Abstract
Many pheromones have very low water solubility, posing experimental difficulties for quantitative binding measurements. A new method is presented for determining thermodynamically valid dissociation constants for ligands binding to pheromone-binding proteins (OBPs), using β-cyclodextrin as a solubilizer and transfer agent. The method is applied to LUSH, a Drosophila OBP that binds the pheromone 11-cis vaccenyl acetate (cVA). Refolding of LUSH expressed in E. coli was assessed by measuring N-phenyl-1-naphthylamine (NPN) binding and Förster resonance energy transfer between LUSH tryptophan 123 (W123) and NPN. Binding of cVA was measured from quenching of W123 fluorescence as a function of cVA concentration. The equilibrium constant for transfer of cVA between β-cyclodextrin and LUSH was determined from a linked equilibria model. This constant, multiplied by the β-cyclodextrin-cVA dissociation constant, gives the LUSH-cVA dissociation constant: ~100 nM. It was also found that other ligands quench W123 fluorescence. The LUSH-ligand dissociation constants were determined to be ~200 nM for the silk moth pheromone bombykol and ~90 nM for methyl oleate. The results indicate that the ligand-binding cavity of LUSH can accommodate a variety ligands with strong binding interactions. Implications of this for the pheromone receptor model proposed by Laughlin et al. (Cell 133: 1255–65, 2008) are discussed.
Keywords: olfaction, 11-cis vaccenyl acetate, LUSH, pheromone-binding proteins, cyclodextrin, tryptophan fluorescence
Introduction
Although insects and vertebrates share features of olfactory signal processing (Hildebrand and Shepherd 1997), the molecular details of olfactory reception are quite different. The odorant-binding proteins (OBPs) of insects and vertebrates belong to different protein families, and insect olfactory receptors (ORs) were recently found to be unusual ligand-gated ion channels (Sato et al. 2008; Smart et al. 2008; Wicher et al. 2008), in contrast to vertebrate G-protein-coupled olfactory receptors. These unique features of insect olfaction offer appealing targets for controlling insect disease vectors and agricultural and urban pests. However, many details remain to be determined, including the roles of OBPs and ORs in specificity of signaling. Grosse-Wilde et al. (Grosse-Wilde et al. 2006) showed that bombykol, the sex pheromone of the silkmoth Bombyx mori, activates BmOr-1 pheromone receptors more strongly when the pheromone-binding protein is also present. Furthermore, Laughlin et al. (Laughlin et al. 2008) found that a Drosophila pheromone receptor, Or67d, could be activated by the OBP known as LUSH in the complete absence of the pheromone 11-cis vaccenyl acetate (cVA) if LUSH was in a particular conformational state. These experiments suggest an important role for OBPs in signaling. Nevertheless, Carlson and co-workers showed that most of the Drosophila ORs are active when expressed in sensilla lacking the proper OBP (Hallem and Carlson 2006; Hallem et al. 2004), and Leal and co-workers expressed an activatable silkmoth pheromone receptor in various Drosophila sensilla (Syed et al. 2006; Syed et al. 2010). Remarkably, when the silkmoth pheromone receptor was substituted for the native Or67d receptor in the Drosophila T1 sensilla, a native-like response to bombykol was observed, despite the fact that the T1 sensilla contained LUSH instead of the silkmoth pheromone-binding protein. Is it possible that bombykol coincidentally binds to LUSH? If so, what would that imply about the mechanism by which the presence of the correct pheromone is signaled to the brain?
One way of answering this question would be to compare the binding of cVA and bombykol to LUSH. Unfortunately, there are serious technical problems in measuring pheromone binding. Most insect pheromones have very low solubility in water. Some studies have detected the amount of pheromone that binds to the protein when the insoluble pheromone is suspended in water (Leal et al. 2005; Plettner et al. 2000; Vogt and Riddiford 1981). Because the pheromone is presented to the binding protein in a separate liquid phase, these methods do not measure thermodynamically valid binding constants. The amount of soluble free pheromone in equilibrium with the binding protein under these conditions is unknown. Phase separation can be used to rigorously measure ligand binding (Davanloo and Crothers 1976), but typically when applied to pheromone-binding proteins this method requires radioactive ligands (Du and Prestwich 1995; Ferrari et al. 1997). We have now developed a new single-phase method to quantitatively measure binding of water-insoluble ligands to OBPs. We have applied this method to directly show that the Drosophila OBP LUSH binds both the Drosophila pheromone cVA and, more weakly, to the silkmoth pheromone bombykol. This result raises questions about the role of LUSH in pheromone reception.
Results and Discussion
NPN binding to LUSH
Pelosi and co-workers (Ban et al. 2002) showed that N-phenyl-1-naphthylamine (NPN) is a useful reagent for studies of hydrophobic ligand-binding proteins, because the protein-dye complex greatly enhances the quantum yield of the dye. We found that NPN fluorescence in the presence of LUSH displayed a saturable fluorescence change suggesting formation of a complex with a dissociation constant of 0.65 μM (Figure 1A). This result is similar to the previous work by Zhou et al. (Zhou et al. 2004), who reported a dissociation constant of 1.5 μM. However, in contrast to Zhou et al., we observed evidence for Förster resonance energy transfer (FRET) between NPN and Trp 123, as indicated by the 280 nm peak in the fluorescence excitation spectrum (Figure 1B, upper curve) and the quenching of tryptophan emission by NPN (Figure 1C). As noted by Jones and co-workers, LUSH expressed in E. coli inclusion bodies must be refolded by reducing and reoxidizing the cysteines in the presence of ethanol (Kruse et al. 2003). When this step was omitted, we found that LUSH did not correctly refold, because Trp-NPN energy transfer was absent (Figure 1B, lower curve), and there was little change in the tryptophan quantum yield after unfolding in the presence of urea (Figure S1). Misfolded LUSH did bind NPN, but it was not saturable in the μM concentration range (Figure S2).
