Abstract
Fluorescent protein (FP) technologies suitable for use within the eukaryotic secretory pathway are essential for live cell and protein dynamic studies. Localization of FPs within the endoplasmic reticulum (ER) lumen has potentially significant consequences for FP function. All FPs are resident cytoplasmic proteins and have rarely been evolved for the chemically distinct environment of the ER lumen. In contrast to the cytoplasm, the ER lumen is oxidizing and the site where secretory proteins are post-translationally modified by disulfide bond formation and N-glycosylation on select asparagine residues. Cysteine residues and N-linked glycosylation consensus sequences were identified within many commonly utilized FPs. Here, we report mTagBFP is post-translationally modified when localized to the ER lumen. Our findings suggest these modifications can grossly affect the sensitivity and reliability of FP tools within the secretory pathway. To optimize tools for studying events in this important intracellular environment, we modified mTagBFP by mutating its cysteines and consensus N-glycosylation sites. We report successful creation of a secretory pathway-optimized blue FP, secBFP2.
Keywords: Fluorescent proteins, BFP, N-glycosylation, Disulfide bonds, Endoplasmic reticulum
1. Introduction
The intrinsic characteristics of fluorescent proteins (FPs) must be considered to effectively use FP technologies within distinct cellular environments, such as the endoplasmic reticulum (ER). The amino acid sequences of commonly used FPs contain residues that have the potential to be post-translationally modified [1]. The sequence of mTagBFP [2], (commercial available from Evrogen as TagBFP), a blue FP, includes three N-linked glycosylation consensus sequences, N-X-S/T, (N73, N129, N223) and three cysteine residues (C27, C115, C223) (Fig. 1S). Within the cytoplasm these amino acids should not be problematic, since neither disulfide bond formation nor N-glycosylation typically occur in the cytoplasm. When nonnative secretory proteins, such as FPs, are targeted to the ER lumen, they are exposed to the uniquely oxidizing environment and ER-specific chaperones.
A correctly folded FP forms a barrel structure composed of 11-βstrands connected by flexible loop regions [3,4]. Barrel formation provides an internal environment which permits chromophore relevant residues to undergo an autocatalytic reaction, to produce a fluorescent chromophore [2,5,6]. (For detailed description of red and blue chromophore structure and chemical transitions refer to recent publications [7,8]). Chromophore formation and stability are influenced by surrounding amino acids within the β barrel. In mTagBFP, 23 surrounding residues form contacts via hydrogen bond formation or van der Waals forces [5]. None of the potentially post-translationally modified residues are known to contact or directly influence mTagBFP chromophore maturation. The positions of the mTagBFP cysteines are too distant to permit intra-molecular disulfide bonds in the rigid barrel native protein conformation. Asparagine residues of the N-glycosylation consensus sequences are on the surface of the folded FP. Together these analyses suggest, (1) if cysteine residues form inappropriate disulfide bonds, the β-barrel would not properly form and therefore will be nonfluorescent, and (2) attachment of sugars at N-linked glycosylation consensus sequences would significantly increase the hydrodynamic radius (Rh) of a correctly folded, fluorescent FP.
Currently, two green FPs (superfolder GFP [9,10], and cfSGFP2 [11]) and the mFruit family [12] of FPs are available for use within the secretory pathway, however no unmodified blue variants exist. Creation of a secretory pathway blue FP will increase investigators’ ability to assay a greater number of reporters simultaneously and could provide a potential FRET pair [13]. In this study, we report mTagBFP is detrimentally post-translationally modified in the ER lumen. We rationally engineered six amino acid substitutions to successfully create a secretory pathway optimized, secBFP2 that is no longer modified when localized to the ER. In addition to the secretory pathway, the first secretory pathway optimized blue FP should be suitable for other oxidizing environments including the mitochondrial inner membrane space and the periplasm of gram negative bacteria.
2. Materials and Methods
2.1 Chemicals
Dithiothreitol (DTT; Fisher Scientific, Pittsburgh, PA) was diluted to the indicated concentrations from a 1 M stock solution in water. Tunicamycin (Tm; Calbiochem, La Jolla, CA) was diluted to the indicated concentrations from a 5 mg/ml stock solution in DMSO.
