Summary
HIV-1 reverse transcription is achieved in the newly infected cell before viral DNA (vDNA) nuclear import. Reverse transcriptase (RT) has previously been shown to function as a molecular motor, dismantling the nucleocapsid complex that binds the viral genome as soon as plus-strand DNA synthesis initiates. We first propose a detailed model of this dismantling in close relationship with the sequential conversion from RNA to double-stranded (ds) DNA, focusing on the nucleocapsid protein (NCp7). The HIV-1 DNA-containing preintegration complex (PIC) resulting from completion of reverse transcription is translocated through the nuclear pore. The PIC nucleoprotein architecture is poorly understood but contains at least two HIV-1 proteins initially from the virion core, namely Integrase (IN) and the viral protein r (Vpr). We next present a set of electron micrographs supporting that Vpr behaves as a DNA architectural protein, initiating multiple DNA bridges over more than 500 base pairs (bp). These complexes are shown to interact with NCp7 bound to single-stranded nucleic acid regions that are thought to maintain IN binding during dsDNA synthesis, concurrently with nucleocapsid complex dismantling. This unexpected binding of Vpr conveniently leads to a compacted but filamentous folding of the vDNA that should favor its nuclear import. Finally, nucleocapsid-like aggregates engaged in dsDNA synthesis appear to efficiently bind to F-actin filaments, a property that may be involved in targeting complexes to the nuclear envelope. More generally, this article highlights unique possibilities offered by in vitro reconstitution approaches combined with macromolecular imaging to gain insights into the mechanisms that alter the nucleoprotein architecture of the HIV-1 genome, ultimately enabling its insertion into the nuclear chromatin.
Keywords: HIV-1, nucleocapsid, Viral protein r, reverse transcription, pre-integration complex, integrase
I. Introduction
During the early steps of human immunodeficiency virus type-1 (HIV-1) replication, the viral core complex, e.g. the conical structure termed “capsid” containing the viral ribonucleoprotein (RNP) complex termed “nucleocapsid” is injected into the cytoplasm of the host cell.
The reverse transcription complex (RTC) is derived from this core complex. It contains about 25–125 copies of RT, 12–60 of integrase (IN; tetramer form), hundreds of viral protein r (Vpr) copies (Briggs et al., 2004; Müller et al., 2000) and a dynamic and condensed RNA:protein architecture composed of a pair of single-stranded (ss-) positive 9200nt.-long RNA molecules (vRNA) covered by about 1500–2500 nucleocapsid proteins (NCp) (Thomas and Gorelick, 2008). This RNP assembly is where reverse transcription takes place, converting the dimerized vRNA into a single 9.8kbp.-long viral dsDNA (vDNA) (Darlix et al., 2011; Levin et al., 2010). Along with reverse transcription, the RTC also matures into a preintegration complex (PIC). The PIC is a nucleoprotein complex assembled on the fully reverse transcribed vDNA, responsible for preparing and inserting the vDNA into the host’s chromatin (Suzuki and Craigie, 2007). Within the PIC, the protein content derived from the viral core is rearranged while it utilizes the cellular nuclear import, chromatin remodeling, and DNA repair processes to achieve completion of the early steps of HIV-1 infection. RTC to PIC transition is associated with uncoating of the core, while these complexes use the cellular cytoskeleton and the nuclear pore complexes for routing viral nucleoprotein complexes to the cell’s nucleus. Due to its fundamental importance in the virus life cycle, the molecular events describing the early journey of HIV-1 within the cell have hence been at the heart of active, but complicated and controversial research over the last two decades (Arhel, 2010; Goff, 2001; Nermut and Fassati, 2003; Shah and Aiken, 2010; Warrilow et al., 2009). Our current understanding of these steps and their spatiotemporal coordination remain essentially descriptive rather than mechanistic despite the impressive body of data obtained over the years.
In this report, we present the events that occur during the transition from the virion core to the PIC in order to highlight a mechanistic perspective that remains poorly defined in the literature: the remodeling per se of the HIV-1 nucleoprotein assemblies from the nucleocapsid complex to the PIC, taking into account the characterized interactions and transactions between the related macromolecular components, while the genome is converted from RNA to dsDNA. Due to the dynamic interactions between RTC components involving a high degree of coordination and precision, we speculate that RT could act as the molecular motor that drives the remodeling of HIV-1 cores, changing both the nature of its nucleic acids (NA) template (RNA to dsDNA) and consequently its nucleoprotein architecture, according to the concepts of macromolecular machines (Alberts, 1998). In the first section, we focus on the trio formed by the viral genome, RT and NCp, taking into account both the architectural and functional role of NCp throughout the reverse transcription process. HIV-1 nucleocapsid-to-PIC remodeling is presented in a rather simple model where the nucleocapsid complex, comprised of single stranded regions condensed by NCp7 is progressively dismantled during RT-driven dsDNA synthesis, allowing (i) the nascent dsDNA to become a new template for binding of additional proteins and (ii) the concentration of NCp7 and RT on the last ss-regions of vDNA (the central DNA flap and the Long Terminal Repeats (LTRs)). Next, we present some astonishing images of non-specific Vpr-, Vpr-NCp7- and IN-Vpr-DNA interactions observed from our RTC-PIC molecular reconstitution experiments observed by transmission electron microscopy (TEM) (Hameau et al., 2001; Mirambeau et al., 2007; Mirambeau et al., 2006). These micrographs present for the first time a convincing scenario of the stepwise coordination of nucleocapsid disassembly and PIC assembly, with an unexpected, but clearly evident dsDNA architectural role for Vpr. Finally, several micrographs are presented at the end of this article where our synthetic RTC-like complexes bind to actin filaments, opening new intriguing questions regarding the interactions between actin and the HIV-1 nucleocapsid/RTC.
II. The Reverse Transcription Complex as a macromolecular machine delivered from within the HIV-1 core
HIV-1 reverse transcription is a complex series of sequential biochemical reactions involving NA-NA interactions to virtually guide RT enzymes along vRNA and ssDNA templates in a coordinated manner. This elegant mechanism proceeds through interaction/exchange/annealing of RNA or ssDNA among complicated stem-loop structures between the 5′ and 3′ UTRs of the viral genome, as vDNA sequences are synthesized by RT. This process helps RT switch from strand to strand by two strand transfers, which involve circularized intermediates of vRNA/vDNA and strand displacement events, whereas the RNA genome appears to naturally adopt a circularized state due to a U3R-Gag RNA interaction (Beerens and Kjems, 2010; Ooms et al., 2007).
To perform such a complex mechanism, HIV-1 reverse transcription uses a finely tuned partnership between RT and NC proteins. The latter are small basic polypeptides closely associated with viral NA with a dual function: (i) non-specific aggregation/condensation with/of NA into a dynamic RNP complex, which offers the optimal context (e.g. plasticity and macromolecular crowding effects) for the pool of RT enzymes to complete dsDNA synthesis (Lener et al., 1998); (ii) specific binding, destabilization and annealing of RNA and DNA stem-loops structures (i.e. the nucleic acid chaperone function of NC), which accelerate the strand transfers and template switching followed by continued reverse transcription. This partnership between RT, NCp7 and viral NA is a key element of HIV-1 infection and is presented below. Recent in-depth reviews are available for more information on the reverse transcription mechanism assisted by NCp7 (Darlix et al., 2011; Godet and Mély, 2010; Levin et al., 2010; Mori et al., 2010).
II.1 Complexes formed between NC proteins and nucleic acids
As noted above, reverse transcription occurs in and is dependent on the RNP complex formed between NCp, RT and vRNA. The RNP complex itself is assembled during virus maturation through the action of the various forms of NCp (Mirambeau et al., 2010). During packaging, the NC domain of Pr55Gag(Gag) acts as a sensor for selective capture of vRNA among the pool of cellular RNA and as the “loading” domain for Gag and Gag-Pol on vRNA; the vRNA functions as a scaffold for the promotion of particle assembly (Muriaux and Darlix, 2010; Muriaux et al., 2001). During virus maturation, the NC domain is released from Gag during the first wave of proteolysis by the viral protease (PR), and then shortened at its C-terminus in two subsequent steps releasing successively the p6 protein and a small 16-residue peptide called p1 (spacer peptide 2). Cleavage of the NC domain from Gag results in the 55 amino acid polypeptide called NCp7, the mature form of NCp that is a protein with highly efficient NA chaperone activity (Cruceanu et al., 2006; Henderson et al., 1992). This ultimate peptide is composed of two small zinc fingers (ZF) connected by a basic linker sequence of few residues and flanked by a flexible N-terminal tail, rich in Arg and Lys.
NCp7 binds nonspecifically to virtually any RNA or ssDNA of sufficient length (5–8 nt.) with an additional specific binding component depending on the nucleotide sequence; a reverse binding polarity occurs with NC binding between RNA and ssDNA (Bazzi et al., 2011; Bourbigot et al., 2008). A residue contained in each ZF (F16 and W37) play a key role in the formation of a hydrophobic plateau which stacks preferentially with unpaired guanosines. The most recent models propose that this contact positions other hydrophobic residues of the ZFs for interactions with the C2′ atoms of the ribose/deoxyribose, while the basic N-terminal tail and the linker regions engage in non-specific electrostatic interactions with NA backbones. A single NCp7 molecule has multiple nucleic acid-binding sites that also allow bridging interactions (Fisher et al., 2006; Fisher et al., 1998): therefore, the binding of multiple units of NCp7 on ss-NA induce strong ss-NA:NCp7 co-aggregation (Le Cam et al., 1998; Mirambeau et al., 2007). The ZFs cooperate with the N-terminal basic region in this highly dynamic macromolecular crowding context to strongly accelerate/stabilize annealing of complementary sequences (matchmaker activity, Rajkowitsch et al., 2007). These NCp7-mediated NA condensing and chaperoning activities are dependent upon the degree of sequence occupancy within the RNP (Darlix et al., 2011). At occupancies that match both optimal activity (chaperone and aggregation) in vitro and the estimated in virio NCp7/nt ratio (1:15 to 1:7), NCp7 binds NA both specifically and non-specifically, driving the most stable and compact nucleoprotein structure. Such high NCp7 occupancy is a major factor for the matchmaking activity that allows for intermolecular RNA interactions to occur upon NCp binding, which accelerates major changes in several structured motifs clustered in the UTR to properly shape the RNA genome. The NA binding and chaperone activities of NCp7 in vitro are also strongly dependent on ionic strength, especially with respect to divalent cations that efficiently compete with NCp for electrostatic binding (Athavale et al., 2010; Grohmann et al., 2008; Lapadat-Tapolsky et al., 1995). It is worth noting that NCp7 is present at an extremely high concentration within virions (close to l00 mM) based on the NCp7:RNA occupied volume (De Marco et al., 2010). The resulting RNP complex in virions, the starting version of the RTC, is injected in the cytoplasm of the newly infected cell. The RNP “cage” results in macromolecular crowding, which tends to increase the processivity of RT, as recently presented (Grohmann et al., 2008). Direct physical interactions between RT and NCp7 have been suggested for quite a while (Cameron et al., 1997; Druillennec et al., 1999; Lener et al., 1998). However, attempts to map such potential sites of interaction have never been successful; it has been speculated that NCp7 maintains RT in close contact with the NA substrate.
II.2 Reverse transcription and RT-NCp7 transactions
Initiation of reverse transcription requires proper annealing of tRNALys,3 with the primer binding site (PBS) site on the vRNA. NCp7 is critical for optimizing productive annealing (Saadatmand and Kleiman, this issue; Sleiman et al., this issue). Then, after synthesis of (-) strong stop DNA (-SSDNA), RT requires the minus-strand transfer event to extend this DNA in order to produce a full-length minus-strand DNA copy of the vRNA genome. This step involves annealing of the newly synthesized antisense TAR DNA domain (cTAR) with its TAR RNA complement at the 3′ ends of vRNA; NCp7 performs one of its major roles in promoting (i.e. chaperoning) this annealing (Piekna-Przybylska et al., 2010). NCp7’s helix destabilizing activity is also required to block non-specific self-priming reactions that compete with the transfer reaction. After minus-strand transfer, RT continues synthesis of the minus-strand DNA. Since the vRNA template is highly structured, the nucleic acid chaperone activity of NCp7 alleviates the resulting pausing in these structured regions, thereby allowing RT to translocate unimpeded along a template. This DNA synthesis is coupled with the subsequent degradation of vRNA by the RNase H domain of RT (Champoux and Schultz, 2009). RNase H is crucial in promoting complete digestion of the reverse transcribed RNA template with its three modes of RNA degradation (3′ DNA end-directed, 5′ RNA end-directed, and internal) that benefit from the numerous available RT copies, suggesting a limited life-time for RNA-DNA hybrid intermediates.
Remarkably, two polypurine tract (PPT) RNA primers derived from the genome, located near the 3′ UTR end and in a central region of the vRNA are resistant to RNase H degradation. With the support of NCp7, they are selectively used as the two primers for (+) strand DNA synthesis (Götte et al., 2010; Hergott et al., this issue; Post et al., 2009). The 3′ PPT primer is extended by RT and the resulting DNA extension product is termed (+) strand strong-stop DNA (+SSDNA). Plus-strand transfer is then mediated by base pairing of the 18-nt PBS region at the 3′-terminus of +SSDNA, termed (+) PBS and the complementary region, termed (-) PBS, at the 3′-terminus of minus-strand DNA, resulting in the formation of a new circular intermediate. Both the (-) and (+) PBS regions are short but stable stem loops. NCp7 stimulates their interaction by accelerating annealing kinetics and switches the reaction towards another pathway involving loop-loop recognition (Bourbigot et al., 2008; Darlix et al., 2011). Additionally, NCp7’s nucleic acid chaperone activity has been shown to be crucial for efficient removal of the tRNALys,3 primer (Levin et al., 2010).
Reverse transcription next proceeds using the cPPT primer to initiate the second (+) strand DNA fragment that terminates at the U5 extremity after a 650 nt-long strand displacement of 3′ PPT primed (+) strand DNA. The 3′PPT primed DNA fragment terminates at the center of the genome after a 100 nt-long strand displacement of the cPPT primed (+) strand DNA fragment generating the central DNA flap, with a stimulatory effect of NCp7 (Hameau et al., 2001). This three branched DNA assembly captures several molecules of NCp7, and may also form unusual DNA structures due to its guanines repeats (Lyonnais et al., 2003; Lyonnais et al., 2002). NCp7 appears also to stimulate, although poorly, the strand displacement required to duplicate the LTR sequence at the end of vDNA (Amacker et al., 1997; Fuentes et al., 1996). NCp7 participates to remove the non PPT vRNA fragments, when RT translocates along the (-) DNA template, which helps in maintaining the fidelity of (+) strand DNA initiation (Jacob and Destefano, 2008; Post et al., 2009). Finally, NCp7 may protect vDNA ends from degradation by nucleases (Buckman et al., 2003).