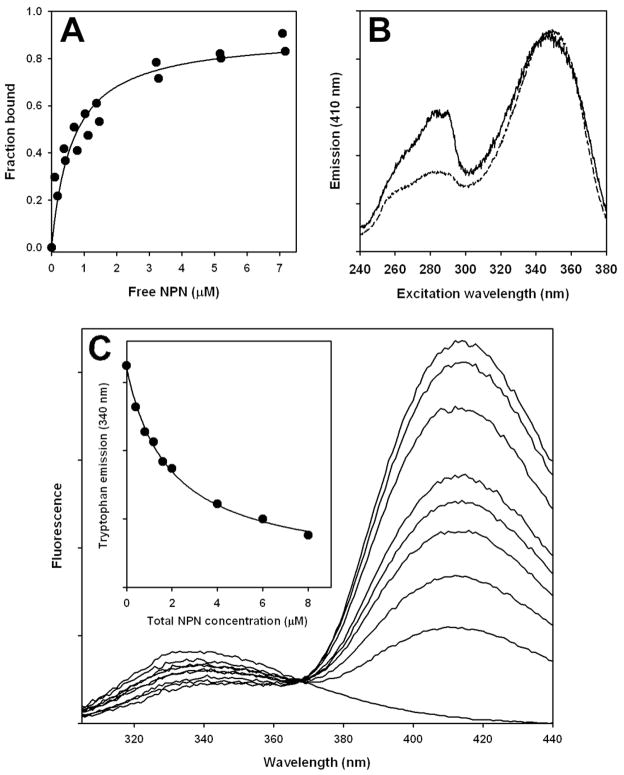
Figure 1.
NPN binding to LUSH. Conditions: 20 mM Tris, pH 8.4; 1 μM LUSH. A. Enhancement of NPN fluorescence emission at 410 nm (350 nm excitation) by binding to LUSH. Line fit with dissociation constant of 0.65 μM (correlation coefficient = 0.97). B. Comparison of fluorescence excitation spectra of 2 μM NPN in the presence of 1 μM refolded LUSH (solid line) or 1 μM non-refolded LUSH (dashed line). C. Tryptophan quenching by NPN bound to LUSH. Emission measured at NPN concentrations from 0 to 8 μM. Excitation wavelength: 280 nm. Inset: Emission at 340 nm plotted against NPN concentration. Line fit to hyperbolic decay function (correlation coefficient = 0.998).
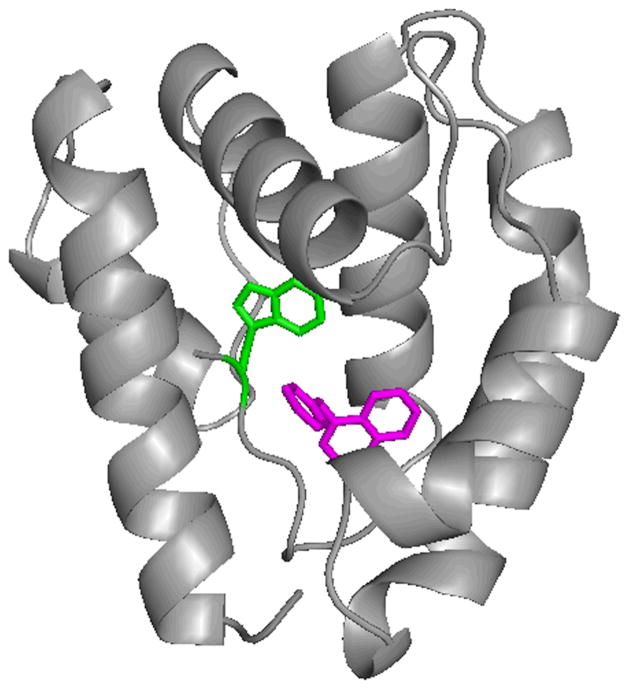
Quenching of LUSH Trp 123 fluorescence emission at 340 nm as a function of NPN concentration was fit with a hyperbolic decay function (f = yo + ab/(b+x)) to obtain the limiting intensity of tryptophan emission (yo) and the maximum span of tryptophan quenching (a). On the assumption that the extent of tryptophan quenching was proportional to the concentration of LUSH with NPN bound, the data were converted to fraction of LUSH with NPN bound, Y, using the equation Y = 1 − (f − yo)/a. The limiting tryptophan quenching by NPN (Figure 1C, inset) was 92.4%, indicating an energy transfer efficiency of 0.924. We modeled the binding of NPN to LUSH with AutoDock (Morris et al. 2009), using the structure of the cVA-LUSH complex (Protein Data Bank structure 2GTE) from which we removed the cVA molecule. The three lowest energy model clusters all placed NPN in the cVA binding pocket near Trp 123 (Figure 2 and Table 1). In models resulting from 44 of the 50 computational runs, the NPN naphthalene group occupied the same region as carbons 9–18 of cVA. The dissociation constant calculated from the lowest-energy model, representing the largest cluster of models, was 1.9 μM. Using the models to calculate the dipole orientation factor (Table 1), we obtained NPN to Trp 123 distances of 12–14 Å from the FRET data, compared with 6–8 Å from the models. We also used AutoDock with the lowest-energy LUSH-NPN model (model 1) to search for additional NPN binding sites. About half of the second NPN molecules added to the ligand-binding site of LUSH, with calculated binding constants in the μM range, and the other sites were on the protein surface and had weak binding constants. The strong-binding models clustered in two different geometries and contained the first NPN molecule as part of the site. However, the first NPN molecule blocked access to the second molecule’s sites, so it seems unlikely that an NPN molecule could actually occupy the second site. Furthermore, there is no evidence of cooperativity in NPN binding to LUSH, unlike what we observed for NPN binding to chemosensory proteins (Gonzalez et al. 2009). Therefore, we conclude that the stoichiometry of the LUSH-NPN complex is 1:1.
Figure 2.
Lowest energy NPN binding site fit by AutoDock. LUSH coordinates from Protein Data Bank file 2GTE. Trp 123, green. NPN, magenta. AutoDock-generated coordinates for bound NPN (model 1, Table 1) are given in Supporting Information (Table S1). Figure generated with PyMOL.
Table 1.