2.2 Mammalian plasmids
mTagBFP [2] was previously described. Secretory optimized FP, secBFP1 contains cysteine to serine and asparagine within N-linked glycosylation consensus sequences to aspartate mutations were synthesized by GenScript (Piscataway, NJ). To create cytoplasmic and ER-localized versions, FP encoding sequences were amplified using the following primers:
ER-mTagBFP
F5′ GCAATGGGCGGTAGGCG
R5′ GATCGCGGCCGCGTTACAATTCATCCTTATTAAGTTTGTGCCC
Cytoplasmic-mTagBFP
R5′ GATCGCGGCCGCTTAATTAAGCTTGTGCCC
ER-secBFP
F5′ GATCACCGGTCGCCAGCAGCGAGGAGCTGATTAAGGAAGAAC
R5′ GATCGCGGCCGCTTACAGCTCGTCCTTATTAAGCTTGTGCCC
Cytoplasmic-secBFP
F5′ GATCACCGGTCGCCACCATGAGCGAGGAGCTGATTAAGGAGAAC
R5′GATCGCGGCCGCTTAATTAAGCTTGTGCCC
FP fragments were cloned into the AgeI/NotI sites of N1 GFP (Clontech, Mountain View, CA) or ER-RFP [14] creating ER-mTagBFP. To investigate specific amino acid substations consequences on FP fluorescence, reverting mutations were reintroduced into FPs. Site-directed mutagenesis primers include:
secBFP
S29C- 5′ GACAACCATCACTTCAAGTGTACATCCGAGGGCGAAGGC
S29V- 5′ GACAACCATCACTTCAAGGTGACATCCGAGGGCGAAGGC
S29G- 5′ GACAACCATCACTTCAAGGGCACATCCGAGGGCGAAGGC
S29A- 5′ GACAACCATCACTTCAAGGCAACATCCGAGGGCGAAGGC
D93N- 5′ CTCTACGGCAGCAAGACCTTCATTAATCACACCCAGGGCATCCCC
D129N- 5′ GTCAAGATCAGAGGGGTTAACTTCACATCCAACGGC
mTagBFP
C118S- 5′ GATCTTGACGTTGTAGATCAAGCTTCCGTCCTGGAGGCTGGTGTC
C29S- 5′ CATCACTTCAAGAGCACATCCGAGGGCGAAGGC
All constructs were confirmed by sequencing.
2.3 Bacterial cloning, expression and protein purification
mTagBFP and secBFP2 sequences were amplified using the following primers:
5′ GATCCCATGGGTATGAGCGAGGAGCTG
5′ GATCCTGCAGCATTAAGCTTGTGCCCCAG
FP fragments were cloned into pEcoli-Cterm 6xHN vector (Clontech) at NcoI and PstI restriction sites. Vectors were expressed in CodonPlus competent, BL-21 RP (Stratagene, La Jolla CA) cells. 5 ml cultures were grown at 37°C, 225 rpm overnight in LB under of chloramphenicol and ampicillin drug selection. 1 ml of overnight cultures were added to 50 ml of drug free LB for 2 h at 37°C, 225 rpm. For induction, 1 mM IPTG was added to cultures, and returned to 37°C, 225 rpm shaker for 4 h. Cultures were pelleted at 3000 rpm for 15 min at 4°C. For protein purification procedure refer to manufactures protocol for His60 Ni Superflow™ Resin & Gravity Column manual (Clontech).