Of note is that our description of reverse transcription does not take into account the dimeric nature of the vRNA. This property allows a high potential of recombination especially during (-) strand DNA synthesis, by an NCp-facilitated RNA template exchange during DNA elongation (Negroni and Buc, 2001). It has been proposed that only one vDNA molecule is formed at the end of the process. The fate of the residual vRNA that has not been used by RT is unknown, as well as of the other RNA molecules present in the virion.
II.3 A model for NC reshuffling throughout the reverse transcription process
We previously reported the large-scale co-aggregation of NCp7 with RNA and ssDNA in a Mg2+-dependent manner, which could be visualized by TEM (Le Cam et al., 1998; Mirambeau et al., 2006). We observed that these aggregates could be dispersed by adding either parallel DNA G-quadruplexes, which are DNA structures that NCp7 binds with high affinity (Lyonnais et al., 2003), or an excess of T4 gene 32 protein, a prototypical SSB protein with strong cooperative binding. Critically, these competition experiments demonstrated that such aggregates are in equilibrium with the components in the solution. Using a circular ssDNA template hybridized with an oligonucleotide primer, we next reconstituted a reverse transcription assay in the context of ssDNA:NCp7 aggregates in the presence of 6 mM Mg2+ (optimal for large templates) and adding deoxynucleotide triphosphates (dNTPs) to convert the primed ssDNA to circular dsDNA (Mirambeau et al., 2007). In this simple system, disruption of ssDNA-NCp7 aggregates was observed as ssDNA was converted into dsDNA. The HIV-1 RT activity was almost as efficient with ssDNA aggregated with NCp7 as in an aqueous solution of naked ssDNA. Apart from DNA synthesis per se, the presence of Mg2+, necessary for RT function, resulted in a strong and selective reduction of NCp7:dsDNA aggregation. At the end of the assay, some residual NCp7 copies remained aggregated to the last single-stranded segment of the circular DNA, the central DNA flap, which was shown previously to be efficiently produced by RT with the assistance of NCp7 under the same conditions. These results demonstrated that RT under such conditions is able to virtually disrupt NCp7:ss-DNA aggregates by tracking along the minus strand DNA template, releasing dsDNA that is a poor substrate for NCp7 aggregation. We therefore propose a model for RTC remodeling where HIV-1 RT molecules act as molecular motors to dismantle the overall nucleocapsid architecture (Mirambeau et al., 2007), similar to the dismantling of Rec-like nucleofilaments by certain DNA helicases (Veaute et al., 2003).
The optimal magnesium concentration used here for RT with a large template (6 mM) is much higher compared to intracellular free magnesium concentrations (estimated to be 0.5–0.7 mM) but is close to total intracellular concentrations (Romani, 2011). It is also close to the optimum generally required for dsDNA-associated Mg2+-dependent enzymes (Hartwig, 2001). Taking into account the high efficiency of magnesium to stabilize dsDNA in its hydrated B-form, we propose this intrinsic property of dsDNA to be relevant to the in vivo situation providing weak aggregative dsDNA-NCp7 contacts within a crowded environment.
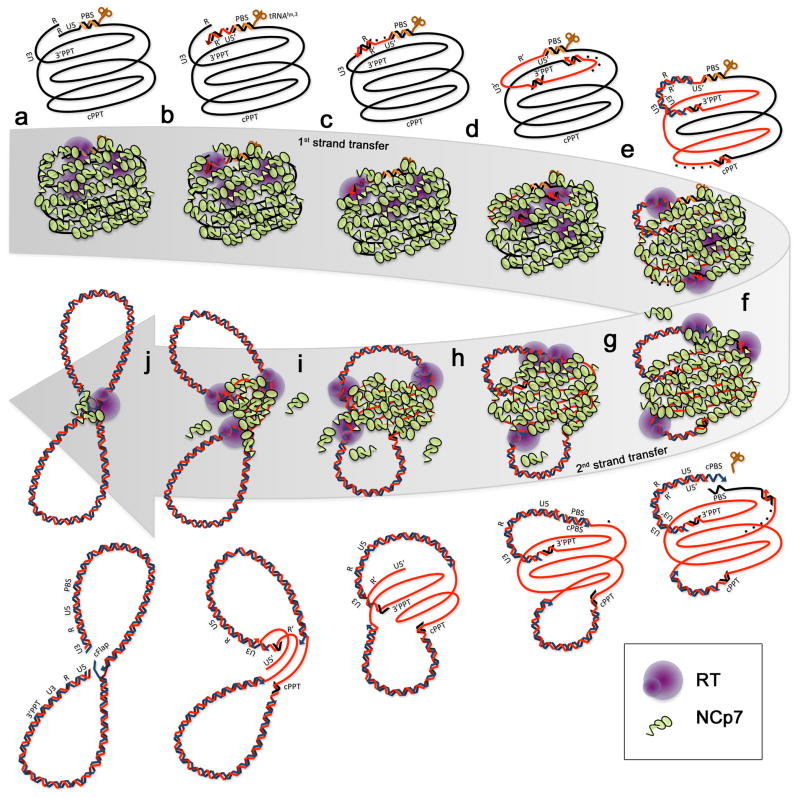
Fig. 1 presents our proposed model in more detail, by focusing schematically on the successive interactions between the NCp7 population and the various forms of viral NAs encountered during reverse transcription. The upper and lower sections focus on the vDNA being synthesized in the reverse transcription reaction itself. The middle portion, contained in the large curved grey arrow attempts to illustrate the remodeling of the RNP architecture, showing changes in the NCp7 occupancy on the NA. Viral ss-NAs are represented as being folded into several large loops and co-axial strands bridged together with NCp7 in order to sketch some possible modes of ss-NA aggregation. This arrangement is not necessarily supported by experimental evidence but provides a useful way to represent large-scale association of NCp with ss-NA, taking into account NA backbone repulsion that is reduced upon NCp binding and NA bridging. Ss-NAs are seen “extended” but are obviously highly structured molecules with numerous stem and loops (Watts et al., 2009). The scheme also postulates some continuous contacts between NCp7 and RT to illustrate our suggestion of macromolecular machinery centered on viral ss-NAs. As we hypothesized in a previous animated model, the timing of the reaction proceeded with a regular progression of RTs along their templates suggesting that (+) strand DNA central- and LTR-termination occurs within the same time frame (Mirambeau et al., 2007).
Figure 1. Remodeling of HIV-1 nucleocapsid complex upon reverse transcription.
The upper and lower parts focus on the vDNA being synthesized in the reverse transcription reaction itself. The middle portion contained in the large curved, grey arrow is a schematic representation of the changes in the NCp7 occupancy on the NA on the same molecules. For simplicity, the model focuses on the progression of one unique RNA molecule being converted to a ds-vDNA. tRNAlys,3 is brown, vRNA is black, (-)DNA is red, (+) DNA is blue. For clarity, NCp7 molecules are represented as exclusively specific to ss-NA to better emphasize their concentration on vRNA and (-)ss-DNA versus dsDNA. Attention has been also focused on the following points: double helices and the three primers (tRNALys,3, 3′PPT and cPPT) are oversized for clarity; the two NA ends are in close proximity throughout reverse transcription, in order to show a continuous process at the time of the strand transfers. RNA fragments released by the RNaseH activity of RT are shown as dashed points behind RT along the elongating (-) strand DNA template. (a) reverse transcription initiation from the tRNAlys,3 primer. (b) synthesis of (-) SSDNA. (c) minus strand transfer and RT elongation. (d) (-) strand DNA synthesis, with RNAse-H activity releasing the 3′PPT. (e) Release of the cPPT upon (-) strand DNA synthesis and (+) SSDNA synthesis from the 3′PPT. (f) Removal of the tRNALys,3 upon (+) SSDNA synthesis and (+) strand synthesis starting from the cPPT. (g) (+) strand transfer by base pairing of the PBS and cPBS sequences, elongation of the (+) strand DNA. (h-i) Synthesis of (+) strand DNA, with strand displacement of the U5 extremity. (j) Termination of (+) strand synthesis with LTR duplication and strand displacement to generate the central DNA Flap (cFlap).
From a to e, the model proposes that the RNP complex switches from RNA-based (nucleocapsid) to ssDNA-based complexes (RTC) without significant changes in shape and assembly: given the high dynamics of NCp7 binding, RT probably frontally ejects the NCp7 proteins loaded on their RNA template whereas unbound NCp7 has plenty of opportunities to re-associate with the newly synthesized (-) DNA strand, or remaining vRNA that is in close proximity. The reverse NCp7 binding polarity between DNA and RNA fits well into our proposed model (Bazzi et al., 2011). The first step of (-) strand DNA synthesis is coupled with a rather efficient RNase H digestion of the RNA template within RNA-DNA heteroduplexes (Champoux and Schultz, 2009). This suggests that networks of aligned NCp7 bound to vRNA should easily rebind to (-) DNA without significant rearrangement or orientation due to the antiparallelism between the vRNA and (-) DNA strands. This should allow NCp7 re-aggregation along the (-) DNA immediately following RT translocation, thus maintaining a compact NCp7:(-) DNA:vRNA complex. Degradation of vRNA into small fragments and the behavior of NCp7 prior to formation of RNA-DNA heteroduplexes require further study. Nevertheless, the nucleoprotein cage formed between NCp7 and ssDNA remains nearly unchanged: such a model helps to maximize nucleoprotein complex condensation and obligatorily allocate the chaperone activities combined with the macromolecular crowding effects where they are needed, up to the second strand-transfer.
Next, from f to j (Fig. 1), the model shows the release of the newly synthesized dsDNA from the initial RNP with a progressive reduction in the size of the nucleoprotein complex upon ssDNA template shortening. This process results in concentration of NCp7 and RT on ss-template regions, which in turn enhances the aggregative properties of NCp7 (by increasing the protein to nt. ratio) with (-) strand ssDNA. Concomitantly, nascent dsDNA helices extrude progressively, an effect that could be reinforced by the profound changes in rigidity and bending properties of dsDNA compared to that of ss-NAs, but also the consecutive binding of other dsDNA specific proteins, as we show next with Vpr. To visually reinforce our hypothesis of molecular machinery, we represent RT trapped in the diminishing NCp7:(-)ssDNA complex so that it appears virtually “immobile” while the dsDNA chains extrude from the complex. We propose that this phenomenon accompanies an important loss of NCp7 at a moment where only a limited pool of NCp7 (no more than one hundred molecules) is required for the final strand-displacement steps that complete reverse transcription. Consequently, the “flip-flop” flap strand(s) would be the last locus to stably retain NCp7 at the RT central termination step resulting in the ultimate residual nucleocapsid complex that we have positioned in Fig. 1 at the crossover of the figure eight-folded vDNA, close to the two DNA ends. We will discuss at the end of the next section the advantage of such a topological arrangement in sequestering the IN pool. Another obvious advantage of such a small complex centered on the last single-stranded parts of the completed vDNA is to enhance protection from nuclease degradation.
II.4 The special case of NC H23C and H44C ZF mutants
HIV-1 reverse transcription is a highly regulated process that occurs only under specific conditions. Typically, there is very little vDNA contained in HIV-1 particles that have just been released from producer cells; 1:800 to 1:6000 for R-U5 vDNA:genomes for HIV-1 or MLV (Lori et al., 1992; Thomas et al., 2008; Trono, 1992; Zhang et al., 1993). However several mutant viruses were found to contain very high levels of intravirion DNA due to premature initiation of reverse transcription and not surprisingly, these viruses are replication defective. One would predict that improper timing of reverse transcription initiation would be deleterious to viral replication. Situations where virions are found to contain high levels (up to 1 R-U5 vDNA per genome) have been observed when certain regions of the Gag protein have been altered by site directed mutagenesis. This phenomenon, called premature reverse transcription, or also termed late reverse transcription (due to its timing during the late – assembly – stage of the virus life cycle) has been observed in NCH23C- NCH44C-, NCp ZF deletion- and N-terminal basic amino acid region-mutants of HIV-1 (Didierlaurent et al., 2008; Houzet et al., 2008; Thomas et al., 2008). In addition, the budding-defective HIV-1 mutant p6PTAP! LIRL mutant (Huang et al., 1995) also contains elevated levels of vDNA (Thomas et al., 2008). It is presumed that virions with high levels of vDNA accumulate while the mutant virions are still attached to producer cells (Didierlaurent et al., 2008; Houzet et al., 2008; Thomas et al., 2008). Intravirion DNA is not a result of natural endogenous reverse transcription since the levels are maximal at the start of incubations of ZF deletion mutants with dNTPs (Houzet et al., 2008). The levels of R-U5 are maximal at the earliest time points in an infection of HOS cells with the NCH23C and NCH44C mutants (Thomas et al., 2006). Additionally, R-U5 levels are maximal at the onset of endogenous reverse transcription reactions initiated by the addition of non-ionic detergent, dNTPs and Mg2+ whereas maximal levels with wild-type HIV-1 are obtained at later times (Thomas et al., 2008).
As mentioned above, the budding defective HIV-1 mutant (p6PTAP! LIRL) also contains high levels of intravirion DNA (1 R-U5 per 10 genomes) suggesting that a delay in virion release could be a cause for premature reverse transcription (Thomas et al., 2008). In support of this are recent observations that certain NCp regions interact with ESCRT components as does the PTAP motif of p6, thus NCp also appears to be involved in the regulation of virion release (Dussupt et al., 2009; Popov et al., 2008). Additionally, mutation of the N-terminal basic amino acid region of NCp disrupts budding to a certain extent (Dussupt et al., 2010). Thus altering NCp in ways that result in viruses with high levels of intravirion DNA, also disrupts interactions that can result in budding defects/delays. Consequently, if budding is delayed, the collection of all the components necessary for the initiation of reverse transcription (mature NCp7, RT and dNTPs) have time to accumulate and interact before separation from the producer cell.
The accumulation of intravirion DNA in NCp-mutant HIV-1 was certainly thought to be the direct cause of replication defects observed in these viruses. However, at least with the NCH23C and NCH44C mutants, this conclusion is not the case. Thomas and co-workers were able to prevent premature reverse transcription in these mutants by treating producer cultures with high levels of a combination of two RT inhibitors, resulting in particles that had very low levels of intravirion DNA, even below that found in wild-type HIV-1 (Thomas et al., 2011). These treatments result in the very effective maintenance of infectivity and replication of wild-type HIV-1 once the inhibitors were removed (Thomas et al., 2011).