AutoDock models of NPN binding to LUSH
| Model (% of runs)a | Dissociation constant (μM)b | κ2 | Ro (Å) | R (FRET)c (Å) | R (model) (Å) |
|---|---|---|---|---|---|
| 1 (56) | 1.9 | 0.487 | 21.2 | 14.0 | 8.0 |
| 2 (32) | 7.9 | 0.334 | 19.9 | 13.1 | 8.3 |
| 3 (8) | 12.7 | 0.165 | 17.7 | 11.7 | 5.8 |
Fifty AutoDock runs were combined into clusters of nearly identical geometry.
Dissociation constants calculated from free energy computed for each model.
Calculated from the measured transfer efficiency, E = 0.924 (see figure 1C).
The difference between our binding results and those of Zhou et al. (Zhou et al. 2004) may be explained by different methods of preparing LUSH. We solubilized LUSH from E. coli inclusion bodies and refolded it. However, Zhou et al. used soluble LUSH extracted from the periplasm. Their protein seems folded, as saturable binding of NPN was observed. However, they did not observe any quenching of tryptophan by NPN. A possible explanation could be that LUSH picked up a ligand from E. coli, as Oldham et al. (Oldham et al. 2001) found for the B. mori pheromone-binding protein. The acquired ligand, if non-polar, could bind near Trp 123 of LUSH without altering its quantum yield. NPN could still associate with this partially occupied ligand-binding pocket, but now the distance and orientation of NPN would be unfavorable for detectable FRET. In our preparation method, any ligands picked up by LUSH from E. coli would be removed by the guanidinium unfolding step (Kruse et al. 2003; Prestwich 1993) prior to the NPN binding studies.
Pheromone binding to LUSH
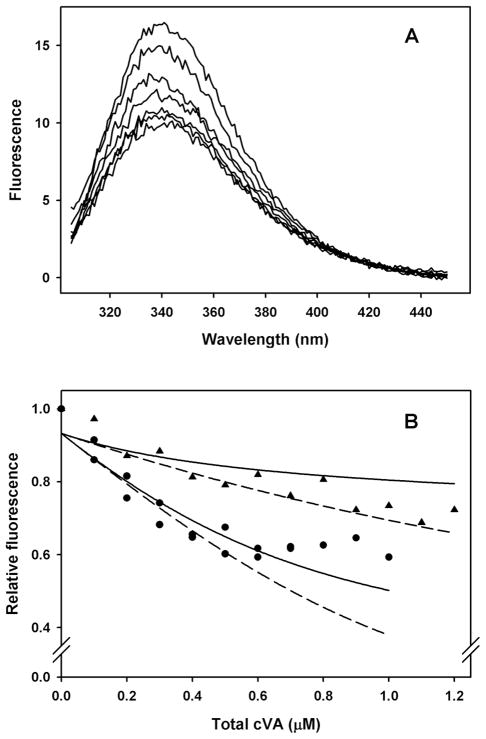
Consistent with the quenching of Trp 123 by NPN binding to correctly folded LUSH, Laughlin et al. (Laughlin et al. 2008) showed, in supplementary data, that addition of cVA quenches LUSH Trp 123 in a cVA-concentration-dependent, saturable manner. The quenching effect occurs at sub-micromolar cVA concentrations, suggesting a very strong interaction with LUSH. Because Laughlin et al. did not report a quantitive analysis of cVA binding to LUSH, we repeated these experiments (Figure 3), with the modification that we included β-cyclodextrin as a solubilizer and transfer agent for cVA (Gonzalez et al. 2009).
Figure 3.
Binding of cVA to LUSH measured by quenching of tryptophan fluorescence. A. Tryptophan emission spectra. Concentrations: LUSH, 1.0 μM; β-cyclodextrin, 170 μM; cVA, 0–1μM. Buffer: 0.02 M Tris, pH 8.5. Excitation, 280 nm. B. LUSH-cVA dissociation constant measured from tryptophan fluorescence quenching. Circles: same conditions as A. Triangles: 1.7 mM β-cyclodextrin. Solid lines calculated from eq 7 and 9 using the same parameters for both lines, except for the difference in β-cyclodextrin concentrations: K = 1.08 × 10−3, a = 0.153, b = 0.933, correlation coefficient = 0.91. Dashed lines calculated with the same constants but omitting the effect of ethanol competition with cVA.
The mechanism of tryptophan fluorescence quenching by cVA is not clear. Comparison of the structures of the cVA-LUSH complex and the apo-protein does not indicate any large-scale change in protein side chain orientations near Trp 123 that would cause a large decrease in the tryptophan quantum yield. In the apo-protein, the side chain oxygen of Ser 52 is close to the indole ring η carbon, and a C-terminal carboxyl oxygen is near the indole nitrogen, providing a moderately polar environment. The positions of Trp 123 and these two polar atoms change very little between ligand-free LUSH and the cVA-LUSH complex (Laughlin et al. 2008). However, the ester oxygen of cVA is positioned 3.40 Å from the tryptophan η carbon (average of A and B molecules in the asymmetric unit). Although this might be expected to increase the polarity of the Trp 123 environment, the emission spectrum (Figure 3A; see also Figures S3A and B in Laughlin et al. 2008) does not show a red shift as cVA is added. Similar quenching of tryptophan fluorescence was reported for Antheraea polyphemus PBP1 (Bette et al. 2002) and for the honeybee pheromone-binding protein ASP1 (Pesenti et al. 2009). For several ligands essentially no shifts occurred in the emission wavelength maxima. Callis and co-workers have developed a theory of protein tryptophan quantum yields based on the strength of charge transfer interactions between the indole ring and the backbone amide nitrogen (Callis and Liu 2004). The lower tryptophan quantum yield for LUSH with cVA bound could be caused by the cVA ester oxygen enhancing excited-state charge transfer.
Quantitative analysis of ligand binding
It is not a simple matter to obtain the dissociation constant of the LUSH-cVA complex using the tryptophan fluorescence quenching data in Figure 3. There is a long-standing problem involved with measurements of equilibria between proteins and water-insoluble ligands. For example, the dissociation constant of palmitic acid from bovine serum albumin was found to be 34 nM (Bojesen and Bojesen 1992), but the water solubility of palmitic acid was reported to be less than 0.1 nM (Vorum et al. 1992). Clearly there are other equilibria involved in this type of ligand-binding study, such as equilibria between the saturated aqueous solution and ligand molecules adsorbed on the walls of the container, or aggregations of ligand molecules into small suspended droplets, particles or micelles. Except at very low total ligand concentration, these other equilibria will dominate binding studies. In order to suppress other ligand equilibria, we dispersed cVA in β-cyclodextrin, which we previously demonstrated is effective at transferring water-insoluble ligands to an insect chemosensory protein (Gonzalez et al. 2009).