2.4 Cell culture and transfection
U2OS cells were routinely cultured in RPMI medium (Mediatech, Manassas, VA), supplemented with 5 mM glutamine, penicillin/streptomycin (Invitrogen, Carlsbad, CA), and 10% heat inactivated fetal bovine serum (Hyclone from Thermo Scientific, Rockford, IL) at 37°C in 5% CO2. All constructs were transiently transfected for 16–20 h into cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
2.5 Live cell fluorescence imaging
For imaging experiments, cells were grown in 8 well LabTek coverglass chambers (Nunc, Rochester, NY). Cells were imaged in phenol red-free RPMI freshly supplemented with 10 mM Hepes (Fisher Scientific) and 10% fetal bovine serum. Live cells were imaged on a 37°C environmentally controlled chamber of a confocal microscope system (Zeiss LSM 5 LIVE microscope with Duoscan attachment; Carl Zeiss MicroImaging, Inc., Thornwood, NY.) with a 63X/1.4 NA oil objective and a 405 nm 50 mW diode laser with a 415–505 nm bandpass or 510 nm long pass filter for mTagBFP, or a 489 nm 100mW diode laser with a 495–555 or 520–555 nm bandpass filter for Alexa anti-488. Image analysis and composite figures were prepared using ImageJ (National Institutes of Health; Bethesda, MD), Photoshop CS4 and Illustrator CS4 software (Adobe Systems, San Jose, CA).
2.6 Immunoblots
Total cell lysates for immunoblotting were prepared in 1% SDS, 0.1 M Tris, pH 8.0 with 100 mM DTT (reducing conditions) or no DTT (nonreducing conditions) using cells in 24 well plates at 80–90% confluence. For the reducing and nonreducing immunoblots, cells were first treated with 20 mM NEM in PBS for 15 min at room temperature. Proteins were separated using either 5 or 12% Tris-tricine gels, transferred to nitrocellulose, probed with the indicated antibodies, and developed using enhanced chemiluminescent reagents (Pierce, Rockford, IL), and exposed to X-ray film. Antibodies used included anti-tRFP (Evrogen, Moscow, Russia), anti-β actin (Sigma Aldrich, St. Louis, MO), and horseradish peroxidase-labeled anti-mouse or anti-rabbit secondary antibodies (Jackson Immunoresearch Laboratories, West Grove, PA).
2.7 Immunofluorescence
Cells were fixed with freshly diluted 3.7% formaldehyde in PBS at room temperature for 15 min and permeabilized with 0.1% Triton X-100 in PBS. Blocking was preformed with 10% fetal bovine serum in PBS for 30 min. Subsequently, cells were labeled with anti-tRFP, followed by Alexa-Fluor-488 conjugated anti-rabbit IgG secondary antibody (Invitrogen). Images were acquired using an Axiovert 200 widefield fluorescence microscope (Carl Zeiss Microimaging Inc.) with a 63x oil immersion 1.4 NA objective, and 470/40 excitation, 525/50 emission bandpass filter for Alexa Fluor 488, 565/30 excitation, or 365 excitation, 445/50 emission bandpass filter for mTagBFP.
2.8 Photobleaching analysis
Fluorescence Recovery after Photobleaching (FRAP) was performed by photobleaching a small region of interest (ROI) and monitoring fluorescence recovery or loss over time, as described previously [15,16]. Fluorescence intensity plots and D measurements were calculated as described previously [15,16]. P values were calculated using a Student’s two-tailed t test in Prism 5.0c (GraphPad Software, San Diego, CA). The relatively large spread of D values for ER proteins likely reflects differences in ER geometry between cells [17]. Rh values were calculated using the Stokes-Einstein Equation, D= κBT/6πηRh [18].
2.9 Protein characterization
Absorbance was measured using Hitachi U-2000 spectrophotometer. Excitation and emission spectra were measured with FluoroMax-3 spectrofluorometer. Purified protein samples were measured in phosphate buffered saline (PBS).
To assess photobleaching sensitivity under comparable imaging parameters, secBFP2 and mTagBFP were transiently expressed in U2OS cells. Cells were imaged using live cell imaging conditions (Section 2.5). Images were acquired at 5 frames/s for 400 frames.
2.10 Statistical analysis
Two-tailed Student’s t-tests with Prism software (GraphPad) were used to compare the different conditions. Variances of data sets were compared using an F-test (GraphPad) to establish whether to use equal or nonequal variance t-tests.