Endogenous reverse transcription (ERT) was examined in these RT inhibitor-treated NCp7 mutants by the addition of non-ionic detergent, dNTPs and Mg2+, and surprisingly reverse transcription intermediate levels and kinetics could be restored to WT levels. When inhibitor-treated NC-mutant virions were used to infect cells in culture, the initial kinetics could be restored to wild-type levels with the maximal vDNA intermediate levels occurring several hours into the infection, rather than at the onset, which occurred with untreated NC-mutant infections. However, at later times vDNA intermediate levels dropped below wild-type levels indicating that there are still issues with vDNA stability or synthesis in NC-mutant infections (Buckman et al., 2003; Thomas et al., 2006; Thomas et al., 2011). Thus key observations that refute the conclusion that premature reverse transcription is the predominant factor accounting for the replication defects observed in these RT inhibitor-treated NCp7-mutant viruses are that i) single-round TZM-bl cell infectivity assays of RT inhibitor-treated mutants showed absolutely no increase in titer whereas RT inhibitor-treated wild-type virus titer is maintained, ii) there is no rescue of integration upon RT inhibitor treatment (wild-type integration is not significantly affected by the RT inhibitor treatment; J. A. Thomas, personal communication), and iii) there remain issues with vDNA stability/synthesis in NCp mutant infections as mentioned above.
There are undoubtedly other factors associated with these NCp mutant viruses that contribute to their replication defectiveness. These could include a more rapid dissociation of the mutant NC proteins from the RTC, which could account for the reduction in vDNA synthesis. The binding constants of these mutant NC proteins for RNA is reduced (Déméné et al., 1994; Urbaneja et al., 1999; Williams et al., 2002); in an infection, if the genomes become prematurely unprotected due to more rapid loss of NCp7, they would be susceptible to nucleases, reducing the levels of vDNA that survive prior to integration. Another issue could be that, as NCp7 dissociates from RTCs during conversion to PICs, the effective concentration of NCp7 is reduced, and it has been shown that the nucleic acid chaperone activities of these NCp mutant proteins are reduced compared to wild-type NCp7 (Bampi et al., 2004; Guo et al., 2002; Lee et al., 2003; Levin et al., 2005; Thomas and Gorelick, 2008; Williams et al., 2002). Thus NA chaperoning by the remaining NCp7 that is attached to the evolving RTC/PICs is not as effective and may result in incomplete vDNA synthesis in the cell. This is in contrast to what is observed by ERT with RT inhibitor-treated NCp-mutant virions as reverse transcription defects can be rescued under these conditions, but can be explained since NCp is maintained at high concentrations within the viral membrane, shifting the equilibrium toward completion of reverse transcription and masking any apparent nucleic acid chaperone defects (Thomas et al., 2011). Additionally, cellular nucleases are excluded under ERT conditions. Part of vDNA stability can be attributed to the formation of the provirus, thus if the RTC has not been completely converted to a PIC by the time it reaches the nucleus, it will not be functional for the integration event. Certainly, there may be problems with the proper architectural conversion of the nucleocapsid to form the functional PIC structure as has been presented in this and other studies (Mirambeau et al., 2007; Mirambeau et al., 2010). More research is certainly needed to further define the properties associated with these NCp mutants that contribute to their replication defectiveness, which could shed light on additional antiviral interventions.
III. Architectural plasticity: From HIV-1 virions to Pre Integration Complexes
III.1 A brief description of HIV-1 core’s assembly and maturation
Focusing on HIV-1 particle assembly, immature particles are comprised of Pr55Gag and Gag-Pol polyprotein precursors that cooperatively assemble with vRNA at their C-terminal end (NC domain), bind to membranes of either internal vesicles or the plasma membrane at their N-terminal end (Matrix, MA) and further associate through capsid-capsid contacts from their central domain (Capsid, CA). Membrane microdomains directly promote the self-assembly by clustering the individual precursors. Immature viruses are composed of a Gag and Gag-Pol binding along the genome lattice via NC domain contacts, with the Pol moieties of Gag-Pol being positioned at the center of the complex. These particles are characterized by their high stability (Briggs and Kräusslich, 2011).
HIV-1 next becomes infectious through a maturation process during which the Gag/Gag-Pol bound to the vRNA lattice is sequentially cleaved into its constituent components by PR, which are autoactivated initially from Gag-Pol:Gag-Pol dimers during or just after budding. Maturation results in a profound change in virus architecture in an overall destabilization process whereby the RNP complex comprised of vRNA, the pool of NCp, the RT and IN enzymes (processed from Gag-Pol) folds and condenses within the capsid, shaping the reverse transcription machine into an active configuration (Ganser-Pornillos et al., 2012).
The viral core contains all of the elements required for completion of reverse transcription, which can be initiated within particles by supplementing Mg2+ and dNTPs (Zhang et al., 1996) as noted before. An estimated 2000 NCp7 molecules coating the vRNA dimer are found within the nucleocapsid complex, together with RT, IN, Vpr, Nef, Vif, cellular RNAs (tRNAlys,3, ribosomal RNAs, 7S and other small RNAs) as well as cellular proteins. Proteolysis/maturation is thought to be sequential and initially separates the RNP complex from the MA-CA shell, which in turn is cleaved into the mature MA and CA proteins. The CA proteins self-assemble and form the conical protein shell of the capsid. The nascent nucleocapsid complex condenses and is enclosed within this capsid. A pool of about 25–125 RT and IN proteins is also generated from the co-assembled Gag-Pol. At least one RT molecule forms a stable ternary RNP initiation complex with vRNA and the tRNAlys,3 primer. IN, intrinsically a nucleic acid binding protein, has been shown to stably associate with the HIV-1 RNP complex and interacts with RT, which is necessary for the initiation of reverse transcription (Hehl et al., 2004; Zhu et al., 2004). Essential for its function during integration, the IN oligomerization state within the HIV-1 core is not known. Vpr is also present in the RNP complex, at a significant copy number (1 for 7 Gag molecules; (Müller et al., 2000)), whereas p6, the processed product of Vpr’s initial viral recruiter, is not (Welker et al., 2000). More generally, a realistic macromolecular model of the core still needs to be determined.
III.2 A puzzling route for reverse transcription complexes leading to preintegration complexes
The general process of HIV-1 reverse transcription occurs in the cytoplasm of the infected cell after virus-cell fusion and injection of the HIV-1 core in the cytoplasm, but its localization remains obscure, as well as its coordination with capsid uncoating. Even so, both processes are shown to be interconnected (Arhel, 2010; Dismuke and Aiken, 2006; Goff, 2001; Hulme et al., 2011; Wacharapornin et al., 2007). Trafficking of complexes has been observed along the cytoskeleton with the involvement of both actin and microtubules. For example, observation of enzymatically active RTCs by Vpr-associated fluorescence and correlation microscopy revealed that they were associated with HIV-1 CA proteins and tracked along cellular microtubules (McDonald et al., 2002). Similarly, using fluorescently labeled IN, a combination of slow and fast movements of RTCs/PICs have been highlighted within the cytoplasm of infected cells (Arhel et al., 2006). Again, any coordination between reverse transcription and core trafficking is still unclear at this time. HIV-1’s journey to the nucleus may also be shorter than previously thought since viral entry through endocytosis has been proposed recently as a normal pathway to internalize the HIV-1 core after endosomal migration within the infected cell (Miyauchi et al., 2009). Whatever the constraints for the spatiotemporal coordination that allows the core/RTC/PIC to dock with the nuclear envelope, maturation of HIV-1 RTCs to PICs is preferentially associated with a prerequisite for capsid uncoating and the PIC is located in close proximity with a nuclear pore complex. This critical step allows infection of metabolically active non-dividing cells, a conserved feature of lentiviruses, unique among orthoretroviruses and of crucial importance in HIV-1 tropism and AIDS acquisition (Weinderg et al., 1991).
Completion of HIV-1 reverse transcription leads to a linear dsDNA that includes the central DNA flap and proper DNA ends for productive IN binding. This DNA product (≈9.8 kbp) also serves as the scaffold for the PIC within a new nucleoproteic architecture competent for integration and additionally capable of translocation through the nuclear pore of interphasic cells (Katz et al., 2003). This involves the exploitation of nuclear localization signals from components of the PIC, found on IN, Vpr and MA proteins (Bukrinsky, 2004). On the other hand, the CA protein has been shown to promote the selective capture of several host proteins (nucleoporin 153, nucleoporin 358 and transportin 3), in order for the PIC to couple capsid uncoating and active translocation through the nuclear pore followed by routing to specific chromatin domains (Matreyek and Engelman, 2011; Ocwieja et al., 2011; Schaller et al., 2011; Zhou et al., 2011). Synthesis of the central DNA flap has also been shown to be involved in coupling capsid uncoating and PIC nuclear import (Ao et al., 2004; Zennou et al., 2000), with two alternate hypotheses: i) a direct role for the flap structure or ii) more rapid dsDNA synthesis due to the corresponding (+) strand DNA double initiation-termination strategy. Despite these observations, mechanisms for capsid uncoating as well as for PIC translocation are still not clear. The vDNA has to be maintained with the remaining IN molecules in a competent assembly for DNA integration (Bowerman et al., 1989; Iordanskiy et al., 2006). Recruitment of cellular proteins, for example the Barrier-to-autointegration factor, BAF (Lin and Engelman, 2003; Suzuki and Craigie, 2007), or the transcriptional activator LEDGF 75 within the cytoplasm, has also been shown critical in preparing for intranuclear routing (Ciuffi et al., 2005; Emiliani et al., 2005; Llano et al., 2004). Several other cellular proteins have been proposed to be included in the PIC, but require more in-depth characterization (Warrilow et al., 2009).
Extraction procedures to isolate macromolecular complexes that are heterogeneous by nature, in a metastable posture and engaged in a large set of contacts with intracellular networks from the host cell are strong impediments to obtain a clear picture of the PIC maturation process. However, some RTCs that lead to a nearly complete reverse transcription event (e.g. and could thus be considered as PICs) have been analyzed by diverse biochemical approaches. Viral proteins essentially observed include Vpr and IN, whereas NCp7 and RT were nearly undetectable (Fassati and Goff, 2001; Karageorgos et al., 1993; Miller et al., 1997). TEM visualization by negative staining of purified RTC/PIC 3–4h post-infection revealed filamentous DNA-protein assemblies of 6 nm in width and few hundred nm in length covered by Vpr and IN (Nermut and Fassati, 2003). Small cellular DNA-binding proteins were also proposed to cover the vDNA, but were not further characterized (Nermut and Fassati, 2003). These studies also emphasized a strong morphological change during the maturation from the core/RTC to the PIC (Warrilow and Harrich, 2007; Warrilow et al., 2009). Loss of the capsid shell is largely complete at this juncture. Given a length of 3.3μm for the extended 9.8 kbp-long HIV-1 DNA suggests that the PIC architecture is presumably a closely packed structure, which is coincident with an increase in the nuclease resistance of the cDNA product (Khiytani and Dimmock, 2002). Interestingly, HIV-1 replication can be reduced before integration by intracellular expression of single-chain variable antibody fragments against RT and IN, indicating that these proteins are accessible in the complex (Levy-Mintz et al., 1996; Shaheen et al., 1996).
With the PIC being dedicated to carrying out the integration step of the completed vDNA, it is supposed that most of the 60–240 IN monomers remain associated with the vDNA. At a minimum, an IN tetramer must bind to a pair of DNA ends that leads to a pre-activated “intasome”, the minimal HIV-1 DNA/protein complex that must enter into the nucleus, routed by appropriated cellular partners through the pore and next towards advantageous chromatin regions (Delelis et al., 2008; Li et al., 2012). Importin α3 is the last known IN partner to be involved in PIC nuclear import (Jayappa et al., 2011).
III.3 Vpr as the most abundant protein component in the Pre-Integration Complexes
The multifunctional Vpr protein is required for efficient viral replication in non-dividing cells such as macrophages, and promotes, to some extent, viral replication in proliferating CD4+ T cells. The specific activities of Vpr include modulation of the fidelity of reverse transcription, transactivation of the HIV-1 LTR promoter, induction of G2 cell cycle arrest and cell death via apoptosis. Vpr may also modulate the HIV-1 mutation rate through its association with the uracil-DNA-glycosylase UNG2 and is associated with the DNA-damage response pathway. Recently, a role as a major mediator in early replication has been highlighted by a genome-wide siRNA screen that linked Vpr to a number of host-cell proteins important for reverse transcription. Another important function of Vpr is to counteract host innate, cell- or humoral-mediated immune responses. For recent reviews on Vpr, see (Fritz et al., 2010; Romani and Engelbrecht, 2009; Zhao et al., 2011).
An estimated 275 to 700 Vpr molecules are specifically packaged in the virion by an interaction with the p6 region of Gag (Müller et al., 2000; Paxton et al., 1993). While p6 appears to be released from the core during viral maturation, Vpr is thought to remain associated with the nucleocapsid complex (Accola et al., 2000). It is able to interact at least with NCp7 and RNA (De Rocquigny et al., 1997; Zhang et al., 1998), whereas peptides of Vpr have been shown to modulate the activities of RT and IN (Bischerour et al., 2003). It is not known whether the presence of Vpr plays a direct role during reverse transcription, but several pieces of evidence show that Vpr is maintained throughout core remodeling, including the PIC: Vpr contains a non-canonical nuclear localization sequence and possesses the ability to shuttle between the cytoplasm and the nucleus (Heinzinger et al., 1994), co-localizes with HIV-1 RTCs in cells within 4–16 h after the onset of viral infection (Fassati and Goff, 2001) and has been suggested to be directly implicated in the nuclear import of the PIC. Here, two explanations have been proposed: an importin-dependent pathway (Takeda et al., 2011) or direct interactions with the nuclear pore complex including the hCG1 nucleoporin (Jacquot et al., 2007; Le Rouzic et al., 2002). The recent characterization of a CyclophilinA (CypA)-binding site in the Vpr N- and C-terminal domains might be taken into consideration with the CypA-binding site of the HIV-1 CA protein in the localization of the PIC to the nucleoporins (Schaller et al., 2011; Solbak et al., 2010; Solbak et al., 2011; Zander et al., 2003)
Vpr is a small basic 96 amino acid protein with a three-dimensional structure composed of three amphipathic α-helices mutually oriented to form a central hydrophobic core (Morellet et al., 2003). This promotes the formation of Vpr oligomers in HeLa cells and their targeting to the nuclear envelope. Assembly into multimers, from dimers to decamers also exposes the hydrophilic faces of the α-helices to present the interaction domains of Vpr to their partners (for review: (Morellet et al., 2009). As noted above, Vpr is also a NA-binding protein (Bischerour et al., 2003; De Rocquigny et al., 2000; De Rocquigny et al., 1992; Zhang et al., 1998). This function is located in a small region rich in basic residues in the C-terminal part of the α-helix III and is modulated by the N-terminal part of the protein (De Rocquigny et al., 2000). Thus, complete Vpr and synthetic peptides containing a portion of the α-helix III have been shown to interact directly with RNA and various dsDNA molecules, without detectable sequence or structure preference, although the affinity of Vpr should be higher for RNA than for DNA (in the μmolar range). Vpr binding induces NA aggregation and condensation, whereas discrete complexes have only been observed by gel electrophoresis in one case, between full-length Vpr and a 492bp dsDNA fragment (De Rocquigny et al., 2000). NA binding properties of Vpr were also enhanced by the presence of NCp7 (De Rocquigny et al., 1997; Li et al., 1996). Observed by gel shift assays and dark-field TEM, Vpr52–96 induced the compaction of a 7kb DNA plasmid, resulting in formation of aggregates with irregular sizes and shapes, lamellar structures and rods (Kichler et al., 2000). Vpr77–96 induced an ordered and reversible condensation without collapse of the DNA molecules, suggesting possible implications of the Vpr oligomerization state in its interactions with NA. Indeed, the region 60–80 has been shown to be necessary for both Vpr dimerization/oligomerization and nucleic acid binding. This could explain the apparent cooperative binding onto DNA and the aggregates that form. Interestingly, Vpr52-96 can act as an excellent carrier for DNA transfection of mammalian cells with an efficiency equivalent to the best transfection reagents (Coeytaux et al., 2003; Kichler et al., 2000). These NA binding activities have not yet been shown to be associated with any particular function, but unequivocally suggest Vpr to be a DNA partner within the PIC. This prompted us to carefully re-examine this putative partnership in the context of RTC to PIC remodeling discussed in the previous section.
