh-Barbat J, Nakamura A, Miao Z-H, Davies SL, Sacca B, Hickson ID, Bensimon A, Pommier Y. Endogenous {gamma}-H2AX-ATM-Chk2 Checkpoint Activation in Bloom’s Syndrome Helicase Deficient Cells Is Related to DNA Replication Arrested Forks. Mol Cancer Res. 2007;5:713–724. doi: 10.1158/1541-7786.MCR-07-0028. [DOI] [PubMed] [Google Scholar]
- 16.Davies SL, North PS, Hickson ID. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat Struct Mol Biol. 2007;14:677–679. doi: 10.1038/nsmb1267. Epub 2007 Jun 2024. [DOI] [PubMed] [Google Scholar]
- 17.Franchitto A, Pirzio LM, Prosperi E, Sapora O, Bignami M, Pichierri P. Replication fork stalling in WRN-deficient cells is overcome by prompt activation of a MUS81-dependent pathway. J Cell Biol. 2008;183:241–252. doi: 10.1083/jcb.200803173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ammazzalorso F, Pirzio LM, Bignami M, Franchitto A, Pichierri P. ATR and ATM differently regulate WRN to prevent DSBs at stalled replication forks and promote replication fork recovery. EMBO J. 2010;29:3156–3169. doi: 10.1038/emboj.2010.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sidorova JM, Li N, Folch A, Monnat RJ., Jr The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle. 2008;7:796–807. doi: 10.4161/cc.7.6.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pichierri P, Rosselli F, Franchitto A. Werner’s syndrome protein is phosphorylated in an ATR/ATM-dependent manner following replication arrest and DNA damage induced during the S phase of the cell cycle. Oncogene. 2003;22:1491–1500. doi: 10.1038/sj.onc.1206169. [DOI] [PubMed] [Google Scholar]
- 21.Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Phosphorylation of the Bloom’s Syndrome Helicase and Its Role in Recovery from S–Phase Arrest. Mol Cell Biol. 2004;24:1279–1291. doi: 10.1128/MCB.24.3.1279-1291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pichierri P, Nicolai S, Cignolo L, Bignami M, Franchitto A. The RAD9-RAD1-HUS1 (9.1.1) complex interacts with WRN and is crucial to regulate its response to replication fork stalling. Oncogene. 2011 doi: 10.1038/onc.2011.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng W-H, Muftic D, Muftuoglu M, Dawut L, Morris C, Helleday T, Shiloh Y, Bohr VA. WRN Is Required for ATM Activation and the S-Phase Checkpoint in Response to Interstrand Crosslink-induced DNA Double Strand Breaks. Mol Biol Cell. 2008:E07-07–0698. doi: 10.1091/mbc.E07-07-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patro BS, Frohlich R, Bohr VA, Stevnsner T. WRN helicase regulates the ATR-CHK1-induced S-phase checkpoint pathway in response to topoisomerase-I-DNA covalent complexes. J Cell Sci. 2011;124:3967–3979. doi: 10.1242/jcs.081372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mao FJ, Sidorova JM, Lauper JM, Emond MJ, Monnat RJ. The Human WRN and BLM RecQ Helicases Differentially Regulate Cell Proliferation and Survival after Chemotherapeutic DNA Damage. Cancer Research. 2010;70:6548–6555. doi: 10.1158/0008-5472.CAN-10-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swanson C, Saintigny Y, Emond MJ, Monnat J, Raymond J. The Werner syndrome protein has separable recombination and survival functions. DNA Repair. 2004;3:475–482. doi: 10.1016/j.dnarep.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Dhillon KK, Sidorova JM, Albertson TM, Anderson JB, Ladiges WC, Rabinovitch PS, Preston BD, Monnat RJ., Jr Divergent cellular phenotypes of human and mouse cells lacking the Werner syndrome RecQ helicase. DNA Repair (Amst) 2009;9:11–22. doi: 10.1016/j.dnarep.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sidorova JM, Li N, Schwartz DC, Folch A, Monnat RJ., Jr Microfluidic-assisted analysis of replicating DNA molecules. Nature Protocols. 2009;4:849–861. doi: 10.1038/nprot.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fenech M. The in vitro micronucleus technique. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2000;455:81–95. doi: 10.1016/s0027-5107(00)00065-8. [DOI] [PubMed] [Google Scholar]