3. Results/Discussion
3.1 mTagBFP is post-translationally modified in the ER
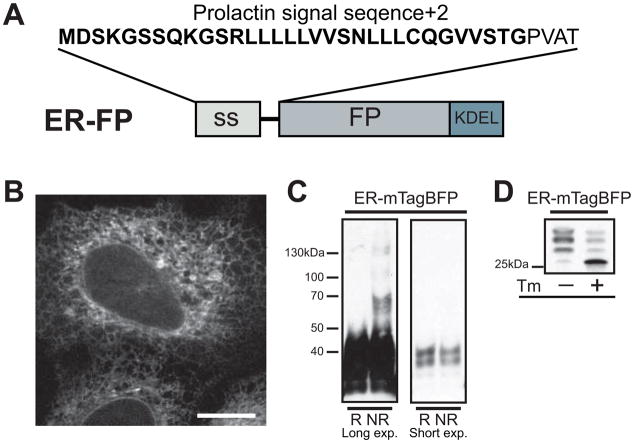
To localize the FP to the ER, the prolactin signal sequence was fused upstream of mTagBFP and a –KDEL ER retrieval motif was fused downstream (Fig. 1A), [14]. ER-mTagBFP correctly localizes and fluoresces (Fig. 1B), however immunoblots reveal a proportion of the FP is post-translationally modified in the ER. Under nonreducing conditions, where disulfide bonds are maintained, a laddering pattern reveals both an expected monomeric form of mTagBFP (~25 kDa) and problematic higher molecular weight FP oligomers (~50,100, 130 kDa, Fig. 1C). Analysis of the native FP structure confirm the three cysteine residues (C27, C115, C223) are too far apart (≫0.23 nm) to form intra-molecular disulfide bonds. Additionally, C27 and C115 are orientated towards the interior of the folded β-barrel, preventing thiol groups in correctly folded barrels from participating in disulfide bonds. If either C27 or C115 form a nonnative disulfide bond between FP molecules, the FPs would have to be unfolded and therefore nonfluorescent. The third cysteine residue, C223 is within the structurally unresolved C-terminus of the protein. This residue could theoretically form dimers between folded ER-mTagBFP molecules. Regardless of which form of oligomers formed, the oligomers are undesirable for creating fusion proteins. Furthermore, unfolded nonfluorescent FPs will decrease the signal and under represent the levels of fusion protein in the ER.
Fig. 1.
ER-localized mTagBFP is post-translationally modified. (A) Schematic representation of ER-localized FP (ER-FP), including required prolactin signal sequence and KDEL retrieval motif. (B) Representative images of transiently transfected U2OS cells expressing ER-mTagBFP, scale bar = 10 μm. Immunoblot (C) under reducing (R)/nonreducing (NR) conditions (long exposures and short exposures of the same blot) illustrate the propensity of mTagBFP to oligomerize under nonreducing conditions or (D) treated 5 h −/+ 5 μg/ml Tm. The lower molecular weight bands (~25 kDa) denote nonglycosylated population of FPs.
Next, we investigated the impact of N-linked glycosylation on ER-mTagBFP. Asparagine residues (N73, N129, N223) are each contained within glycosylation consensus sequences and could be glycosylated as the protein emerges from the ER translocation channel. The addition of an N-glycan would increase the relative size and molecular weight of ER-mTagBFP. ER-mTagBFP migrates higher than predicted (~25 kDa) on a reducing SDS-PAGE and this could be due to glycosylation. Inhibition of N-glycosylation in cells via treatment with tunicamycin (Tm), a GlcNAc phosphotransferase inhibitor, caused the shift to the lower expected molecular weight of nonglycosylated, ER-mTagBFP (Fig. 1D). Consequently, unfolded FPs and glycosylated FPs are potential targets of the ER quality control machinery, including chaperones and this interaction could titrate native chaperones and possibly induce ER stress.
When utilizing FP technologies, the goal is to introduce functional, but inert, tools to probe live cells. Furthermore, the optimal FP is as small as possible. At 5 nm, FPs are already large protein tags. N-linked glycosylation could potentially sterically hinder interactions between an FP fusion protein and partner proteins. For these reasons, it is imperative to improve and protect FPs for environmental post-translational modifications environments.