IV. Experimental evidence for a DNA architectural function for Vpr within the Pre-Integration Complexes
IV.1 DNA bridging by Vpr
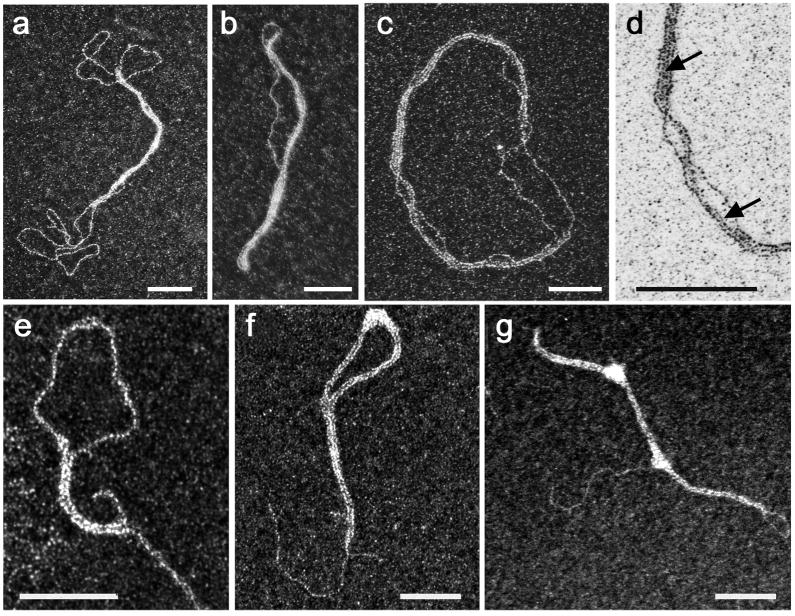
Taking into account the number of Vpr copies in the virion, the maximal Vpr/dsDNA ratio for the PIC is in the range of one Vpr for 20 bp (Briggs et al., 2004; Müller et al., 2000). This ratio is much lower than those used in the studies describing Vpr-NA interactions. We thus addressed the capability of Vpr to form complexes with diverse dsDNA molecules, either circular plasmids or linear chimeric fragments containing HIV-1 sequences (e.g. parts of the LTRs and the cPPT-central termination site (CTS) region) using a Vpr:DNA ratio of one for 10 to 20 bp, with the same experimental setup used previously to probe NCp7 assemblies with DNA. Buffer composition corresponded to those used in our RT in vitro circular ssDNA assay, as described previously (Mirambeau et al., 2007). Agarose gel electrophoresis was also used for this purpose but we failed to detect any relevant and reproducible binding events, except a slight shift of the DNA band when Vpr was incubated with negatively supercoiled plasmids at low ionic strength (not shown). In contrast, positive staining and dark-field TEM imaging of DNA incubated with these limiting amounts of Vpr unambiguously revealed very similar Vpr:DNA architectures with all the DNA molecules tested; a few examples are presented in Fig. 2. (a–d) that show TEM images of relaxed (form Ir) pTZ18R plasmids DNA (2.86 kbp) incubated with Vpr (1:10 – 1:20 Vpr:bp ratio). They revealed a new property of Vpr: bridging two or more DNA helices into synaptic and stretched nucleofilaments. Rather than being covered by a continuous coat of proteins, the DNA helices appear bridged by a smooth-bright granular layer of proteins (arrows in d), associated in co-axial arrangements suggesting a long-range but labile “zipper”-like structure, where a strand of DNA can sometimes detach from the complex. This arrangement leads to intramolecular folding of the plasmid into “double-lasso” structures (Fig. 2a, see also Fig. 6), where two strands associate in a synaptic filament surrounded by two unbound large loops. Growth of the nucleofilament up to the full length of the plasmid leads to its compaction by two-fold, with formation of lamellar structures (b–c). These structures are imperfect and often present one DNA strand unzipped from the nucleofilament (d). Taking into account the technical constraints associated with DNA fixation and staining required for TEM imaging, these defects suggest limited stability of the DNA bridges. Formation of the nucleofilaments appears to be significantly cooperative, as we often observed a mixture of naked and Vpr-bound plasmids with synaptic contacts along 500 bp or more (see also Fig. 6). Association of two DNA helices induces a local stiffening of the complex, but the nucleofilaments appear dynamic enough to support significant bending and circularization (c). Under identical conditions, linear DNA fragments incubated with Vpr formed complexes with the same arrangements (e–g). We found linear molecules folded in two, with partial formation of nucleofilaments presenting naked terminal segments and loops (e), but also complexes with nucleofilament bent into loops (f). Compaction also induced formation of bright spots that appear to be accumulations of proteins, often associated with bending of the nucleofilaments (f–g).
Figure 2. Complexes formed between Vpr and dsDNA observed by TEM in dark field mode.
Reaction were carried out for 15 min. at 37 °C in 25mM HEPES (pH7.5), 50mM sodium acetate, 6mM magnesium acetate. a-d) Vpr:dsDNA nucleofilaments formed after incubation of pTZ18R DNA (2.5nM) and Vpr (1μM). (a) typical double-lasso structures bearing a synaptic joint; b) wire-like structure upon full coverage; c) plasmid spread by Vpr. d) is a zoom of e) with inverted contrast to better see the protein shell on DNA strands (arrows). e-g) Complexes obtained with Vpr and 1.68kbp linear dsDNA, for respectively a bp/Vpr ratio of 16, 12 and 10. The scale bars represent 80 nm.
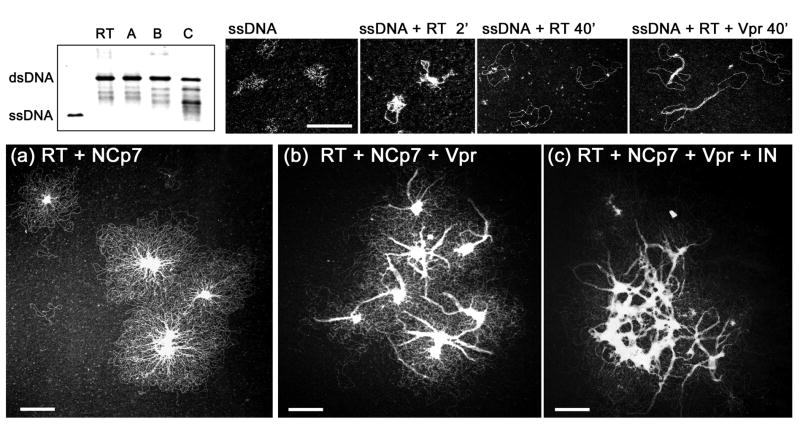
Figure 6. Progression of dsDNA synthesis by HIV-1 RT on circular ssDNA templates in the presence of NCp7, Vpr and IN.
The polymerase reaction was carried out in a RT buffer containing 50 mM Tris-acetate pH 7.8, 50 mM sodium acetate, 6 mM magnesium acetate and 0.5 mM DTT at 37°C for 40 min. (see (Hameau et al., 2001) and (Mirambeau et al., 2007) for more details). Upper left panel: comparison by agarose gel electrophoresis of DNA synthesis by RT in the absence (labeled RT) and presence of NCp7 (A), NCp7 and Vpr (B), NCp7, Vpr and IN (C). Concentrations of primed-ssDNA circles, RT and NCp7 were, respectively, 5 nM, 100nM, 3.4 μM. In B and C, the Vpr concentration was 2 μM. In C, the IN concentration was 0.5 μM. ssDNA and proteins were premixed for 4 min. at 37°C, followed by incubation with RT for 2 min. before addition of dNTPs (100 μM each) to start the reaction. DNA products were heated to 70°C for 10 min. in the presence of 1% (w/v) SDS and 20 mM EDTA before gel electrophoresis on a 1% agarose gel in 0.5x TBE. Upper right 4 panels: TEM images of ssDNA control followed by dsDNA produced by RT alone (at either 2 or 40 min.) or with Vpr (same concentrations as in B) analyzed after 40 min. at 37°C. Lower panels, a-c) RT polymerization observed by TEM at 40 min. under polymerization conditions described for the agarose gel analysis described above. 5 μl-aliquots of the polymerization assay were diluted 5-fold in the reaction buffer without DTT and next deposited onto the EM grid. The scale bars correspond to 250 nm.
A more detailed study on theses complexes between Vpr and DNA is under investigation to understand the underlying mechanism of the architectures observed here for the first time, to our knowledge. They are however strikingly similar to the nucleofilaments formed between DNA and the family of histone-like nucleoid structural proteins such as E.Coli H-NS or StpA (Dame et al., 2006; Lim et al., 2011; Liu et al., 2010). These structures are also comparable to those formed between DNA and the eukaryotic chromatin remodeling factor, ISW1a (De Cian et al., 2012). These stable, bridged oligomers formed in solution led us to speculate that Vpr may assemble with vDNA in an equivalent manner leading to a yet undiscovered function as a vDNA-condensing agent within the PIC. The quasi rigid nucleoprotein wires observed in Fig. 2b (see also Fig. 3, 5 and 6) appear particularly interesting in the context of nuclear import but more detailed analyses are required to make firm conclusions regarding the function of such wire-like structures for the PIC. We note however that the nucleofilaments share the same width as those observed from RTC/PIC extracted from cells (less than 10 nm). Such an assembly could easily assist with vDNA folding, compaction and transient protection of dsDNA. Vpr coating could also connect vDNA to the nuclear envelope and the related factors (like Nucleoporin 358 and its CypA-domain) in order to facilitate the entry and to direct the progressive translocation of the vDNA through the nuclear pore complex by the way of these synaptic nucleofilaments. Of importance are both the apparent limited stability and the cooperative assembly of these DNA:Vpr bridges, cooperativity providing a key benefit due to the reduced amounts of Vpr copies.
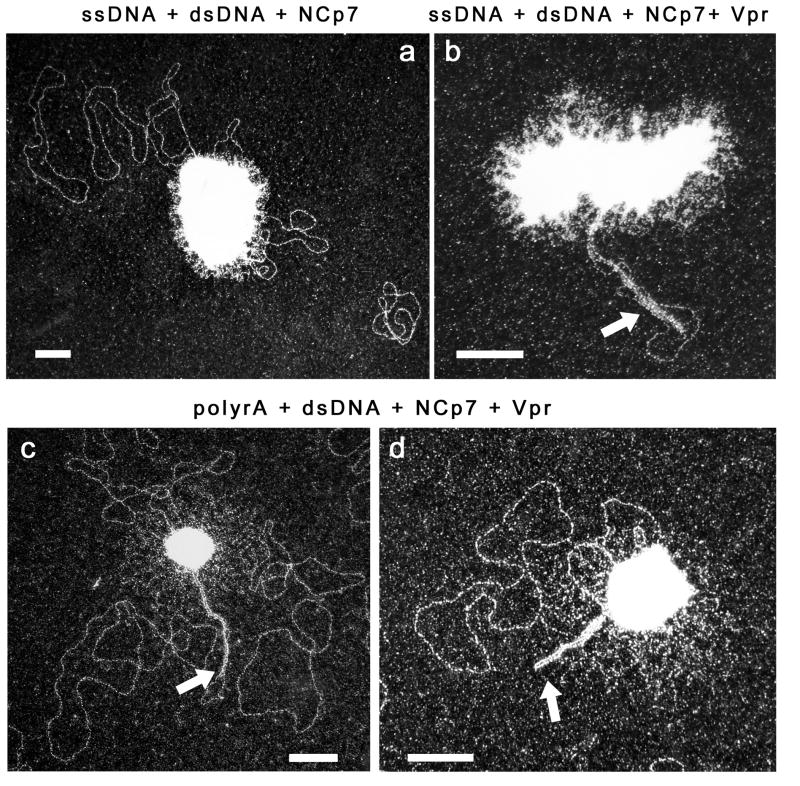
Figure 3. TEM visualization of RNA-dsDNA or ssDNA-dsDNA mixtures, incubated with NCp7 or with NCp7 and Vpr.
Reaction were carried out for 15 min. at 37 °C in 25mM HEPES (pH7.5), 50mM sodium acetate, 6mM magnesium acetate. (a) Incubation of 2.5 nM dsDNA (pTZ18R) and 5 nM ssDNA (circular, 3.3 kbp) with 3.4μM NCp7 (nucleotide/NCp7 ratio: 10). (b) Incubation of 3.4μM NCp7 plus 3μM Vpr with 2.5 nM dsDNA and 5nM ssDNA (nucleotide/NCp7 and /Vpr ratio: 10). (c-d) Incubation 30min. at 37°C of 3.4μM NCp7 plus 3μM Vpr with 2.5 nM dsDNA and 5nM polyrA (nucleotide/NCp7 and/Vpr ratio: 10). Complexes were diluted 5-fold in the reaction buffer before deposition on the TEM grid. Arrows point Vpr:dsDNA nucleofilaments. The scale bars represent 80 nm.