- 30.Bryant HE, Petermann E, Schultz N, Jemth A-S, Loseva O, Issaeva N, Johansson F, Fernandez S, McGlynn P, Helleday T. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009;28:2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu L, Davies SL, Levitt NC, Hickson ID. Potential Role for the BLM Helicase in Recombinational Repair via a Conserved Interaction with RAD51. Journal of Biological Chemistry. 2001;276:19375–19381. doi: 10.1074/jbc.M009471200. [DOI] [PubMed] [Google Scholar]
- 32.Otterlei M, Bruheim P, Ahn B, Bussen W, Karmakar P, Baynton K, Bohr VA. Werner syndrome protein participates in a complex with RAD51, RAD54, RAD54B and ATR in response to ICL-induced replication arrest. J Cell Sci. 2006;119:5137–5146. doi: 10.1242/jcs.03291. [DOI] [PubMed] [Google Scholar]
- 33.Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes & Development. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bugreev DV, Mazina OM, Mazin AV. Bloom Syndrome Helicase Stimulates RAD51 DNA Strand Exchange Activity through a Novel Mechanism. Journal of Biological Chemistry. 2009;284:26349–26359. doi: 10.1074/jbc.M109.029371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ouyang KJ, Woo LL, Zhu J, Huo D, Matunis MJ, Ellis NA. SUMO Modification Regulates BLM and RAD51 Interaction at Damaged Replication Forks. PLoS Biol. 2009;7:e1000252. doi: 10.1371/journal.pbio.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rassool FV, North PS, Mufti GJ, Hickson ID. Constitutive DNA damage is linked to DNA replication abnormalities in Bloom’s syndrome cells. Oncogene. 2003;22:8749–8757. doi: 10.1038/sj.onc.1206970. [DOI] [PubMed] [Google Scholar]
- 37.Lahkim Bennani-Belhaj K, Rouzeau S, Buhagiar-Labarchede G, Chabosseau P, Onclercq-Delic R, Bayart E, Cordelières F, Couturier J, Amor-Gueret M. The Bloom Syndrome Protein Limits the Lethality Associated with RAD51 Deficiency. Molecular Cancer Research. 2010;8:385–394. doi: 10.1158/1541-7786.MCR-09-0534. [DOI] [PubMed] [Google Scholar]
- 38.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-Stalled Replication Forks Become Progressively Inactivated and Require Two Different RAD51-Mediated Pathways for Restart and Repair. Molecular Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hashimoto Y, Puddu F, Costanzo V. RAD51- and MRE11-dependent reassembly of uncoupled CMG helicase complex at collapsed replication forks. Nat Struct Mol Biol. 2011 doi: 10.1038/nsmb.2177. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su X, Bernal JA, Venkitaraman AR. Cell-cycle coordination between DNA replication and recombination revealed by a vertebrate N-end rule degron-Rad51. Nat Struct Mol Biol. 2008;15:1049–1058. doi: 10.1038/nsmb.1490. [DOI] [PubMed] [Google Scholar]
- 41.Wilsker D, Petermann E, Helleday T, Bunz F. Essential function of Chk1 can be uncoupled from DNA damage checkpoint and replication control. Proceedings of the National Academy of Sciences. 2008;105:20752–20757. doi: 10.1073/pnas.0806917106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naim V, Rosselli F. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat Cell Biol. 2009;11:761–768. doi: 10.1038/ncb1883. [DOI] [PubMed] [Google Scholar]
- 43.Naim V, Rosselli F. The FANC pathway and mitosis: a replication legacy. Cell Cycle. 2009;8:2907–2911. doi: 10.4161/cc.8.18.9538. [DOI] [PubMed] [Google Scholar]