3.2 Cysteine 29 mutagenesis
In the initial round of protein engineering to create a secretory pathway optimized mTagBFP, we substituted all cysteines with serines and N-glycosylation consensus asparagines with aspartates. However, the engineered FP, secBFP1 was no longer fluorescent when transiently expressed, even in the cytoplasm of cells (Fig. 2S). Expression of the nonfluorescent protein was confirmed by immunofluorescence (Fig. 2S).
To restore fluorescence to the engineered FP, we performed structural analyses, which predicted mutations most likely responsible for diminishing secBFP1 fluorescence: C29S, N73D, C118S, and N129D. None of these amino acids form contacts with the chromophore. However, mutations C29S, C118S, and N129D are located within β-strands, while N73D is found at the end of the internal α-helix. The N73D mutation could alter the helix rigidity and as a consequence disrupt chromophore formation. The additional mutations, N214D and C229S are within flexible loop regions at the C-terminal of the protein and therefore unlikely to play roles in altering fluorescence. Each mutated residue was individually reverted to the original amino acid and transiently expressed in cells to identify the amino acid substitutions that decreased fluorescence. The normalized mean fluorescence intensities (MFI) of cells expressing the various mutant constructs were compared (Fig. 2A). Re-introduction of either asparagine residues (N73 or N129) or C118S had no affect on FP fluorescence. However, the S29C mutation restored fluorescence comparable to the parental mTagBFP. Introduction of the converse mutation, C29S, within mTagBFP background attenuated fluorescence, confirming the importance of this cysteine. We expressed ER-secBFP1 S29C and examined the protein under nonreducing conditions. An immunoblot (Fig. 3S) revealed a higher molecular weight laddering pattern, with a distinct population corresponding to a potential dimer species (50 kDa). These data indicate not only the importance of the residue to maintaining the proteins fluorescence but also the ability of C29 to participate inappropriate disulfide bonds.
Fig. 2.
Amino acid substitution, C29A, resulted in a fluorescent secBFP that was no longer post-translationally in the ER. (A) Normalized relative MFI comparisons identified the cysteine to serine mutation at position 29 as the residue responsible for the absence of fluorescence of secBFP1. Immunoblots of cells transfected with ER-mTagBFP or -secBFP2 confirmed optimized FP under (B) reducing/nonreducing conditions does not participate in inappropriate disulfide bonds and (C) was not N-glycosylated. (D) Representative image of transiently transfected U2OS cells expressing ER-secBFP2, scale bar = 10 μm.
Although serines are frequently used as an amino acid substitution for cysteines, these mutations are not necessarily structurally conservative. Serine can even induce bending in a helices at positions normally occupied by cysteines [19]. Also, cysteine can exhibit a hydrophobic character [20]. More hydrophobic and structurally similar amino acids, valine (S29V) and alanine (S29A) were introduced to restore fluorescence in secBFP1. S29A produced the first fluorescent secretory optimized blue FP, secBFP2 (Fig. 2B). See Supplementary Figure 4 for characterization of secBFP2’s properties. The S29G had no affect on protein fluorescence demonstrating the significance of maintaining the structural integrity of the amino acid at critical residues (Fig. 2A).
Immunoblots comparing ER-mTagBFP and –secBFP2 verified secBFP2 localized to the ER lumen without being post-translationally modified. Under nonreducing conditions, ER-secBFP2 did not form higher molecular weight oligomers (Fig. 2B). Additionally, ER-secBFP2 is not glycosylated as indicated by a single species band at the expected FP size (Fig. 2C).
3.3 Enhancing folding of secBFP2
To improve the utility of secBFP2, we introduced the enhanced mTagBFP2point mutation, I174A. This mutation increases photostability and chemical stability (Fig. 3A) [13]. Incorporation of the I174A mutation improved the yield of fluorescent secBFP2, as measured by overall increased fluorescence intensity compared to secBFP2 (Fig. 3B, top).
Fig. 3.
Enhanced mTagBFP2 mutation and lower temperatures improves folding of secBFP2. (A) U2OS cell transiently expressing ER-secBFP2 I174A displays a typical ER tubular network pattern. (B) U2OS cells expressing cytoplasmic-secBFP2 I174A or cyto-secBFP2 at different temperatures were imaged under identical conditions. At physiologic temperature for mammalian cells, 37°C secBFP2 I174A has significantly increased detectable fluorescence, most likely reflecting improved maturation. Cells transiently transfected at 25°C for 18 h expressing either cyto-secBFP2 I174A or -secBFP2 have comparable bright fluorescence indicating that secBFP2 maturation improves at lower temperatures Scale bar = 10 μm.