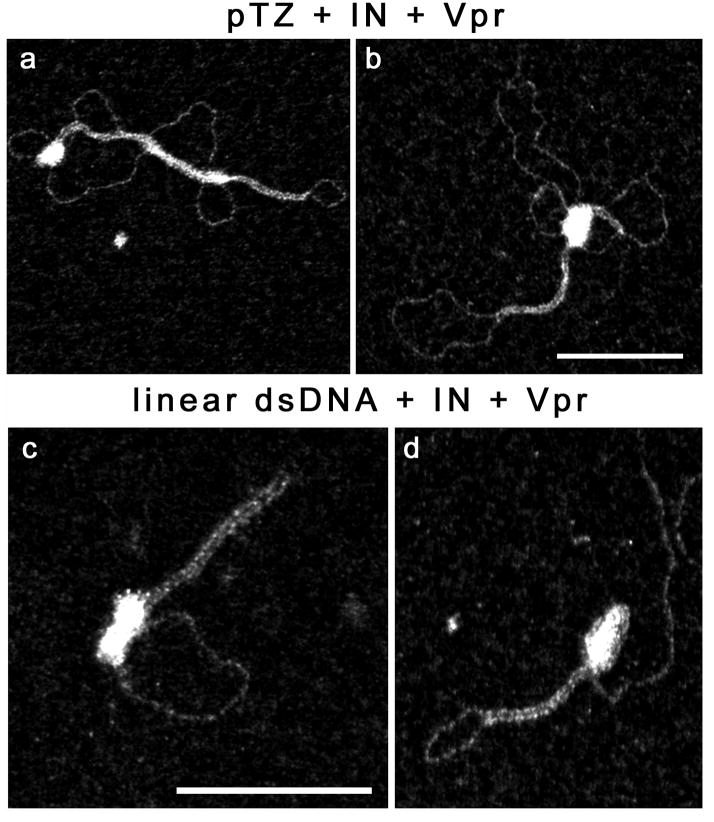
Figure 5. TEM visualization of complexes formed between Vpr, IN and dsDNA.
Reaction were carried out for 40 min. at 37 °C in 25mM HEPES (pH7.5), 50mM sodium acetate, 6mM magnesium acetate. (a-b) Incubation of 2 nM dsDNA (pTZ18R) with 500nM Vpr (bp/Vpr ratio:12) and 100nM IN. (c-d) Incubation of 2nM dsDNA (linear, 1.68kbp) with 250nM Vpr (bp/Vpr ratio:15) and 40nM IN. The scale bars represent 100nm and are the same sizes for a-b and c-d.
IV.2 Cross-talk between IN, Vpr, NCp7 and Nucleic Acids
Due to these remarkable properties of Vpr binding to dsDNA, we next addressed whether NCp7 and Vpr could co-assemble on different NA intermediates of reverse transcription. In Fig. 3, a mixture of ss-NA (ssDNA circles in a-b, RNA in c-d) and dsDNA circles was incubated with NCp7 and Vpr under conditions previously used with NCp7 alone and imaged with TEM. Each electron micrograph shows the previously reported typical dense globular complexes with spheroid shapes, reflecting co-aggregation of NCp7 with either RNA (Le Cam et al., 1998) or ssDNA (Mirambeau et al., 2006), surrounded by dsDNA molecules, some of them being attached to the dense network of NCp7 and ss-NA. Upon addition of Vpr (b-d), some nucleofilaments exactly comparable to those of Fig. 2 were observed, generally attached to the NCp7:ss-NA spheroids. Because of Vpr cooperative binding, some other dsDNA regions were found to be uncoated without noticeable structures or arrangements. These results surprisingly show that Vpr could easily capture and fold vDNA into nucleofilaments while NCp7 should remain strongly bound to the RNP complex, in agreement with our proposal in Fig. 1.
In vitro, IN catalyzes the key steps of DNA integration, 3′ processing and strand transfer, in a concerted reaction with simple DNA substrates that mimic the two ends of the vDNA with either Mg2+ or Mn2+ as a cofactor (5–10mM range). Under in vitro reaction conditions, most of the integration products contain a single vDNA end branched into a strand of target DNA (half-site integration). Concerted integration of pairs of vDNA ends can be catalyzed by HIV-1 IN in vitro, but the reaction conditions are far more restrictive than for half-site integration: (i) the vDNA substrates must be much longer than the 20 bp that are sufficient for near-maximal efficiency of half-site integration, (ii) the presence of high concentrations of the crowding agent PEG (polyethylene glycol) is required and (iii) there is a high sensitivity to both the concentration and the stoichiometry of IN and DNA substrates. When the correct conditions were obtained, stable complexes of IN with two viral DNA ends that mimic the association of IN with vDNA in the PIC are formed (Li and Craigie, 2009; Pandey et al., 2007). They have been recently imaged with atomic force microscopy (AFM, (Kotova et al., 2010)) showing the formation of stable synaptic complexes where IN tetramers bridge DNA ends into intermediates on the integration reaction pathway. These intermediates are rapidly converted into concerted strand-transfer complexes in the presence of target DNA. AFM also revealed salt-sensitive but very stable self-association of the IN:DNA complexes into aggregates joining IN and the DNA extremities together.
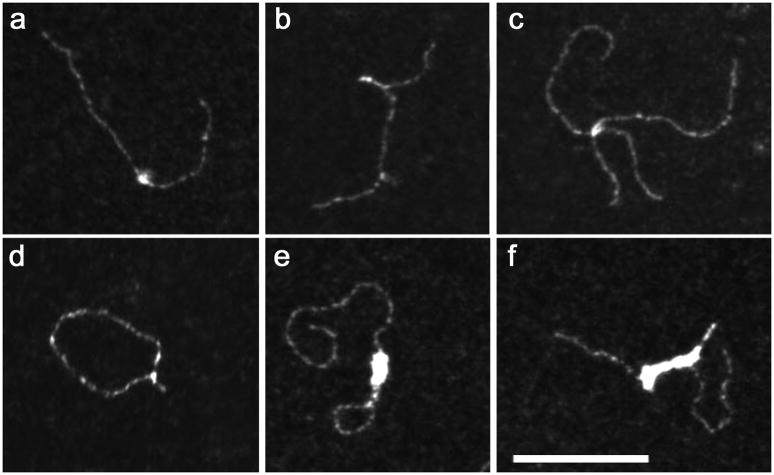
Fig. 4 shows TEM micrographs of complexes obtained upon incubation of IN and linear DNA molecules of about 800bp containing a 400bp-long segment corresponding to the vDNA cPPT-CTS locus, flanked by two 200bp regions that include the 5′ U3-terminal and 3′ U5-terminal LTR ends. These experiments were performed in the buffer used for Vpr and NCp7 binding, which are considered “low ionic strength conditions” for IN, favoring IN:DNA aggregation and half-site integration according to the AFM study by (Kotova et al., 2010). Importantly, we observed heterogeneity of complexed DNA with a high yield of DNA:protein aggregates very close or identical to those observed by the Kotova’s AFM study. Preferential fixation of IN on one or both extremities of the DNA fragments was observed in no more than 10% of the subset of molecules, which resulted in formation of synaptic complexes or DNA dimers (not shown), also comparable to those previously described. We also identified different fixation events on target DNA such as IN clusters (a), which can induce bridging of two molecules into X-shape assemblies (c); hairpin-like dsDNA structures (b) or DNAs closed into circles (d). The same experiments were carried out at 4°C to decrease the kinetics of IN oligomerization, which thereby lowered aggregate formation to some extent. The same arrangements were observed, with several new structures such as DNA center + extremity bridging (e), causing loop formation and a coat of IN on the DNA strands, suggesting polymerization along the DNA (f). In conclusion, in excellent agreement with the independent AFM analysis, IN has a high propensity (under our experimental conditions for optimal RT, NCp7 and Vpr activities) to oligomerize and establish intermolecular interactions with preferential binding to DNA extremities and also a minor but noticeable specific binding to a segment of DNA containing the cPPT-CTS sequence without the central flap.
Figure 4. Complexes formed between IN and dsDNA observed by TEM.
Reactions were carried out for 10 min. in 10mM HEPES (pH 7.5), 100mM NaCl, 7.5mM MgCl2 using 3.8nM of a HIV-1 derived DNA fragment (800bp) and 125nM IN monomer (nucleotide/IN ratio: 32). The most representative individual IN-DNA complexes have been selected once formed at 37°C (a-d) or at 4°C (e-f). The scale bar is the same for all panels and represents 100 nm.
We next present images in Fig. 5 of puzzling complexes obtained when IN, Vpr and pTZ18R plasmid DNA or the 1.6kbp fragment used in Fig. 2 were combined. Again we observed the typical nucleofilaments formed between Vpr and dsDNA, but these nucleofilaments were connected to bright dense complexes that were similar to the IN clusters shown in the Fig. 4e-f. An interpretation is that Vpr can synaptically bridge dsDNA segments whereas IN can form oligomer clusters directed to the DNA ends or to preferential internal sites acting as promoters of the IN cluster nucleation. These assemblies are also shown to be in contact with the Vpr-associated nucleofilaments. This may be an important function during the dsDNA synthesis via RT translocation: a general deposition of Vpr along dsDNA to promote assembly of bridged and compacted nucleofilaments to relocate IN at the DNA ends and the central flap locus, noting that a DNA flap is related to a strand transfer intermediate in the integration reaction. Interestingly, a previous report by (Bischerour et al., 2003) has highlighted that Vpr (52–96) can stabilize the complexes formed following the binding of IN to its cognate high affinity sites at the extremities of a mini-viral DNA. This effect is strongly reminiscent of what we observed here and obviously needs further characterization.
IV. 3 Reverse transcription leads to dsDNA-coating and bridging by Vpr and IN
In order to expand this emerging model of RTC to PIC conversion in a more physiologically relevant setting, we added Vpr and IN to our RT circular ssDNA assay in addition to RT and NCp7 (Fig. 6). Adding Vpr at a ratio of 1 for 20 bp does not affect the elongation efficiency, whereas IN slows the reaction when added in a limited excess compared to RT (IN monomer:RT = 10:1). This weak inhibition confirms previous results (Tasara et al., 2001). Despite this, substantial elongation is observed and the reaction appeared to be delayed due to some pausing at specific loci on our Bluescript-derived ssDNA (not shown). Upon addition of NCp7 only, TEM imaging of the nucleoprotein complexes already lead to our model presented in section II,3. Residual aggregates of NCp7 and ssDNA are joined to extruding dsDNA to generate huge networks of hundreds of DNA circles (6,a). Each DNA circle is attached to the residual aggregates at least by the central DNA flap where the reaction terminates. TEM controls of ssDNA alone, ssDNA plus RT and dNTP leading to circular dsDNA, or the same plus Vpr are shown in the upper part of Fig. 6. Note that Vpr induced the same effect as shown in the simple dsDNA:Vpr mixture: wire-like/synaptically bridged dsDNA nucleofilaments over hundreds of base pairs. Therefore, we imaged the samples where Vpr and IN were added along with RT and NCp7. A strikingly unique picture was obtained when Vpr (6,b) or Vpr plus IN (6,c) was added, whereas IN alone did not profoundly affect the complicated aspect of the aggregates (not shown). In the case of Vpr alone, a clear phenomenon of dsDNA bridging occurred with some of the numerous dsDNA extruded regions exposed from the NCp7:ssDNA aggregates, confirming that Vpr is able to trap dsDNA once it is formed in the presence of NCp7, a situation relevant to the RTC complex engaged in the plus-strand DNA synthesis step. In the case of Vpr plus IN, these nucleofilaments were reinforced: they appeared both longer and broader, whereas the residual NCp7-associated aggregates appeared also to be affected by the presence of IN. As the DNA herein does not provide the high affinity DNA ends (att sites) for IN binding, thus we cannot attribute a specific location to IN but clearly IN contributes in strengthening the Vpr effect on binding to dsDNA. Clearly, more experiments are required to better appreciate these assemblies and the protein ballet that mediates nucleocapsid to PIC remodeling via the RTC. TEM and also AFM are excellent tools to gain insights into this ballet and to unravel the corresponding mechanisms that allow functional deposition of both IN and Vpr along the vDNA.
V. Concluding remarks
Our primary goal in this special issue intended to focus the virologists’ attention on the progression of the HIV-1 nucleoprotein complex from the nucleocapsid to the preintegration architecture, a still poorly documented phenomenon. Consequently, we have provided additional details to our previous model of nucleocapsid dismantling (section II) and we incorporated a set of new TEM micrographs of Vpr-DNA and IN-DNA complexes (section IV) that, for the first time, emphasize a critical role for Vpr as an HIV-1 DNA architectural protein engaged in the promotion of its nuclear import as a component of the PIC.
Dismantling of the nucleocapsid complex can be considered a direct consequence of the second step of reverse transcription, i. e. the synthesis of dsDNA, where the combination of RT translocation with the reduction of the number of bound NCp7 molecules is the key. By contrast, the RNase-containing RT pool that efficiently digest vRNA within RNA-DNA heteroduplexes, as well as the remarkable inverted binding polarity of NCp7 for DNA vs. RNA that should allow fast re-association of displaced NCp7 along the (-) DNA strand are favorable properties that maintain the nucleocapsid assembly during the initial minus-strand DNA synthesis step. Such a proposal requires validation with experiments similar to those presented in Fig. 6, using RNA and RNA-DNA heteroduplexes instead. The relevance of the magnesium effect (i. e. reduction NCp7:dsDNA contacts) must also be examined in vivo. At the end of the process, a limited amount of NCp7 molecules remains associated with the central DNA flap region, while this assembly should be close to the DNA ends at the time the intasome formed and RT mostly dissociates (fig. 1, i-j). It is tempting to speculate that such an assembly could i) prevent degradation of the DNA flap by nucleases and ii) favour the intasome assembly due to their spatial proximity.
DNA-coating and bridging by Vpr is unambiguously demonstrated by our TEM analysis. Moreover, the related DNA-Vpr assemblies are strikingly similar to those shown with the bacterial protein H-NS and STpA, known to regulate the transcriptional activity of the bacterial nucleoid by evoking such nucleoprotein architectures (Dame et al., 2006; Lim et al., 2011; Liu et al., 2010). We also suspect that the local concentration of Vpr in the RTC is much higher than in our assays and should more easily drive the formation of synaptically bridged DNA. One question was raised here: what is the reason for such a DNA architectural activity for Vpr? As we discussed in the section III, vDNA compaction is necessary for a correct positioning of the remaining HIV-1 core onto the nuclear envelope in a coordinated way with karyophilic signaling, independent of the capsid status. Vpr may be required here to regulate stresses on the capsid since dsDNA that extrudes from the initial condensed nucleocapsid complex will induce mechanical and destabilizing stresses on the capsid shell. On the other hand, vDNA must translocate through the nuclear pore and its synaptic bridging by Vpr could provide a great advantage. Several dsDNA fragments are shown here to be bridged together, making it possible to simplify the direct vDNA translocation by runs of 2, 3 or 4 bridged DNA strands, in a closed organization with the nucleoporin partners of Vpr, like h-CG1 or others. The diameter of such a synaptic DNA filament is compatible with the diameter of the dilated pore (nearly 25 nm). Binding of both Vpr and IN can be seen in our assay as IN clusters along the DNA in a more disordered way than Vpr, but the IN-Vpr-DNA assembly requires further study with appropriate DNA constructs to clarify the architecture and function associated with these complexes. Additionally, the central DNA flap synthesis could be proposed to be an efficiency factor promoting the coupling between termination of reverse transcription and convenient exposure of the vDNA to the nuclear pore complex by the way of an accurate Vpr, IN deposition and assembly along the DNA, allowing the best-fit PIC compaction. Such proposals again require further studies.