- 44.Fenech M, Kirsch-Volders M, Natarajan AT, Surralles J, Crott JW, Parry J, Norppa H, Eastmond DA, Tucker JD, Thomas P. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis. 2011;26:125–132. doi: 10.1093/mutage/geq052. [DOI] [PubMed] [Google Scholar]
- 45.Saintigny Y, Makienko K, Swanson C, Emond MJ, Monnat RJ., Jr Homologous Recombination Resolution Defect in Werner Syndrome. Mol Cell Biol. 2002;22:6971–6978. doi: 10.1128/MCB.22.20.6971-6978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Opresko PL. Telomere ResQue and preservation--Roles for the Werner syndrome protein and other RecQ helicases. Mechanisms of Ageing and Development. 2008;129:79–90. doi: 10.1016/j.mad.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 47.Chabosseau P, Buhagiar-Labarchede G, Onclercq-Delic R, Lambert S, Debatisse M, Brison O, Amor-Gueret M. Pyrimidine pool imbalance induced by BLM helicase deficiency contributes to genetic instability in Bloom syndrome. Nat Commun. 2011;2:368. doi: 10.1038/ncomms1363. [DOI] [PubMed] [Google Scholar]
- 48.Pichierri P, Franchitto A, Mosesso P, Palitti F. Werner’s Syndrome Protein Is Required for Correct Recovery after Replication Arrest and DNA Damage Induced in S-Phase of Cell Cycle. Mol Biol Cell. 2001;12:2412–2421. doi: 10.1091/mbc.12.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lahkim Bennani-Belhaj K, Buhagiar-Labarchede G, Jmari N, Onclercq-Delic R, Amor-Gueret M. BLM Deficiency Is Not Associated with Sensitivity to Hydroxyurea-Induced Replication Stress. J Nucleic Acids. 2010;2010:1–8. doi: 10.4061/2010/319754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ralf C, Hickson ID, Wu L. The Bloom’s Syndrome Helicase Can Promote the Regression of a Model Replication Fork. J Biol Chem. 2006;281:22839–22846. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- 51.Machwe A, Xiao L, Groden J, Orren DK. The Werner and Bloom Syndrome Proteins Catalyze Regression of a Model Replication Fork. Biochemistry. 2006;45:13939–13946. doi: 10.1021/bi0615487. [DOI] [PubMed] [Google Scholar]
- 52.Machwe A, Xiao L, Lloyd RG, Bolt E, Orren DK. Replication fork regression in vitro by the Werner syndrome protein (WRN): Holliday junction formation, the effect of leading arm structure and a potential role for WRN exonuclease activity. Nucl Acids Res. 2007;35:5729–5747. doi: 10.1093/nar/gkm561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Machwe A, Karale R, Xu X, Liu Y, Orren DK. The Werner and Bloom Syndrome Proteins Help Resolve Replication Blockage by Converting (Regressed) Holliday Junctions to Functional Replication Forks. Biochemistry. 2011;50:6774–6788. doi: 10.1021/bi2001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Machwe A, Lozada E, Wold MS, Li G-M, Orren DK. Molecular Cooperation between the Werner Syndrome Protein and Replication Protein A in Relation to Replication Fork Blockage. Journal of Biological Chemistry. 2011;286:3497–3508. doi: 10.1074/jbc.M110.105411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nimonkar AV, Özsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proceedings of the National Academy of Sciences. 2008;105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes & Development. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tomimatsu N, Mukherjee B, Deland K, Kurimasa A, Bolderson E, Khanna KK, Burma S. Exo1 plays a major role in DNA end resection in humans and influences double-strand break repair and damage signaling decisions. DNA Repair. 2012;11:441–448. doi: 10.1016/j.dnarep.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao S, Guay C, Toczylowski T, Yan H. Analysis of MRE11’s function in the 5′--> 3′ processing of DNA double-strand breaks. Nucleic Acids Research. 2012;40:4496–4506. doi: 10.1093/nar/gks044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu L, Bachrati CZ, Ou J, Xu C, Yin J, Chang M, Wang W, Li L, Brown GW, Hickson ID. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proceedings of the National Academy of Sciences. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chan K-L, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26:3397–3409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pillaire MJ, Betous R, Conti C, Czaplicki J, Pasero P, Bensimon A, Cazaux C, Hoffmann JS. Upregulation of error-prone DNA polymerases beta and kappa slows down fork progression without activating the replication checkpoint. Cell Cycle. 2007;6:471–477. doi: 10.4161/cc.6.4.3857. Epub 2007 Feb 2019. [DOI] [PubMed] [Google Scholar]
- 62.Rey L, Sidorova JM, Puget N, Boudsocq F, Biard DSF, Monnat RJ, Jr, Cazaux C, Hoffmann J-S. Human DNA Polymerase {eta} Is Required for Common Fragile Site Stability during Unperturbed DNA Replication. Mol Cell Biol. 2009;29:3344–3354. doi: 10.1128/MCB.00115-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bétous R, Rey L, Wang G, Pillaire M-J, Puget N, Selves J, Biard DSF, Shin-ya K, Vasquez KM, Cazaux C, Hoffmann J-S. Role of TLS DNA polymerases eta and kappa in processing naturally occurring structured DNA in human cells. Molecular Carcinogenesis. 2009;48:369–378. doi: 10.1002/mc.20509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McIlwraith MJ, Vaisman A, Liu Y, Fanning E, Woodgate R, West Human SC. DNA Polymerase [eta] Promotes DNA Synthesis from Strand Invasion Intermediates of Homologous Recombination. Molecular Cell. 2005;20:783–792. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 65.McIlwraith MJ, West SC. DNA Repair Synthesis Facilitates RAD52-Mediated Second-End Capture during DSB Repair. Molecular Cell. 2008;29:510–516. doi: 10.1016/j.molcel.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 66.Kamath-Loeb AS, Lan L, Nakajima S, Yasui A, Loeb LA. Werner syndrome protein interacts functionally with translesion DNA polymerases. Proceedings of the National Academy of Sciences. 2007;104:10394–10399. doi: 10.1073/pnas.0702513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Julias JG, Pathak VK. Deoxyribonucleoside Triphosphate Pool Imbalances In Vivo Are Associated with an Increased Retroviral Mutation Rate. J Virol. 1998;72:7941–7949. doi: 10.1128/jvi.72.10.7941-7949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hakansson P, Hofer A, Thelander L. Regulation of Mammalian Ribonucleotide Reduction and dNTP Pools after DNA Damage and in Resting Cells. Journal of Biological Chemistry. 2006;281:7834–7841. doi: 10.1074/jbc.M512894200. [DOI] [PubMed] [Google Scholar]
- 69.Mathews CK. DNA precursor metabolism and genomic stability. The FASEB Journal. 2006;20:1300–1314. doi: 10.1096/fj.06-5730rev. [DOI] [PubMed] [Google Scholar]
- 70.Flanagan SA, Robinson BW, Krokosky CM, Shewach DS. Mismatched nucleotides as the lesions responsible for radiosensitization with gemcitabine: a new paradigm for antimetabolite radiosensitizers. Molecular Cancer Therapeutics. 2007;6:1858–1868. doi: 10.1158/1535-7163.MCT-07-0068. [DOI] [PubMed] [Google Scholar]
- 71.Lazzaro F, Novarina D, Amara F, Watt Danielle L, Stone Jana E, Costanzo V, Burgers Peter M, Kunkel Thomas A, Plevani P, Muzi-Falconi M. RNase H and Postreplication Repair Protect Cells from Ribonucleotides Incorporated in DNA. Molecular Cell. 2012;45:99–110. doi: 10.1016/j.molcel.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hashimoto Y, Chaudhuri AR, Lopes M, Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol. 2010;17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-Strand Break Repair-Independent Role for BRCA2 in Blocking Stalled Replication Fork Degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.