In addition, we investigated whether we could improve secBFP2 fluorescence without further mutagenesis. Other FPs (early GFP variants [21] and coral-derived FPs, such as mEos [22]) have been described which mature poorly at 37°C. FPs originally isolated from jelly fish or coral, both cold water organisms, exhibited improved folding and fluorescence at cooler temperatures (25°C). These suboptimal features have been addressed by evolving FPs that efficiently fold and mature in mammalian systems (37°C). We asked whether the sea anemone-derived secBFP2 might mature better at lower temperatures. Transfected cells were cultured at 25°C overnight and now exhibited comparable fluorescence for both secBFP2 and secBFP2 I174A (Fig. 3B). Thus, protein folding and maturation of secBFP2 can be improved at either lower temperatures or with the additional I174A mutation.
3.4 Consequences of mTagBFP glycosylation in the ER
Finally, we asked whether mutating post-translational modifications in mTagBFP could improve the size and mobility of secBFP2 in the ER in live cells. Mobility and size can be readily assessed by measuring the FP’s effective diffusion coefficient (Deff) by Fluorescence Recovery after Photobleaching (FRAP) in live cells [15,16]. FRAP analysis (Fig. 4A and B) revealed a significant difference in ER-mTagBFP and –secBFP2 mobilities (Fig. 4C). ER-mTagBFP (3.0 μm2/s) had a 1.5x slower mean Deff compared to ER-secBFP2 (4.6 μm2/s) (Fig. 4C). This difference in Deff values is consistent with a significant decrease in the Rh of the improved ER-secBFP2 as calculated using the Stokes-Einstein Equation [18]. While a typical FP Rh is 2.3 nm (Fig. 4D, dashed circle) [23], our measurements establish ER-mTagBFP has a 1.5x larger Rh (Fig. 4D, solid circle). This size increase is consistent with the size and flexibility of the N-glycans on ER-mTagBFP. The change could also reflect bind and release interactions of the glycosylated FP with the ER lectin chaperones calnexin and calreticulin [14]. Aside from potential utility as a reporter for glycosylated secretory proteins, a glycosylated FP is generally undesirably larger and, most importantly, not inert.
Fig. 4.
Analysis of protein dynamics in live cells revealed the consequences of post-translational modifications and the requirement for secretory pathway optimized FPs. Time series of representative FRAP analysis of cells transiently expressing (A) ER-mTagBFP or (B) -secBFP2. White box indicates FRAP bleach region. Scale bar = 10 μm. (C) Quantitation of Deff values revealed ER-mTagBFP had a significantly lower Deff value compared with –secBFP2, 3.04 μm2/s and 4.6 μm2/s, respectively. p=0.0004 (D) The average calculated Rh of ER-mTagBFP was 3.6 nm, solid circle. Unmodified mTagBFP (PDB 3M24) (Rh dashed circle) is represented with ribbon structure and N-glycosylation consensus sequences are red ball and stick.
4. Conclusions
To improve the utility of FPs within multiple cellular environments, we have successfully engineered an inert mutant of mTagBFP for the ER and other oxidizing environments. The secretory optimized, secBFP2 is the first blue FP that is protected from post-translational modifications when localized to the ER. We propose this method for creating FPs suitable for the ER environment can be applied to other FPs and we are currently developing a tool box of other improved oxidizing environment friendly FPs.
Supplementary Material
Amino acid sequence alignment and secondary structure of mTagBFP. FP sequence including the relative location of amino acids with secondary structures. Grey shading specify inward facing residues within a correctly folded 11-β-strand barrel. Underlined residues denote location of cysteine and N-glycosylation consensus sequences.
Synthesized secretory optimized FP is misfolded in the ER. Immunofluorescence of transiently transfected U2OS cells expressing ER-mTagBFP or -secBFP1. No detectable fluorescence was observed in cells expressing secBFP1, anti-tRFP staining indicates secBFP1 was expressed and ER localized.