Over the next few years, a better understanding of the biological properties of the HIV-1 preintegration steps is expected through the use of imaging and biochemical approaches. On a related topic, Fig. 7 is presented in order to show additional information that has been obtained by TEM imaging, highlighting the great potential of in vitro reconstitution approaches coupled with macromolecular visualization. In this figure, we show the results of incubating our RT-NCp7 in vitro assay in the presence of F-actin. The two images on the left side (a, b) are reaction products obtained from very large initial NCp7:ssDNA aggregates and we can see dense NCp7 aggregates separated by filamentous dsDNA, similar to that shown in Fig. 6a. The three other images (c-e) show the same reaction in the presence of F-actin. All of the dense NCp7 aggregates are attached to the F-actin. As NCp7 has been shown to bind to F-actin by its N-terminal domain, these images are not totally surprising (Liu et al., 1999). RT could also be involved in similar interactions since the large subunit of HIV-1 RT was shown to interact with beta-actin (Hottiger et al., 1995). As F-actin has been shown to be involved in the motility of HIV-1 PICs especially in the vicinity of the nuclear envelope (Arhel et al., 2006) such images suggest that RTCs may move toward the nuclear pore complex through its binding to F-actin by NCp7, or by the RT-NCp7 duo, or may be stabilized by F-actin onto the nuclear envelope close to a pore. But what about the capsid shell? And when does reverse transcription really occur?
Figure 7. DNA synthesis by HIV-1 RT on circular ssDNA template in the presence of NCp7 and F-actin.
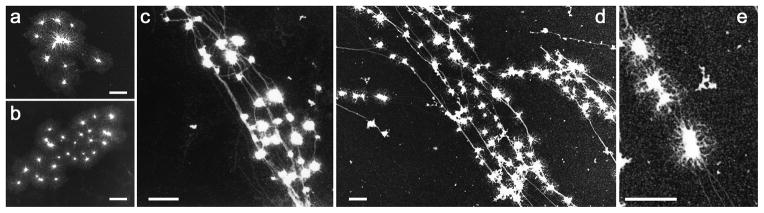
The polymerase reaction was carried out over 40 min. as in Fig. 6. Concentrations of primed-ssDNA circles, RT and NCp7 were, respectively, 5 nM, 100nM, 3.4 μM. F-actin filaments were added to the assay at a concentration close to 0.1 μM (monomer). NCp7 and primed-ssDNA circles were premixed for 4 min. at 37°C, followed by incubation with RT for 2 min., F-actin for 2 min. before addition of dNTPs (100 μM each) to start the reaction. a and b: controls without F-actin showing clusters of dismantling NCp7-ssDNA aggregates due to dsDNA extrusion. c, d and e: reactions in the presence of F-actin showing binding of the NCp7-RT-DNA complexes all along the F-actin filaments. 5 μl-aliquots of the assay were diluted 5-fold in the reaction buffer without DTT and next deposited onto the EM grid. The scale bars correspond to 0.5 μm.
The spatiotemporal coordination between RTC-PIC remodeling and trafficking, combined with capsid uncoating leading to HIV-1 DNA translocation into the nucleus is clearly a fascinating and challenging system to be defined in virology. Fortunately, it has benefited from the huge numbers of studies that have been engaged in the worldwide HIV field since its discovery in 1983. We hope that the scientific community will continue to follow this route to fully characterize this process, paying close attention to the interplay between viral and cellular constituents as well as the related molecular mechanisms involved.
Materials and Methods
Proteins
Recombinant HIV-1 RT (p66/p51 heterodimer) was prepared as described (Müller et al., 1989). Recombinant HIV-1 wild-type NCp(1–55) (i.e. NCp7) based on the pNL4-3 sequence from GenBank, accession number AF324493 were expressed and purified according to (Carteau et al., 1999; Wu et al., 1996). Peptide Vpr(1–96) from HIV-1 strain LAI (GenBank accession number K02013), with the sequence NH2-MEQAPEDQG PQREPYNDWT LELLEELKNE AVRHFPRIWL HSLGQHIYET YGDTWTGVEA LIRILQQLLF IHFRIGCRHS RIGIIQQRRT RNGASKS-COOH, was used. The peptides were prepared by automated solid phase synthesis using the FMOC strategy and then purified by reversed-phase HPLC, using procedures already reported (De Rocquigny et al., 1997). Vpr and NCp7 were suspended at a concentration of 1 mg/ml in 25 mM Tris–acetate (pH 7.5), 30 mM sodium acetate, 3 mM DTT, and 20% (v/v) ethylene glycol, and stored in aliquots at −80 °C. HIV-1 IN was purified from a yeast expression system using the INHybrid methods described previously (Lesbats et al., 2008). Purified IN was kept at −80°C in 300 mM NaCl. F-Actin was provided by Marie-France Carlier (CNRS, Gif-sur-Yvette, France).
Nucleic Acids
The pTZ-18R dsDNA plasmid was relaxed using purified human topoisomerase I (Topogen), then purified using MonoQ anion-exchange resin (AP Biotech). The circular 3,352 nt single-stranded DNA, that contains the HIV-Bru cPPT-CTS sequence, as well as its dsDNA plasmid counterpart, were prepared as described in (Hameau et al., 2001). The 800 bp fragment used in Fig. 4 was composed of a 400 bp fragment encompassing the central region of HIV-1 DNA flanked by two 200 bp fragments, one corresponding to the U3 LTR end and the other to the RU5 LTR end. The fragments were produced by PCR starting from the HIV1 pNL4-3 plasmid using primers oligonucleotides designed to produce sticky ends with the Bsa I restriction enzyme, according to the method described in (Escudé et al., 2007)
Reverse Transcription Assay
The RT assay was previously described in (Hameau et al., 2001; Mirambeau et al., 2007; Mirambeau et al., 2006). Briefly, the 3,352-nt circular ssDNA template was hybridized with an oligonucleotide corresponding to the 5′ end of the central DNA flap (5′-TTGGGGGGTACAGTGCA-3′). The polymerase reaction was carried out in the RT buffer (50 mM Tris-acetate pH 7.8, 50 mM sodium acetate, 6 mM magnesium acetate and 0.5 mM DTT) at 37°C. For analysis by agarose gel electrophoresis, DNA products were heated to 70°C for 10 min. in the presence of 1% (w/v) SDS and 20 mM EDTA.
Transmission Electron Microscopy
EM observations were performed with a positive staining procedure as described previously (Le Cam et al., 1998; Mirambeau et al., 2007; Mirambeau et al., 2006). After incubation, 5 μL of the reaction was deposited on a 600-mesh copper grid covered with a thin carbon film, activated by glow-discharge in the presence of pentylamine (Beloin et al., 2003). Grids were washed with aqueous 2% (w/v) uranyl acetate, dried and observed in the annular dark-field mode, using a Zeiss 902 transmission electron microscope. Images were captured at magnifications of 50,000×, 85000× and 140000× with a MegaviewIII CCD camera and iTEM software for acquisition (Olympus Soft Imaging Solution).
A model for dismantling the HIV-1 nucleocapsid by reverse transcriptase is proposed
A HIV-1 DNA architectural role for Vpr is shown
Actin binding to HIV-1 nucleocapsid-reverse transcription complexes is revealed
Spatiotemporal coordination for coupling remodeling and trafficking of HIV-1 genome in the early steps of cellular infection is emphasized
Acknowledgments
S. L., J.-F. M., S. B., E. L. and G.M. are grateful to ANRS-AC 14.2 for its funding, paying a special tribute to Pr. Roger Monier †. S.L. & G. M. are indebted to the European Community’s Seventh Framework Program for the Grant Agreement IEF N° 237738. This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E with SAIC-Frederick, Inc. (RJG). V.P. is grateful to SIDACTION AO2011-2013 for its funding. T.R. acknowledges funding by EC-grant LSHG-CT-2003-503480. G. M. thanks Dr. Kashif Sadiq for a “last minute” editing of this manuscript. We also wish to thank Donald G. Johnson and Catherine V. Hixson of the AIDS and Cancer Virus Program for their assistance in preparing the recombinant nucleocapsid proteins used in this study.
Abbreviations
- (−) SSDNA
minus-strand strong-stop DNA
- (−) ss-DNA
minus-strand single-stranded DNA
- +SSDNA
plus-strand strong-stop DNA
- AFM
atomic force microscopy
- AZT
azidothymidine
- BAF
barrier-to-autointegration factor
- bp
base-pairs
- CA
capsid
- cFlap
central DNA flap
- cPBS
complementary PBS
- cPPT
central polypurine tract
- cTAR
antisense or complementary TAR DNA
- CTS
central termination site
- CypA
CyclophilinA
- dNTP
deoxynucleotide triphosphate
- ds
double-stranded
- ERT
endogenous reverse transcription
- HIV-1
human immunodeficiency virus type 1
- IN
integrase
- LEDGF
lens epithelial growth factor
- LTR
long terminal repeat
- MA
matrix
- MLV
murine leukemia virus
- NA
nucleic acid
- NCp (or NCp7)
nucleocapsid protein
- PBS
primer binding site
- PIC
preintegration complex
- PPT
polypurine tract
- R
repeat region of LTR
- RNP
ribonucleoprotein
- RT
reverse transcriptase
- RTC
reverse transcription complex
- ss
single-stranded
- SSB
single-stranded binding
- TEM
transmission electron microscopy
- U3
3′ untranslated region of LTR
- U5
5′ untranslated region of LTR
- UTR
untranslated regions of the RNA genome
- vDNA
viral DNA
- Vpr
viral protein r
- vRNA
viral RNA
- ZF
zinc finger
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Accola MA, Ohagen A, Gottlinger HG. Isolation of human immunodeficiency virus type 1 cores: retention of Vpr in the absence of p6(gag) J Virol. 2000;74(13):6198–6202. doi: 10.1128/jvi.74.13.6198-6202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92(3):291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- Amacker M, Hottiger M, Mossi R, Hübscher U. HIV-1 nucleocapsid protein and replication protein A influence the strand displacement DNA synthesis of lentiviral reverse transcriptase. AIDS. 1997;11(4):534–536. [PubMed] [Google Scholar]
- Ao Z, Yao X, Cohen EA. Assessment of the role of the central DNA flap in human immunodeficiency virus type 1 replication by using a single-cycle replication system. J Virol. 2004;78(66):3170–3177. doi: 10.1128/JVI.78.6.3170-3177.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arhel N. Revisiting HIV-1 uncoating. Retrovirology. 2010;7(96) doi: 10.1186/1742-4690-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arhel N, Genovesio A, Kim KA, Miko S, Perret E, Olivo-Marin JC, Shorte S, Charneau P. Quantitative four-dimensional tracking of cytoplasmic and nuclear HIV-1 complexes. Nat Methods. 2006;3(10):817–824. doi: 10.1038/nmeth928. [DOI] [PubMed] [Google Scholar]
- Athavale SS, Ouyang W, McPike MP, Hudson BS, Borer PN. Effects of the nature and concentration of salt on the interaction of the HIV-1 nucleocapsid protein with SL3 RNA. Biochemistry. 2010;49(17):3525–3533. doi: 10.1021/bi901279e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bampi C, Jacquenet S, Lener D, Decimo D, Darlix JL. The chaperoning and assistance roles of the HIV-1 nucleocapsid protein in proviral DNA synthesis and maintenance. The international journal of biochemistry & cell biology. 2004;36(9):1668–1686. doi: 10.1016/j.biocel.2004.02.024. [DOI] [PubMed] [Google Scholar]
- Bazzi A, Zargarian L, Chaminade F, Boudier C, De Rocquigny H, René B, Mély Y, Fossé P, Mauffret O. Structural insights into the cTAR DNA recognition by the HIV-1 nucleocapsid protein: role of sugar deoxyriboses in the binding polarity of NC. Nucleic Acids Res. 2011;39(9):3903–3916. doi: 10.1093/nar/gkq1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerens N, Kjems J. Circularization of the HIV-1 genome facilitates strand transfer during reverse transcription. RNA. 2010;16(6):1226–1235. doi: 10.1261/rna.2039610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloin C, Jeusset J, Revet B, Mirambeau G, Le Hégarat F, Le Cam E. Contribution of DNA conformation and topology in right-handed DNA wrapping by the Bacillus subtilis LrpC protein. J Biol Chem. 2003;278(7):5333–5342. doi: 10.1074/jbc.M207489200. [DOI] [PubMed] [Google Scholar]
- Bischerour J, Tauc P, Leh H, De Rocquigny H, Roques BP, Mouscadet JF. The (52–96) C-terminal domain of Vpr stimulates HIV-1 IN-mediated homologous strand transfer of mini-viral DNA. Nucleic Acids Res. 2003;31(10):2694–2702. doi: 10.1093/nar/gkg364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbigot S, Ramalanjaona N, Boudier C, Salgado GFJ, Roques BP, Mély Y, Bouaziz S, Morellet N. How the HIV-1 nucleocapsid protein binds and destabilises the (−)primer binding site during reverse transcription. J Mol Biol. 2008;383(5):1112–1128. doi: 10.1016/j.jmb.2008.08.046. [DOI] [PubMed] [Google Scholar]
- Bowerman B, Brown PO, Bishop JM, Varmus HE. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 1989;3(4):469–478. doi: 10.1101/gad.3.4.469. [DOI] [PubMed] [Google Scholar]
- Briggs JA, Kräusslich HG. The molecular architecture of HIV. J Mol Biol. 2011;410(4):491–500. doi: 10.1016/j.jmb.2011.04.021. [DOI] [PubMed] [Google Scholar]
- Briggs JA, Simon MN, Gross I, Kräusslich HG, Fuller SD, Vogt VM, Johnson MC. The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol. 2004;11(7):672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- Buckman JS, Bosche WJ, Gorelick RJ. Human immunodeficiency virus type 1 nucleocapsid zn(2+) fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J Virol. 2003;77(2):1469–1480. doi: 10.1128/JVI.77.2.1469-1480.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukrinsky M. A hard way to the nucleus. Mol Med. 2004;10(1–6):1–5. [PMC free article] [PubMed] [Google Scholar]
- Cameron CE, Ghosh M, Le Grice SF, Benkovic SJ. Mutations in HIV reverse transcriptase which alter RNase H activity and decrease strand transfer efficiency are suppressed by HIV nucleocapsid protein. Proc Natl Acad Sci USA. 1997;94(13):6700–6705. doi: 10.1073/pnas.94.13.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carteau S, Gorelick RJ, Bushman FD. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J Virol. 1999;73(8):6670–6679. doi: 10.1128/jvi.73.8.6670-6679.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux JJ, Schultz SJ. Ribonuclease H: properties, substrate specificity and roles in retroviral reverse transcription. FEBS J. 2009;276(6):1506–1516. doi: 10.1111/j.1742-4658.2009.06909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman FD. A role for LEDGF/p75 in targeting HIV DNA integration. Nature Med. 2005;11(12):1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- Coeytaux E, Coulaud D, Le Cam E, Danos O, Kichler A. The cationic amphipathic α-helix of HIV-1 viral protein r (Vpr) binds to nucleic acids, permeabilizes membranes, and efficiently transfects cells. J Biol Chem. 2003;278(20):18110–18116. doi: 10.1074/jbc.M300248200. [DOI] [PubMed] [Google Scholar]