For a quantitative analysis of the fluorescence quenching data in Figure 3B, we assume that most of the LUSH-associated cVA is directly transferred to LUSH from cyclodextrin, as described by the following linked equilibria:
| (1) |
| (2) |
where eq 1 gives the transfer of cVA from β-cyclodextrin (free concentration = [D]; β-cyclodextrin-cVA complex concentration = [DV]) to LUSH (free concentration = [L]; LUSH-cVA complex concentration = [LV]); and eq 2 gives the corresponding dissociation of cVA from its complex with β-cyclodextrin (free cVA concentration = [V]). The fraction of LUSH bound to cVA is given by:
| (3) |
where LT is the total LUSH concentration, [L] + [LV]. Under the conditions of our experiments, DT (the total concentration of β-cyclodextrin) is much greater than VT (the total concentration of cVA). Therefore, [D] ≈ DT. Substituting eq 1 into eq 3 gives:
| (4) |
[DV] may be calculated from VT, assuming that [V] ≈ 0:
| (5) |
Eq 4 predicts a hyperbolic relationship between Y and [DV]. The apparent binding constant, K·DT, could be obtained from a plot of Y versus [DV]. K is related to the equilibrium between LUSH and cVA as follows:
| (6) |
Solving eq 4 and eq 5 for Y in terms of VT gives:
| (7) |
Y is an experimentally determined parameter, assumed to be related to the Trp 123 emission at 340 nm at a particular concentration of cVA (Figure 3). Y can be obtained from the fluorescence spectra as follows. The emission at 340 nm, Fi, measured at a particular total concentration of cVA, was normalized to the emission Fo in the absence of cVA to give f = Fi/Fo. Then it follows that:
| (8) |
where a is the limiting normalized emission when all LUSH molecules have cVA bound. The tryptophan emission data as a function of VT (total cVA) (Figure 3B) was fit to eq 8, modified to include the possibility that LUSH is less than fully active:
| (9) |
The fraction of LUSH in the experimental sample that is capable of binding cVA is b. We also included a correction for the weak binding of ethanol to LUSH (Thode et al. 2008), which we used as a solvent for cVA and which competes with cVA for the same binding site. The terms in eq 7 containing K must be multiplied by (1 + [EtOH]/KE), where [EtOH] is the free ethanol concentration, and KE is the dissociation constant for ethanol from LUSH, previously measured to be 107 mM (Thode et al. 2008).
Experimental results for two different total concentrations of β-cyclodextrin, DT (0.17 mM and 1.7 mM), are shown in Figure 3B, with lines fit to equation 9, using equation 7 to evaluate Y. Both lines were fit with measured values of DT and VT. The only adjustable parameters were a and b (eq 9) and K (eq 7), and the same values of a, b and K were used in fitting both lines.
In order to use eq 6 to obtain the apparent dissociation constant of cVA from LUSH, KL, we need to know the dissociation constant of cVA from β-cyclodextrin, KCD. We did not independently measure this, but a large set of β-cyclodextrin affinity data has been collected (Rekharsky and Inoue 1998) and discussed in terms of the thermodynamics of the interaction between cyclodextrin and guest molecules. For molecules like cVA (and other insect pheromones with long hydrocarbon chains), probably the main interaction with the cyclodextrin cavity is through van der Waals attractions. We plotted the solvent excluded surface (approximately the contact surface) versus the logarithm of the association constant (proportional to the free energy of interaction) (Figure S3) for a homologous series of fatty acids, polynuclear aromatics, and several molecules with surfaces similar to cVA. The data shows that the association constant increases with surface up to about 250 Å2. Above this size, the association constants level off, presumably reflecting the size of the cyclodextrin container surface. The entire set of cyclodextrin affinities (Figure S3) can be approximated by a hyperbolic function, thus giving an estimate of the association constant for cVA by interpolation at its solvent excluded surface (360 Å2): 1/KCD = 104 M−1. Combining this with the value of K from figure 3B, using eq 6, gives the dissociation constant for cVA from LUSH as KL = K KCD = 108 nM.
The validity of the linked equilibria approach to measuring pheromone dissociation constants is supported by the fact that experimental data for cVA binding to LUSH at two different β-cyclodextrin concentrations can be fit with the same value for KL (Figure 3B). There is a limit on the useful range of DT: we found that at very high β-cyclodextrin concentrations (16 mM), the cyclodextrin-cVA complex decreases the amount of LUSH-cVA below the level detectable by our fluorescence method. Our analysis ignores the possibility that β-cyclodextrin might form a 2:1 complex with cVA (D2V). If an additional equilibrium were added to the analysis to include D2V, the calculated value of KL would be smaller. A fraction of total cVA would be tied up in D2V, but it is likely that only the 1:1 complex (DV) can transfer cVA to LUSH.
The apparent dissociation constant of cVA from LUSH, 108 nM, indicates a very strong protein-pheromone complex, considerably stronger than for vertebrate pheromone-binding proteins (Ferrari et al. 1997; Lazar et al. 2002; Sharrow et al. 2005), and at the lower end of the range of previous measurements for insect pheromone-binding proteins (Ban et al. 2003; Campanacci et al. 2001; Du and Prestwich 1995; Gong et al. 2009; Honson et al. 2003; Iovinella et al. 2011; Leal et al. 2005; Li et al. 2008; Pesenti et al. 2009; Plettner et al. 2000; Riviere et al. 2003). Although Laughlin et al. (Laughlin et al. 2008) did not report the LUSH-cVA dissociation constant, their data suggests it is approximately 660 nM, much weaker than our measurement. This difference highlights the effect of attempting to measure binding using a water-insoluble ligand.