A single cysteine at position 29 results in higher molecular weight oligomers under nonreducing conditions. Nonreducing/reducing immunoblot of cells transfected with ER-mTagBFP or secBFP1 S29C confirmed that a single cysteine (C29) produced higher molecular weight bands and the formation of inappropriate disulfide bonds under nonreducing conditions.
Spectral characterization of purified mTagBFP and secBFP2. (A) Absorbance of mTagBFP (grey line) and secBFP2 (black line). (B) Excitation and emission spectra (C) Image acquisition under comparable imaging parameters of mTagBFP (grey line) and secBFP2 (black line).
Highlights.
ER-localized FPs are subject to adverse post-translational modifications.
ER-mTagBFP is glycosylated and forms inappropriate inter-chain disulfide bonds.
SecBFP2 is the first blue FP adapted for the ER and other oxidizing environments.
Acknowledgments
We thank the Einstein Analytical Imaging Facility for use of the Zeiss Duoscan. This work was supported by grants from the National Institute of General Medical Sciences (NIGMS) (R01GM086530–01) (E.L.S.), National Institute of Diabetes and Digestive and Kidney Diseases NIDDK (5PO1DK041918) (E.L.S), and NIH Training Program in Cellular and Molecular Biology and Genetics Grant T32 GM007491 (L.M.C.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIGMS or the NIH. E.L.S. and L.M.C conceived and designed the study. V.V.V provided expertise and valuable advice. L.M.C, M.J. and O.M.S. performed the experiments. L.M.C and E.L.S analyzed the data and wrote the manuscript.
Abbreviations
- FP
fluorescent protein
- ER
endoplasmic reticulum
- Tm
tunicamycin
- Deff
effective diffusion coefficient
- Rh
hydrodynamic radius
Footnotes
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Snapp EL. Fluorescent proteins: a cell biologist’s user guide. Trends in Cell Biology. 2009;19:649–55. doi: 10.1016/j.tcb.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Subach OM, Gundorov IS, Yoshimura M, Subach FV, Zhang J, Grüenwald D, et al. Conversion of red fluorescent protein into a bright blue probe. Chemistry & Biology. 2008;15:1116–24. doi: 10.1016/j.chembiol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- 4.Ormö M, Cubitt aB, Kallio K, Gross La, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–5. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 5.Subach OM, Malashkevich VN, Zencheck WD, Morozova KS, Piatkevich KD, Almo SC, et al. Structural characterization of acylimine-containing blue and red chromophores in mTagBFP and TagRFP fluorescent proteins. Chemistry & Biology. 2010;17:333–41. doi: 10.1016/j.chembiol.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pletnev S, Subach FV, Dauter Z, Wlodawer A, Verkhusha VV. Understanding blue-to-red conversion in monomeric fluorescent timers and hydrolytic degradation of their chromophores. Journal of the American Chemical Society. 2010;132:2243–2253. doi: 10.1021/ja908418r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bravaya KB, Subach OM, Korovina N, Verkhusha VV, Krylov AI. Insight into the common mechanism of the chromophore formation in the red fluorescent proteins: the elusive blue intermediate revealed. Journal of the American Chemical Society. 2012;134:2807–14. doi: 10.1021/ja2114568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Subach FV, Verkhusha VV. Chromophore transformations in red fluorescent proteins. Chemical Reviews. 2012;112:4308–27. doi: 10.1021/cr2001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nature Biotechnology. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 10.Aronson DE, Costantini LM, Snapp EL. Superfolder GFP is fluorescent in oxidizing environments when targeted via the Sec translocon. Traffic Copenhagen Denmark. 2011;12:543–548. doi: 10.1111/j.1600-0854.2011.01168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki T, Arai S, Takeuchi M, Sakurai C, Ebana H, Higashi T, et al. Development of Cysteine-Free Fluorescent Proteins for the Oxidative Environment. PLoS ONE. 2012;7:e37551. doi: 10.1371/journal.pone.0037551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature Biotechnology. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 13.Subach OM, Cranfill PJ, Davidson MW, Verkhusha VV. An enhanced monomeric blue fluorescent protein with the high chemical stability of the chromophore. PLoS ONE. 2011;6:e28674. doi: 10.1371/journal.pone.0028674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snapp EL, Sharma A, Lippincott-Schwartz J, Hegde RS. Monitoring chaperone engagement of substrates in the endoplasmic reticulum of live cells. Proceedings of the National Academy of Sciences. 2006;103:6536–41. doi: 10.1073/pnas.0510657103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siggia ED, Lippincott-Schwartz J, Bekiranov S. Diffusion in inhomogeneous media: theory and simulations applied to whole cell photobleach recovery. Biophysical Journal. 2000;79:1761–1770. doi: 10.1016/S0006-3495(00)76428-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snapp EL, Hegde RS, Francolini M, Lombardo F, Colombo S, Pedrazzini E, et al. Formation of stacked ER cisternae by low affinity protein interactions. The Journal of Cell Biology. 2003;163:257–69. doi: 10.1083/jcb.200306020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sbalzarini IF, Mezzacasa A, Helenius A, Koumoutsakos P. Effects of organelle shape on fluorescence recovery after photobleaching. Biophysical Journal. 2005;89:1482–92. doi: 10.1529/biophysj.104.057885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Einstein A. Über die von der molekularkinetischen Theorie der Wärme geforderte Bewegung von in ruhenden Flüssigkeiten suspendierten Teilchen. Annalen Der Physik. 1905;322:549–560. [Google Scholar]
- 19.Ballesteros JA, Deupi X, Olivella M, Haaksma EE, Pardo L. Serine and threonine residues bend alpha-helices in the chi(1) = g(-) conformation. Biophysical Journal. 2000;79:2754–60. doi: 10.1016/S0006-3495(00)76514-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagano N, Ota M, Nishikawa K. Strong hydrophobic nature of cysteine residues in proteins. FEBS Letters. 1999;458:69–71. doi: 10.1016/s0014-5793(99)01122-9. [DOI] [PubMed] [Google Scholar]
- 21.Patterson GH, Knobel SM, Sharif WD, Kain SR, Piston DW. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophysical Journal. 1997;73:2782–90. doi: 10.1016/S0006-3495(97)78307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nienhaus GU, Nienhaus K, Hölzle A, Ivanchenko S, Renzi F, Oswald F, et al. Photoconvertible fluorescent protein EosFP: biophysical properties and cell biology applications. Photochemistry and Photobiology. n.d;82:351–8. doi: 10.1562/2005-05-19-RA-533. [DOI] [PubMed] [Google Scholar]
- 23.Hink MA, Griep RA, Borst JW, van Hoek A, Eppink MH, Schots A, et al. Structural dynamics of green fluorescent protein alone and fused with a single chain Fv protein. The Journal of Biological Chemistry. 2000;275:17556–60. doi: 10.1074/jbc.M001348200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid sequence alignment and secondary structure of mTagBFP. FP sequence including the relative location of amino acids with secondary structures. Grey shading specify inward facing residues within a correctly folded 11-β-strand barrel. Underlined residues denote location of cysteine and N-glycosylation consensus sequences.
Synthesized secretory optimized FP is misfolded in the ER. Immunofluorescence of transiently transfected U2OS cells expressing ER-mTagBFP or -secBFP1. No detectable fluorescence was observed in cells expressing secBFP1, anti-tRFP staining indicates secBFP1 was expressed and ER localized.
A single cysteine at position 29 results in higher molecular weight oligomers under nonreducing conditions. Nonreducing/reducing immunoblot of cells transfected with ER-mTagBFP or secBFP1 S29C confirmed that a single cysteine (C29) produced higher molecular weight bands and the formation of inappropriate disulfide bonds under nonreducing conditions.
Spectral characterization of purified mTagBFP and secBFP2. (A) Absorbance of mTagBFP (grey line) and secBFP2 (black line). (B) Excitation and emission spectra (C) Image acquisition under comparable imaging parameters of mTagBFP (grey line) and secBFP2 (black line).