- Cruceanu M, Urbaneja MA, Hixson CV, Johnson DG, Datta SA, Fivash MJ, Stephen AG, Fisher RJ, Gorelick RJ, Casas-Finet JR, Rein A, Rouzina I, Williams MC. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res. 2006;34(2):593–605. doi: 10.1093/nar/gkj458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame RT, Noom MC, Wuite GJ. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature. 2006;444(7117):387–390. doi: 10.1038/nature05283. [DOI] [PubMed] [Google Scholar]
- Darlix JL, Godet J, Ivanyi-Nagy R, Fossé P, Mauffret O, Mély Y. Flexible nature and specific functions of the HIV-1 nucleocapsid protein. J Mol Biol. 2011;410(4):565–581. doi: 10.1016/j.jmb.2011.03.037. [DOI] [PubMed] [Google Scholar]
- De Cian A, Praly E, Ding F, Singh V, Lavelle C, Le Cam E, Croquette V, Piétrement O, Bensimon D. ATP-Independent cooperative binding of yeast Isw1a to bare and nucleosomal DNA. PLoS One. 2012;7(2):e31845. doi: 10.1371/journal.pone.0031845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marco A, Müller B, Glass B, Riches JD, Kräusslich HG, Briggs JA. Structural analysis of HIV-1 maturation using cryo-electron tomography. PLoS Pathog. 2010;6(11):e1001215. doi: 10.1371/journal.ppat.1001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rocquigny H, Caneparo A, Delaunay T, Bischerour J, Mouscadet JF, Roques BP. Interactions of the C-terminus of viral protein R with nucleic acids are modulated by its N-terminus. Eur J Biochem. 2000;267(12):3654–3660. doi: 10.1046/j.1432-1327.2000.01397.x. [DOI] [PubMed] [Google Scholar]
- De Rocquigny H, Gabus C, Vincent A, Fournie-Zaluski MC, Roques B, Darlix JL. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc Natl Acad Sci USA. 1992;89(14):6472–6476. doi: 10.1073/pnas.89.14.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rocquigny H, Petitjean P, Tanchou V, Decimo D, Drouot L, Delaunay T, Darlix JL, Roques BP. The zinc fingers of HIV nucleocapsid protein NCp7 direct interactions with the viral regulatory protein Vpr. J Biol Chem. 1997;272(49):30753–30759. doi: 10.1074/jbc.272.49.30753. [DOI] [PubMed] [Google Scholar]
- Delelis O, Carayon K, Saib A, Deprez E, Mouscadet J-F. Integrase and integration: biochemical activities of HIV-1 integrase. Retrovirology. 2008;5(114) doi: 10.1186/1742-4690-5-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déméné H, Dong CZ, Ottmann M, Rouyez MC, Jullian N, Morellet N, Mély Y, Darlix JL, Fournié-Zaluski MC, Saragosti S, et al. 1H NMR structure and biological studies of the His23→Cys mutant nucleocapsid protein of HIV-1 indicate that the conformation of the first zinc finger is critical for virus infectivity. Biochemistry. 1994;33(39):11707–11716. doi: 10.1021/bi00205a006. [DOI] [PubMed] [Google Scholar]
- Didierlaurent L, Houzet L, Morichaud Z, Darlix JL, Mougel M. The conserved N-terminal basic residues and zinc-finger motifs of HIV-1 nucleocapsid restrict the viral cDNA synthesis during virus formation and maturation. Nucleic Acids Res. 2008;36(14):4745–4753. doi: 10.1093/nar/gkn474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dismuke DJ, Aiken C. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J Virol. 2006;80(8):3712–3720. doi: 10.1128/JVI.80.8.3712-3720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druillennec S, Caneparo A, De Rocquigny H, Roques BP. Evidence of interactions between the nucleocapsid protein NCp7 and the reverse transcriptase of HIV-1. J Biol Chem. 1999;274(16):11283–11288. doi: 10.1074/jbc.274.16.11283. [DOI] [PubMed] [Google Scholar]
- Dussupt V, Javid MP, Abou-Jaoudé G, Jadwin JA, de La Cruz J, Nagashima K, Bouamr F. The nucleocapsid region of HIV-1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLoS Pathog. 2009;5(3):e1000339. doi: 10.1371/journal.ppat.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussupt V, Sette P, Bello NF, Javid MP, Nagashima K, Bouamr F. Basic residues in the nucleocapsid domain of Gag are critical for late events of HIV-1 budding. J Virol. 2010;85(5):2304–2315. doi: 10.1128/JVI.01562-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emiliani S, Mousnier A, Busschots K, Maroun M, Van Maele B, Tempé D, Vandekerckhove L, Moisant F, Ben-Slama L, Witvrouw M, Christ F, Rain JC, Dargemont C, Debyser Z, Benarous R. Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J Biol Chem. 2005;280(27):25517–25523. doi: 10.1074/jbc.M501378200. [DOI] [PubMed] [Google Scholar]
- Escudé C, Roulon T, Lyonnais S, Le Cam E. Multiple topological labeling for imaging single plasmids. Anal Biochem. 2007;362(1):55–62. doi: 10.1016/j.ab.2006.12.028. [DOI] [PubMed] [Google Scholar]
- Fassati A, Goff SP. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J Virol. 2001;75(8):3626–3635. doi: 10.1128/JVI.75.8.3626-3635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RJ, Fivash MJ, Stephen AG, Hagan NA, Shenoy SR, Medaglia MV, Smith LR, Worthy KM, Simpson JT, Shoemaker R, McNitt KL, Johnson DG, Hixson CV, Gorelick RJ, Fabris D, Henderson LE, Rein A. Complex interactions of HIV-1 nucleocapsid protein with oligonucleotides. Nucleic Acids Res. 2006;34(2):472–484. doi: 10.1093/nar/gkj442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RJ, Rein A, Fivash M, Urbaneja MA, Casas-Finet JR, Medaglia M, Henderson LE. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J Virol. 1998;72(3):1902–1909. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz JV, Briant L, Mély Y, Bouaziz S, De Rocquigny H. HIV-1 viral protein r: from structure to function. Future Virol. 2010;5(5):607–625. [Google Scholar]
- Fuentes GM, Rodriguez-Rodriguez L, Palaniappan C, Fay PJ, Bambara RA. Strand displacement synthesis of the long terminal repeats by HIV reverse transcriptase. J Biol Chem. 1996;271(4):1966–1971. doi: 10.1074/jbc.271.4.1966. [DOI] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, Yeager M, Pornillos O. Assembly and architecture of HIV. Adv Exp Med Biol. 2012;726:441–465. doi: 10.1007/978-1-4614-0980-9_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godet J, Mély Y. Biophysical studies of the nucleic acid chaperone properties of the HIV-1 nucleocapsid protein. RNA Biology. 2010;7(6):687–699. doi: 10.4161/rna.7.6.13616. [DOI] [PubMed] [Google Scholar]
- Goff SP. Intracellular trafficking of retroviral genomes during the early phase of infection: viral exploitation of cellular pathways. J Gene Med. 2001;3(6):517–528. doi: 10.1002/1521-2254(200111)3:6<517::AID-JGM234>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Götte M, Rausch JW, Marchand B, Sarafianos S, Le Grice SF. Reverse transcriptase in motion: conformational dynamics of enzyme-substrate interactions. Biochim Biophys Acta. 2010;1804(5):1202–1212. doi: 10.1016/j.bbapap.2009.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann D, Godet J, Mely Y, Darlix JL, Restle T. HIV-1 nucleocapsid traps reverse transcriptase on nucleic acid substrates. Biochemistry. 2008;47(46):12230–12240. doi: 10.1021/bi801386r. [DOI] [PubMed] [Google Scholar]
- Guo J, Wu T, Kane BF, Johnson DG, Henderson LE, Gorelick RJ, Levin JG. Subtle alterations of the native zinc finger structures have dramatic effects on the nucleic acid chaperone activity of human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 2002;76(9):4370–4378. doi: 10.1128/JVI.76.9.4370-4378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameau L, Jeusset J, Lafosse S, Coulaud D, Delain E, Unge T, Restle T, Le Cam E, Mirambeau G. Human immunodeficiency virus type 1 central DNA flap: dynamic terminal product of plus-strand displacement dna synthesis catalyzed by reverse transcriptase assisted by nucleocapsid protein. J Virol. 2001;75(7):3301–3313. doi: 10.1128/JVI.75.7.3301-3313.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig A. Role of magnesium in genomic stability. Mutat Res. 2001;475(1–2):113–121. doi: 10.1016/s0027-5107(01)00074-4. [DOI] [PubMed] [Google Scholar]
- Hehl EA, Joshi P, Kalpana GV, Prasad VR. Interaction between human immunodeficiency virus type 1 reverse transcriptase and integrase proteins. J Virol. 2004;78(10):5056–5067. doi: 10.1128/JVI.78.10.5056-5067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzinger NK, Bukrinsky MI, Haggerty SA, Ragland AM, Kewalramani V, Lee MA, Gendelman HE, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91(15):7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson LE, Bowers MA, Sowder RCn, Serabyn SA, Johnson DG, Bess JWJ, Arthur LO, Bryant DK, Fenselau C. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J Virol. 1992;66(4):1856–1865. doi: 10.1128/jvi.66.4.1856-1865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergott CB, Mitra M, Guo J, Wu T, Miller JT, Iwatani Y, Gorelick RJ, Levin JG. Zinc finger function of HIV-1 nucleocapsid protein is required for removal of 5′-terminal genomic RNA fragments: A paradigm for RNA removal reactions in HIV-1 reverse transcription. Virus Res. 2012 doi: 10.1016/j.virusres.2012.08.013. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottiger M, Gramatikoff K, Georgiev O, Chaponnier C, Schaffner W, Hubscher U. The large subunit of HIV-1 reverse transcriptase interacts with beta-actin. Nucleic Acids Res. 1995;23(5):736–741. doi: 10.1093/nar/23.5.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houzet L, Morichaud Z, Didierlaurent L, Muriaux D, Darlix JL, Mougel M. Nucleocapsid mutations turn HIV-1 into a DNA-containing virus. Nucleic Acids Res. 2008;36(7):2311–2319. doi: 10.1093/nar/gkn069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Orenstein JM, Martin MA, Freed EO. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69(11):6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulme AE, Perez O, Hope TJ. Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc Natl Acad Sci USA. 2011;108(24):9975–9980. doi: 10.1073/pnas.1014522108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordanskiy S, Berro R, Altieri M, Kashanchi F, Bukrinsky M. Intracytoplasmic maturation of the human immunodeficiency virus type 1 reverse transcription complexes determines their capacity to integrate into chromatin. Retrovirology. 2006;3(4) doi: 10.1186/1742-4690-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob DT, Destefano JJ. A new role for HIV nucleocapsid protein in modulating the specificity of plus strand priming. Virology. 2008;378(2):385–396. doi: 10.1016/j.virol.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquot G, Le Rouzic E, David A, Mazzolini J, Bouchet J, Bouaziz S, Niedergang F, Pancino G, Benichou S. Localization of HIV-1 Vpr to the nuclear envelope: impact on Vpr functions and virus replication in macrophages. Retrovirology. 2007;4(84) doi: 10.1186/1742-4690-4-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayappa KD, Ao Z, Yang M, Wang J, Yao X. Identification of critical motifs within HIV-1 integrase required for importin α3 interaction and viral cDNA nuclear import. J Mol Biol. 2011;410(5):847–862. doi: 10.1016/j.jmb.2011.04.011. [DOI] [PubMed] [Google Scholar]
- Karageorgos L, Li P, Burrell C. Characterization of HIV replication complexes early after cell-to-cell infection. AIDS Res Hum Retroviruses. 1993;9(9):817–823. doi: 10.1089/aid.1993.9.817. [DOI] [PubMed] [Google Scholar]
- Katz RA, Greger JG, Boimel P, Skalka AM. Human immunodeficiency virus type 1 DNA nuclear import and integration are mitosis independent in cycling cells. J Virol. 2003;77(24):13412–13417. doi: 10.1128/JVI.77.24.13412-13417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khiytani DK, Dimmock NJ. Characterization of a human immunodeficiency virus type 1 pre-integration complex in which the majority of the cDNA is resistant to DNase I digestion. J Gen Virol. 2002;83(10):2523–2532. doi: 10.1099/0022-1317-83-10-2523. [DOI] [PubMed] [Google Scholar]
- Kichler A, Pages JC, Leborgne C, Druillennec S, Lenoir C, Coulaud D, Delain E, Lecam E, Roques BP, Danos O. Efficient DNA transfection mediated by the C-terminal domaon of human immunodeficiency virus type 1 viral protein r. J Virol. 2000;74(12):5424–5431. doi: 10.1128/jvi.74.12.5424-5431.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotova S, Li M, Dimitriadis EK, Craigie R. Nucleoprotein Intermediates in HIV-1 DNA Integration visualized by atomic force microscopy. J Mol Biol. 2010;399(3):491–500. doi: 10.1016/j.jmb.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapadat-Tapolsky M, Pernelle C, Borie C, Darlix JL. Analysis of the nucleic acid annealing activities of nucleocapsid protein from HIV-1. Nucleic Acids Res. 1995;23(13):2434–2441. doi: 10.1093/nar/23.13.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Cam E, Coulaud D, Delain E, Petitjean P, Roques BP, Gerard D, Stoylova E, Vuilleumier C, Stoylov SP, Mely Y. Properties and growth mechanism of the ordered aggregation of a model RNA by the HIV-1 nucleocapsid protein: an electron microscopy investigation. Biopolymers. 1998;45(3):217–229. doi: 10.1002/(SICI)1097-0282(199803)45:3<217::AID-BIP4>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Le Rouzic E, Mousnier A, Rustum C, Stutz F, Hallberg E, Dargemont C, Benichou S. Docking of HIV-1 Vpr to the nuclear envelope is mediated by the interaction with the nucleoporin hCG1. J Biol Chem. 2002;277(47):45091–45098. doi: 10.1074/jbc.M207439200. [DOI] [PubMed] [Google Scholar]
- Lee N, Gorelick RJ, Musier-Forsyth K. Zinc finger-dependent HIV-1 nucleocapsid protein-TAR RNA interactions. Nucleic Acids Res. 2003;31(16):4847–4855. doi: 10.1093/nar/gkg679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lener D, Tanchou V, Roques BP, Le Grice SF, Darlix JL. Involvement of HIV-I nucleocapsid protein in the recruitment of reverse transcriptase into nucleoprotein complexes formed in vitro. J Biol Chem. 1998;273(50):33781–33786. doi: 10.1074/jbc.273.50.33781. [DOI] [PubMed] [Google Scholar]