The standard free energy calculated from this dissociation constant is −9.5 kcal/mol. The strength of this complex, along with the previous evidence that cVA is not transferred from LUSH to the olfactory receptor Or67d (Laughlin et al. 2008), places stringent constraints on the shutdown mechanism for cVA signaling. If the cVA binding rate constant is in the 105 M−1 sec−1 range reported for other pheromone-binding proteins (Leal et al. 2005), then the rate constant for dissociation would be 0.01 sec−1. This suggests a rather long lifetime for the active LUSH-cVA complex. Perhaps some type of release mechanism occurs in Drosophila, like the pH-sensitive conformational change that induces pheromone release from in the B. mori pheromone-binding protein (BmorPBP) (Horst et al. 2001). Otherwise, the Drosophila, pheromone signal would probably become saturated at the dendritic membrane of the olfactory receptor neuron.
Bombykol and methyl oleate binding to LUSH
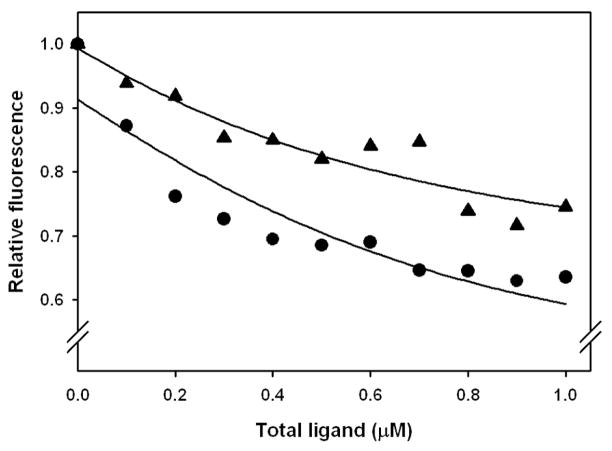
We used β-cyclodextrin as a solubilizer for bombykol to test whether the silkmoth pheromone binds to LUSH. As in the case of cVA, we found that bombykol quenches the fluorescence of LUSH Trp 123 in a very similar way to cVA (Figure 4). Assuming cVA and bombykol have the same affinity for β-cyclodextrin, and using the same analysis as discussed above for cVA, the dissociation constant for bombykol from LUSH is 215 nM, about two-fold weaker than cVA. We also tested methyl oleate, which has a similar structure to cVA. Methyl oleate binds to LUSH with an apparent dissociation constant of 89 nM (Figure 4)---stronger than either bombykol or cVA. By contrast, ethanol had only a small effect on the tryptophan emission of LUSH (Figure S4). Assuming this small fluorescence decrease is a measure of binding, we estimate the dissociation constant to be 21 mM. However, because of the small fluorescence change, the 107 mM dissociation constant measured by Thode et al. (Thode et al. 2008) is also consistent with the data. The ester oxygen of methyl oleate and the alcohol oxygen of bombykol both could be positioned similarly to cVA in complexes with LUSH, explaining why Trp 123 emission is quenched by these two ligands. By contrast, in the crystal structure of the ethanol-LUSH complex, the ethanol oxygen is located 5.09 Å from the nearest Trp 123 atom (η carbon) (Kruse et al. 2003). This may be too far from Trp 123 to affect the tryptophan quantum yield in the same way as the much closer cVA ester oxygen.
Figure 4.
Binding of bombykol (triangles) and methyl oleate (circles) to LUSH. Same conditions as figure 3. Lines fit with eq 7 and 9. Bombykol: K = 2.15 × 10−3, a = 0.362, b = 0.994, correlation coefficient = 0.94; methyl oleate: K = 8.91 × 10−4, a = 0.407, b = 0.914, correlation coefficient = 0.93.
Implications for insect olfactory signaling
Our results show that LUSH is not uniquely specific for binding to the pheromone cVA. We have directly shown that bombykol, as well as cVA, binds to LUSH. This could explain the observation that the silkmoth pheromone receptor expressed in the Drosophila T1 sensilla gives an essentially normal response to bombykol, despite the absence of the silkmoth pheromone-binding protein (Syed et al. 2010). We also found that methyl oleate binds strongly to LUSH. A number of reports have found a similar lack of unique ligand specificity of pheromone-binding proteins. For example, pheromone-binding proteins can bind to different pheromone isomers (Mao et al. 2010; Willett and Harrison 1999; Wojtasek et al. 1998). Oldham et al. (Oldham et al. 2001) found that BmorPBP expressed in E. coli picks up 11-cis vaccenic acid, which mirrors our results with LUSH and bombykol. Lautenschlager et al. (Lautenschlager et al. 2007) showed that BmorPBP can bind to a variety of different ligands. They also noted that the ligand-binding diversity of pheromone-binding proteins should be expected, considering the three dimensional structural similarities between pheromone-binding proteins and general odorant-binding proteins. Previous binding studies using vapor-phase methods showed that general odorant-binding proteins are able to bind a variety of chemically different ligands (Briand et al. 2001), and recent structural work has confirmed and extended that (Lagarde et al. 2011; Lescop et al. 2009).
The variety of ligands capable of binding to a single pheromone-binding protein raises questions about how the specificity of pheromone reception is maintained in the face of background noise. Lautenschlager et al. (Lautenschlager et al. 2007) proposed that the non-pheromone ligands bind more weakly and will be filtered out because they will dissociate from the binding protein before it diffuses to the surface of the olfactory receptor neuron. However, the electrophysiological response to bombykol reported for T1 neurons expressing LUSH and the B. mori pheromone receptor (Syed et al. 2010) seems to indicate this filtering doesn’t always occur. In some insects, the pheromone receptors in the receptor neuron membranes were found to act as filters, because they are specific for a single pheromone isomer, whereas the pheromone-binding proteins in the same sensilla bound several different isomers (Willett and Harrison 1999; Wojtasek et al. 1998). However, specificity determined by receptor binding may not apply to some pheromone signaling mechanisms, including cVA in Drosophila. The results of Laughlin et al. (Laughlin et al. 2008) show that in this system, the pheromone-binding protein conformation determines whether or not the receptor is activated. The cVA molecule itself is not actually necessary to activate the receptor neuron. Therefore, any ligand that stabilizes the active conformation of LUSH could initiate signaling, regardless of the ligand’s specific chemical structure. Further studies will be necessary to clarify this issue. If ligands such as bombykol and methyl oleate stabilize the same LUSH conformation as cVA, then the Drosophila olfactory system must have a mechanism to prevent false positive identification of cVA by T1 sensilla. One type of correction mechanism could use input from olfactory receptors in other sensilla that detect additional chemical features of cVA (e.g. distinguish esters from alcohols). False positives could be controlled either by local interneurons in the antennal lobe (Root et al. 2008) or by processing of projection neuron outputs (Datta et al. 2008).
Experimental Procedures
LUSH was expressed from a pET13a vector provided by Dr. David Jones, U. of Colorado Medical School. The protein was purified by a method similar to that described by Kruse et al. (Kruse et al. 2003). Cell pellets from 500 mL cultures continuously induced with lactose for one day were washed in 0.136 M NaCl, 0.04 M Tris, pH 8, 0.1 M MgCl2, 1% glycerol and then suspended in 25 mL 68 mM NaCl, 20 mM Tris, pH 8, 0.8 M ammonium sulfate, 42.5 mM dithiothreitol. This suspension was extracted twice with 25 mL t-butanol and centrifuged. The pellet was then extracted with a solution containing 0.17% NP40, 0.17% Tween 80, 33 mM dithiothreitol, 33 mM EDTA, 5.3 M urea, and 89 mM glycine, pH 10.6. The extract was chromatographed on DEAE-cellulose and Superdex 75 columns, and the fractions containing LUSH were refolded by the cystine/cysteine method (Kruse et al. 2003; Prestwich 1993). The amino acid sequence of purified LUSH was confirmed by tandem mass spectrometric analysis of tryptic fragments (Gonzalez et al. 2009). The LUSH concentration was measured by UV absorbance, using a calculated extinction coefficient of 8860 M−1cm−1 (Gasteiger et al. 2003). N-phenyl-1-naphthylamine (NPN) was obtained from Invitrogen Molecular Probes (Eugene, OR). 11-cis vaccenyl acetate (cVA), >98%, was obtained from Cayman Chemical Company (Ann Arbor, MI). Methyl cis-9-octadecenoate (methyl oleate), >99%, and β-cyclodextrin were obtained from Sigma-Aldrich (St. Louis, MO). Bombykol was a gift from Dr. Walter Leal, U. of California, Davis. It showed a single peak by HPLC and GC-MS.
Fluorescence spectra were measured on a Photon Technology International (Birmingham, NJ) QM-4 instrument, using a quartz cuvette with a 0.5 cm excitation path and a 1.0 cm emission path. Excitation slits were 1 nm and emission slits were 5 nm. Typical samples for fluorescence spectra contained 1.0 μM LUSH in 20 mM Tris buffer, pH 8.5. Ligands were added from alcohol solutions (0.2 mM or 1.0 mM NPN in methanol; 100 μM cVA, bombykol, or methyl oleate in ethanol). Solvent blanks were recorded for each spectrum and subtracted. For NPN binding, solvent blanks consisted of NPN and methanol without LUSH. For cVA, bombykol or methyl oleate, blanks consisted of an equivalent volume of ethanol added to β-cyclodextrin in buffer. Spectra were measured at 23°C ± 1°. Curve-fitting was done with SigmaPlot or KaleidaGraph software.
Computational modeling of ligand binding to LUSH was done using AutoDock software, version 4.2 (Morris et al. 2009). LUSH coordinates were obtained from Protein Data Bank file 2GTE (molecule A), and cVA was removed. NPN coordinates were composed using the Molefacture routine in VMD (Humphrey et al. 1996). The dipole orientation factor, κ2, was calculated from the position of the tryptophan 1La transition dipole (Lakowicz 2006) and the calculated NPN absorption dipole (Smith and Woody 1976). The Förster critical distance, Ro, was calculated from Ro = 0.211 (κ2 n−4 QD J)1/6, where n, the index of refraction, was taken to be 1.33; QD, the quantum yield of tryptophan, was taken to be 0.12 (Callis and Liu 2004); J, the spectral overlap between tryptophan emission and NPN absorbance, was measured as 6.75 × 1013 cm3 M−1 nm4. The distance, R, was calculated from the transfer efficiency, E, as R = Ro6 (1−E)/E.
Supplementary Material
Acknowledgments
We thank David N. M. Jones for providing the pET13a vector containing the coding sequence for LUSH, and we thank Walter Leal for providing a sample of bombykol. This research was supported in part by grants from the National Institutes of Health (R21 DC010071 and G12 MD007591) and from the Texas Imported Fire Ant Research and Management Project.
ABBREVIATIONS
- BmorPBP
Bombyx mori pheromone-binding protein
- cVA
11-cis vaccenyl acetate
- FRET
Förster resonance energy transfer
- NPN
N-phenyl-1-naphthylamine
- OBPs
odorant- and pheromone-binding proteins
- ORs
olfactory receptors
Footnotes
Additional Supporting Information may be found in the online version of this article.
Figure S1. LUSH tryptophan fluorescence in urea.
Figure S2. NPN binding to unfolded LUSH
Table S1. Atomic coordinates for NPN Model 1
Figure S3. Relationship between β-cyclodextrin-guest affinities and molecular surface area
Figure S4. Effect of ethanol binding to LUSH on tryptophan emission.
References
- Ban L, Scaloni A, D’Ambrosio C, Zhang L, Yahn Y, Pelosi P. Biochemical characterization and bacterial expression of an odorant-binding protein from Locusta migratoria. Cell Mol Life Sci. 2003;60:390–400. doi: 10.1007/s000180300032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban L, Zhang L, Yan Y, Pelosi P. Binding properties of a locust’s chemosensory protein. Biochem Biophys Res Commun. 2002;293:50–4. doi: 10.1016/S0006-291X(02)00185-7. [DOI] [PubMed] [Google Scholar]
- Bette S, Breer H, Krieger J. Probing a pheromone binding protein of the silkmoth Antheraea polyphemus by endogenous tryptophan fluorescence. Insect Biochem Mol Biol. 2002;32:241–46. doi: 10.1016/s0965-1748(01)00171-0. [DOI] [PubMed] [Google Scholar]
- Bojesen IN, Bojesen E. Water-phase palmitate concentrations in equilibrium with albumin-bound palmitate in a biological system. J Lipid Res. 1992;33:1327–34. [PubMed] [Google Scholar]
- Briand L, Nespoulous C, Huet JC, Takahashi M, Pernollet JC. Ligand binding and physico-chemical properties of ASP2, a recombinant odorant-binding protein from honeybee (Apis mellifera L.) Eur J Biochem. 2001;268:752–60. doi: 10.1046/j.1432-1327.2001.01927.x. [DOI] [PubMed] [Google Scholar]
- Callis PR, Liu T. Quantitative Prediction of Fluorescence Quantum Yields for Tryptophan in Proteins. The Journal of Physical Chemistry B. 2004;108:4248–4259. [Google Scholar]
- Campanacci V, Krieger J, Bette S, Sturgis JN, Lartigue A, Cambillau C, Breer H, Tegoni M. Revisiting the specificity of Mamestra brassicae and Antheraea polyphemus pheromone-binding proteins with a fluorescence binding assay. J Biol Chem. 2001;276:20078–84. doi: 10.1074/jbc.M100713200. [DOI] [PubMed] [Google Scholar]
- Datta SR, Vasconcelos ML, Ruta V, Luo S, Wong A, Demir E, Flores J, Balonze K, Dickson BJ, Axel R. The Drosophila pheromone cVA activates a sexually dimorphic neural circuit. Nature. 2008;452:473–77. doi: 10.1038/nature06808. [DOI] [PubMed] [Google Scholar]
- Davanloo P, Crothers DM. Phase partition studies of actinomycin-nucleotide complexes. Biochemistry. 1976;15:4433–8. doi: 10.1021/bi00665a015. [DOI] [PubMed] [Google Scholar]
- Du G, Prestwich GD. Protein structure encodes the ligand binding specificity in pheromone binding proteins. Biochemistry. 1995;34:8726–32. doi: 10.1021/bi00027a023. [DOI] [PubMed] [Google Scholar]
- Ferrari E, Lodi T, Sorbi RT, Tirindelli R, Cavaggioni A, Spisni A. Expression of a lipocalin in Pichia pastoris: secretion, purification and binding activity of a recombinant mouse major urinary protein. FEBS Lett. 1997;401:73–7. doi: 10.1016/s0014-5793(96)01436-6. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–8. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Pace TC, Castillo C, Bohne C, O’Neill MA, Plettner E. Ligand-interaction kinetics of the pheromone- binding protein from the gypsy moth, L. dispar: insights into the mechanism of binding and release. Chem Biol. 2009;16:162–72. doi: 10.1016/j.chembiol.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Gonzalez D, Zhao Q, McMahan C, Velasquez D, Haskins WE, Sponsel V, Cassill A, Renthal R. The major antennal chemosensory protein of red imported fire ant workers. Insect Mol Biol. 2009;18:395–404. doi: 10.1111/j.1365-2583.2009.00883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse-Wilde E, Svatos A, Krieger J. A pheromone-binding protein mediates the bombykol-induced activation of a pheromone receptor in vitro. Chem Senses. 2006;31:547–55. doi: 10.1093/chemse/bjj059. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125:143–60. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Ho MG, Carlson JR. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117:965–79. doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Hildebrand JG, Shepherd GM. Mechanisms of olfactory discrimination: converging evidence for common principles across phyla. Annu Rev Neurosci. 1997;20:595–631. doi: 10.1146/annurev.neuro.20.1.595. [DOI] [PubMed] [Google Scholar]
- Honson N, Johnson MA, Oliver JE, Prestwich GD, Plettner E. Structure-activity studies with pheromone-binding proteins of the gypsy moth, Lymantria dispar. Chem Senses. 2003;28:479–89. doi: 10.1093/chemse/28.6.479. [DOI] [PubMed] [Google Scholar]
- Horst R, Damberger F, Luginbuhl P, Guntert P, Peng G, Nikonova L, Leal WS, Wuthrich K. NMR structure reveals intramolecular regulation mechanism for pheromone binding and release. Proc Natl Acad Sci U S A. 2001;98:14374–9. doi: 10.1073/pnas.251532998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–8. 27–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Iovinella I, Dani FR, Niccolini A, Sagona S, Michelucci E, Gazzano A, Turillazzi S, Felicioli A, Pelosi P. Differential expression of odorant-binding proteins in the mandibular glands of the honey bee according to caste and age. J Proteome Res. 2011;10:3439–49. doi: 10.1021/pr2000754. [DOI] [PubMed] [Google Scholar]
- Kruse SW, Zhao R, Smith DP, Jones DN. Structure of a specific alcohol-binding site defined by the odorant binding protein LUSH from Drosophila melanogaster. Nat Struct Biol. 2003;10:694–700. doi: 10.1038/nsb960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarde A, Spinelli S, Tegoni M, He X, Field L, Zhou JJ, Cambillau C. The crystal structure of odorant binding protein 7 from Anopheles gambiae exhibits an outstanding adaptability of its binding site. J Mol Biol. 2011;414:401–12. doi: 10.1016/j.jmb.2011.10.005. [DOI] [PubMed] [Google Scholar]
- Lakowicz JR. Principles of fluorescence spectroscopy. 3. xxvi. Springer; New York: 2006. p. 954. [Google Scholar]
- Laughlin JD, Ha TS, Jones DN, Smith DP. Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell. 2008;133:1255–65. doi: 10.1016/j.cell.2008.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautenschlager C, Leal WS, Clardy J. Bombyx mori pheromone-binding protein binding nonpheromone ligands: implications for pheromone recognition. Structure. 2007;15:1148–54. doi: 10.1016/j.str.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar J, Greenwood DR, Rasmussen LE, Prestwich GD. Molecular and functional characterization of an odorant binding protein of the Asian elephant, Elephas maximus: implications for the role of lipocalins in mammalian olfaction. Biochemistry. 2002;41:11786–94. doi: 10.1021/bi0256734. [DOI] [PubMed] [Google Scholar]
- Leal WS, Chen AM, Ishida Y, Chiang VP, Erickson ML, Morgan TI, Tsuruda JM. Kinetics and molecular properties of pheromone binding and release. Proc Natl Acad Sci U S A. 2005;102:5386–91. doi: 10.1073/pnas.0501447102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescop E, Briand L, Pernollet JC, Guittet E. Structural basis of the broad specificity of a general odorant-binding protein from honeybee. Biochemistry. 2009;48:2431–41. doi: 10.1021/bi802300k. [DOI] [PubMed] [Google Scholar]
- Li S, Picimbon JF, Ji S, Kan Y, Chuanling Q, Zhou JJ, Pelosi P. Multiple functions of an odorant-binding protein in the mosquito Aedes aegypti. Biochem Biophys Res Commun. 2008;372:464–8. doi: 10.1016/j.bbrc.2008.05.064. [DOI] [PubMed] [Google Scholar]
- Mao Y, Xu X, Xu W, Ishida Y, Leal WS, Ames JB, Clardy J. Crystal and solution structures of an odorant-binding protein from the southern house mosquito complexed with an oviposition pheromone. Proc Natl Acad Sci U S A. 2010;107:19102–7. doi: 10.1073/pnas.1012274107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–91. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham NJ, Krieger J, Breer H, Svatos A. Detection and removal of an artefact fatty acid from the binding site of recombinant Bombyx mori pheromone-binding protein. Chem Senses. 2001;26:529–31. doi: 10.1093/chemse/26.5.529. [DOI] [PubMed] [Google Scholar]
- Pesenti ME, Spinelli S, Bezirard V, Briand L, Pernollet JC, Campanacci V, Tegoni M, Cambillau C. Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release. J Mol Biol. 2009;390:981–90. doi: 10.1016/j.jmb.2009.05.067. [DOI] [PubMed] [Google Scholar]
- Plettner E, Lazar J, Prestwich EG, Prestwich GD. Discrimination of pheromone enantiomers by two pheromone binding proteins from the gypsy moth Lymantria dispar. Biochemistry. 2000;39:8953–62. doi: 10.1021/bi000461x. [DOI] [PubMed] [Google Scholar]
- Prestwich GD. Bacterial expression and photoaffinity labeling of a pheromone binding protein. Protein Sci. 1993;2:420–8. doi: 10.1002/pro.5560020314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekharsky MV, Inoue Y. Complexation Thermodynamics of Cyclodextrins. Chem Rev. 1998;98:1875–1918. doi: 10.1021/cr970015o. [DOI] [PubMed] [Google Scholar]
- Riviere S, Lartigue A, Quennedey B, Campanacci V, Farine JP, Tegoni M, Cambillau C, Brossut R. A pheromone-binding protein from the cockroach Leucophaea maderae: cloning, expression and pheromone binding. Biochem J. 2003;371:573–9. doi: 10.1042/BJ20021877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root CM, Masuyama K, Green DS, Enell LE, Nassel DR, Lee C-H, Wang JW. A presynaptic gain control mechanism fine-tunes olfactory behavior. Neuron. 2008;59:311–21. doi: 10.1016/j.neuron.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–6. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- Sharrow SD, Edmonds KA, Goodman MA, Novotny MV, Stone MJ. Thermodynamic consequences of disrupting a water-mediated hydrogen bond network in a protein:pheromone complex. Protein Sci. 2005;14:249–56. doi: 10.1110/ps.04912605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart R, Kiely A, Beale M, Vargas E, Carraher C, Kralicek AV, Christie DL, Chen C, Newcomb RD, Warr CG. Drosophila odorant receptors are novel seven transmembrane domain proteins that can signal independently of heterotrimeric G proteins. Insect Biochem Mol Biol. 2008;38:770–80. doi: 10.1016/j.ibmb.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Smith JC, Woody RW. Molecular orbital calculations on N-phenylnaphthylamines, fluorescence and circular dichroism probes. The Journal of Physical Chemistry. 1976;80:1094–1100. [Google Scholar]
- Syed Z, Ishida Y, Taylor K, Kimbrell DA, Leal WS. Pheromone reception in fruit flies expressing a moth’s odorant receptor. Proc Natl Acad Sci U S A. 2006;103:16538–43. doi: 10.1073/pnas.0607874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed Z, Kopp A, Kimbrell DA, Leal WS. Bombykol receptors in the silkworm moth and the fruit fly. Proc Natl Acad Sci U S A. 2010;107:9436–9. doi: 10.1073/pnas.1003881107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thode AB, Kruse SW, Nix JC, Jones DN. The role of multiple hydrogen-bonding groups in specific alcohol binding sites in proteins: insights from structural studies of LUSH. J Mol Biol. 2008;376:1360–76. doi: 10.1016/j.jmb.2007.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt RG, Riddiford LM. Pheromone binding and inactivation by moth antennae. Nature. 1981;293:161–3. doi: 10.1038/293161a0. [DOI] [PubMed] [Google Scholar]
- Vorum H, Brodersen R, Kragh-Hansen U, Pedersen AO. Solubility of long-chain fatty acids in phosphate buffer at pH 7.4. Biochim Biophys Acta. 1992;1126:135–42. doi: 10.1016/0005-2760(92)90283-2. [DOI] [PubMed] [Google Scholar]
- Wicher D, Schafer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, Hansson BS. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452:1007–11. doi: 10.1038/nature06861. [DOI] [PubMed] [Google Scholar]
- Willett CS, Harrison RG. Pheromone binding proteins in the European and Asian corn borers: no protein change associated with pheromone differences. Insect Biochem Mol Biol. 1999;29:277–84. doi: 10.1016/s0965-1748(99)00003-x. [DOI] [PubMed] [Google Scholar]
- Wojtasek H, Hansson BS, Leal WS. Attracted or repelled?--a matter of two neurons, one pheromone binding protein, and a chiral center. Biochem Biophys Res Commun. 1998;250:217–22. doi: 10.1006/bbrc.1998.9278. [DOI] [PubMed] [Google Scholar]
- Zhou JJ, Zhang GA, Huang W, Birkett MA, Field LM, Pickett JA, Pelosi P. Revisiting the odorant-binding protein LUSH of Drosophila melanogaster: evidence for odour recognition and discrimination. FEBS Lett. 2004;558:23–6. doi: 10.1016/S0014-5793(03)01521-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.