- Lesbats P, Métifiot M, Calmels C, Baranova S, Nevinsky G, Andreola ML, Parissi V. In vitro initial attachment of HIV-1 integrase to viral ends: control of the DNA specific interaction by the oligomerization state. Nucleic Acids Res. 2008;36(22):7043–7058. doi: 10.1093/nar/gkn796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin J, Mitra M, Mascarenhas A, Musier-Forsyth K. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biology. 2010;7(6):1–21. doi: 10.4161/rna.7.6.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- Levy-Mintz P, Duan L, Zhang H, Hu B, Dornadula G, Zhu M, Kulkosky J, Bizub-Bender D, Skalka AM, Pomerantz RJ. Intracellular expression of single-chain variable fragments to inhibit early stages of the viral life cycle by targeting human immunodeficiency virus type 1 integrase. J Virol. 1996;70(12):8821–8832. doi: 10.1128/jvi.70.12.8821-8832.1996. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Li M, Craigie R. Nucleoprotein complex intermediates in HIV-1 integration. Methods. 2009;47(4):237–242. doi: 10.1016/j.ymeth.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Ivanov V, Mizuuchi M, Mizuuchi K, Craigie R. DNA requirements for assembly and stability of HIV-1 intasomes. Protein Sci. 2012;21(2):249–257. doi: 10.1002/pro.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MS, Garcia-Asua G, Bhattacharyya U, Mascagni P, Austen BM, Roberts MM. The Vpr protein of human immunodeficiency virus type 1 binds to nucleocapsid protein p7 in vitro. Biochem Biophys Res Commun. 1996;218(1):352–355. doi: 10.1006/bbrc.1996.0061. [DOI] [PubMed] [Google Scholar]
- Lim CJ, Whang YR, Kenney LJ, Yan J. Gene silencing H-NS paralogue StpA forms a rigid protein filament along DNA that blocks DNA accessibility. Nucleic Acids Res. 2011;40(8):1–13. doi: 10.1093/nar/gkr1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CW, Engelman A. The barrier-to-autointegration factor is a component of functional human immunodeficiency virus type 1 preintegration complexes. J Virol. 2003;77(8):5030–5036. doi: 10.1128/JVI.77.8.5030-5036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Dai R, Tian CJ, Dawson L, Gorelick R, Yu XF. Interaction of the human immunodeficiency virus type 1 nucleocapsid with actin. J Virol. 1999;73(4):2901–2908. doi: 10.1128/jvi.73.4.2901-2908.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chen H, Kenney LJ, Yan J. A divalent switch drives H-NS/DNA-binding conformations between stiffening and bridging modes. Genes Dev. 2010;24(4):329–344. doi: 10.1101/gad.1883510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano M, Vanegas M, Fregoso O, Saenz D, Chung S, Peretz M, Poeschla EM. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J Virol. 2004;78(17):9524–9537. doi: 10.1128/JVI.78.17.9524-9537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lori F, Di Marzo Veronese F, de Vico AL, Lusso P, Reitz MS, Gallo RC. Viral DNA carried by human immunodeficiency virus type 1 virions. J Virol. 1992;66(8):5067–5074. doi: 10.1128/jvi.66.8.5067-5074.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyonnais S, Gorelick RJ, Mergny JL, Le Cam E, Mirambeau G. G-quartets direct assembly of HIV-1 nucleocapsid protein along single-stranded DNA. Nucleic Acids Res. 2003;31(19):5754–5763. doi: 10.1093/nar/gkg716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyonnais S, Hounsou C, Teulade-Fichou MP, Jeusset J, Le Cam E, Mirambeau G. G-quartets assembly within a G-rich DNA flap. A possible event at the center of HIV-1 genome. Nucleic Acids Res. 2002;30(23):5276–5283. doi: 10.1093/nar/gkf644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matreyek KA, Engelman A. The requirement for nucleoporin NUP153 during human immunodeficiency virus type 1 infection is determined by the viral capsid. J Virol. 2011;85(15):7818–7827. doi: 10.1128/JVI.00325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, Hope TJ. Visualization of the intracellular behavior of HIV in living cells. The Journal of Cellular Biology. 2002;159(3):441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MD, Farnet CM, Bushman FD. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71(7):5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirambeau G, Lyonnais S, Coulaud D, Hameau L, Lafosse S, Jeusset J, Borde I, Reboud-Ravaux M, Restle T, Gorelick RJ, Le Cam E. HIV-1 protease and reverse transcriptase control the architecture of their nucleocapsid partner. PLoS One. 2007;2(7):e669. doi: 10.1371/journal.pone.0000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirambeau G, Lyonnais S, Coulaud D, Hameau L, Lafosse S, Jeusset J, Justome A, Delain E, Gorelick RJ, Le Cam E. Transmission electron microscopy reveals an optimal HIV-1 nucleocapsid aggregation with single-stranded nucleic acids and the mature HIV-1 nucleocapsid protein. J Mol Biol. 2006;364(3):496–511. doi: 10.1016/j.jmb.2006.08.065. [DOI] [PubMed] [Google Scholar]
- Mirambeau G, Lyonnais S, Gorelick RJ. Features, processing states, and heterologous protein interactions in the modulation of the retroviral nucleocapsid protein function. RNA Biology. 2010;7(6):724–734. doi: 10.4161/rna.7.6.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell. 2009;137(3):433–444. doi: 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morellet N, Bouaziz S, Petitjean P, Roques BP. NMR structure of the HIV-1 regulatory protein Vpr. J Mol Biol. 2003;327(1):215–227. doi: 10.1016/s0022-2836(03)00060-3. [DOI] [PubMed] [Google Scholar]
- Morellet N, Roques BP, Bouaziz S. Structure-function relationship of Vpr: biological implications. Curr HIV Res. 2009;7(2):184–210. doi: 10.2174/157016209787581490. [DOI] [PubMed] [Google Scholar]
- Mori M, Dietrich U, Manetti F, Botta M. Molecular dynamics and DFT study on HIV-1 nucleocapsid protein-7 in complex with viral genome. J Chem Inf Model. 2010;50(4):638–650. doi: 10.1021/ci100070m. [DOI] [PubMed] [Google Scholar]
- Müller B, Restle T, Weiss S, Gautel M, Sczakiel G, Goody RS. Co-expression of the subunits of the heterodimer of HIV-1 reverse transcriptase in Escherichia coli. J Biol Chem. 1989;264(24):13975–13978. [PubMed] [Google Scholar]
- Müller B, Tessmer U, Schubert U, Kräusslich HG. Human Immunodeficiency Virus Type 1 Vpr Protein is incorporated into the virion in significantly smaller amounts than Gag and is phosphorylated in infected cells. J Virol. 2000;74(20):9727–9731. doi: 10.1128/jvi.74.20.9727-9731.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriaux D, Darlix JL. Properties and functions of the nucleocapsid protein in virus assembly. RNA Biology. 2010;7(6):744–753. doi: 10.4161/rna.7.6.14065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriaux D, Mirro J, Harvin D, Rein A. RNA is a structural element in retrovirus particles. Proc Natl Acad Sci USA. 2001;98(9):5246–5251. doi: 10.1073/pnas.091000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negroni M, Buc H. Retroviral recombination: what drives the switch? Nat Rev Mol Cell Biol. 2001;2(2):151–155. doi: 10.1038/35052098. [DOI] [PubMed] [Google Scholar]
- Nermut MV, Fassati A. Structural analyses of purified human immunodeficiency virus type 1 intracellular reverse transcription complexes. J Virol. 2003;77(15):8196–8206. doi: 10.1128/JVI.77.15.8196-8206.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocwieja KE, Brady TL, Ronen K, Huegel A, Roth SL, Schaller T, James LC, Towers GJ, Young JA, Chanda SK, König R, Malani N, Berry CC, Bushman FD. HIV integration targeting: a pathway involving Transportin-3 and the nuclear pore protein RanBP2. PLoS Pathog. 2011;7(3):e1001313. doi: 10.1371/journal.ppat.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms M, Abbink TEM, Pham C, Berkhout B. Circularization of the HIV-1 RNA genome. Nucleic Acids Res. 2007;35(15):5253–5261. doi: 10.1093/nar/gkm564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey KK, Bera S, Zahm J, Vora A, Stillmock K, Hazuda D, Grandgenett DP. Inhibition of human immunodeficiency virus type I concerted integration by strand transfer inhibitors which recognize a transient structural intermediate. J Virol. 2007;81(22):12189–12199. doi: 10.1128/JVI.02863-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxton W, Connor RI, Landau NR. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J Virol. 1993;67(12):7229–7237. doi: 10.1128/jvi.67.12.7229-7237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekna-Przybylska D, DiChiacchio L, Mathews DH, Bambara RA. A sequence similar to tRNA Lys3 gene is embedded in HIV-1 U3-R and promotes minus-strand transfer. Nat Struct Mol Biol. 2010;17(1):83–89. doi: 10.1038/nsmb.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov S, Popova E, Inoue M, Gottlinger HG. Human immunodeficiency virus type 1 Gag engages the Bro1 domain of ALIX/AIP1 through the nucleocapsid. J Virol. 2008;82(3):1389–1398. doi: 10.1128/JVI.01912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post K, Kankia B, Gopalakrishnan S, Yang V, Cramer E, Saladores P, Gorelick RJ, Guo J, Musier-Forsyth K, Levin J. Fidelity of plus-strand priming requires the nucleic acid chaperone activity of HIV-1 nucleocapsid protein. Nucleic Acids Res. 2009;37(6):1755–1756. doi: 10.1093/nar/gkn1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani AM. Cellular magnesium homeostasis. Arch Biochem Biophys. 2011;512(1):1–23. doi: 10.1016/j.abb.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani B, Engelbrecht S. Human immunodeficiency virus type 1 Vpr: functions and molecular interactions. J Gen Virol. 2009;90(8):1795–1805. doi: 10.1099/vir.0.011726-0. [DOI] [PubMed] [Google Scholar]
- Saadatmand J, Kleiman L. Aspects of HIV-1 assembly that promote primer tRNALys3 annealing to viral RNA Virus Res. 2012 doi: 10.1016/j.virusres.2012.06.001. http://dx.doi.org/10.1016/j.virusres.2012.06.001. [DOI] [PubMed]
- Schaller T, Ocwieja KE, Rasaiyaah J, Price AJ, Brady TL, Roth SL, Hué S, Fletcher AJ, Lee K, KewalRamani VN, Noursadeghi M, Jenner RG, James LC, Bushman FD, Towers GJ. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog. 2011;7(12):e1002439. doi: 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah VB, Aiken C. HIV Nuclear Entry: Clearing the Fog. Viruses. 2010;2(5):1190–1194. doi: 10.3390/v2051190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen F, Duan L, Zhu M, Bagasra O, Pomerantz RJ. Targeting human immunodeficiency virus type 1 reverse transcriptase by intracellular expression of single-chain variable fragments to inhibit early stages of the viral life cycle. J Virol. 1996;70(6):3392–3400. doi: 10.1128/jvi.70.6.3392-3400.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleiman D, Goldschmidt V, Barraud P, Marquet R, Paillart J-C, Tisné C. Initiation of HIV-1 reverse transcription and functional role of nucleocapsid-mediated tRNA/viral genome interactions. Virus Res. 2012 doi: 10.1016/j.virusres.2012.06.006. http://dx.doi.org/10.1016/j.virusres.2012.06.006. [DOI] [PubMed]
- Solbak SM, Reksten TR, Wray V, Bruns K, Horvli O, Raae AJ, Henklein P, Henklein P, RR, Mitzner D, Schubert U, Fossen T. The intriguing cyclophilin A-HIV-1 Vpr interaction: prolyl cis/trans isomerisation catalysis and specific binding. BMC Struct Biol. 2010;10(31):1–15. doi: 10.1186/1472-6807-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solbak SM, Wray V, Horvli O, Raae AJ, Flydal MI, Henklein P, Henklein P, Nimtz M, Schubert U, TF The host-pathogen interaction of human cyclophilin A and HIV-1 Vpr requires specific N-terminal and novel C-terminal domains. BMC Struct Biol. 2011;11(49):1–16. doi: 10.1186/1472-6807-11-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Craigie R. The road to chromatin - nuclear entry of retroviruses. Nat Rev Microbiol. 2007;5(3):187–196. doi: 10.1038/nrmicro1579. [DOI] [PubMed] [Google Scholar]
- Takeda E, Murakami T, Matsuda G, Murakami H, Zako T, Maeda M, Aida Y. Nuclear exportin receptor CAS regulates the NPI-1-mediated nuclear import of HIV-1 Vpr. PLoS One. 2011;6(11):e27815. doi: 10.1371/journal.pone.0027815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasara T, Maga G, Hottiger MO, Hübscher U. HIV-1 reverse transcriptase and integrase enzymes physically interact and inhibit each other. FEBS Lett. 2001;507(1):39–44. doi: 10.1016/s0014-5793(01)02945-3. [DOI] [PubMed] [Google Scholar]
- Thomas JA, Bosche WJ, Shatzer TL, Johnson DG, Gorelick RJ. Mutations in human immunodeficiency virus type 1 nucleocapsid protein zinc fingers cause premature reverse transcription. J Virol. 2008;82(19):9318–9328. doi: 10.1128/JVI.00583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JA, Gagliardi TD, Alvord WG, Lubomirski M, Bosche WJ, Gorelick RJ. Human immunodeficiency virus type 1 nucleocapsid zinc-finger mutations cause defects in reverse transcription and integration. Virology. 2006;353(1):41–51. doi: 10.1016/j.virol.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Thomas JA, Gorelick RJ. Nucleocapsid protein function in early infection processes. Virus Res. 2008;134(1–2):39–63. doi: 10.1016/j.virusres.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JA, Shatzer T, Gorelick RJ. Blocking premature reverse transcription fails to rescue the HIV-1 nucleocapsid-mutant replication defect. Retrovirology. 2011;8(46) doi: 10.1186/1742-4690-8-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trono D. Partial reverse transcripts in virions from human immunodeficiency and murine leukemia viruses. J Virol. 1992;66(8):4893–4900. doi: 10.1128/jvi.66.8.4893-4900.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaneja MA, Kane BP, Johnson DG, Gorelick RJ, Henderson LE, Casas-Finet JR. Binding properties of the human immunodeficiency virus type 1 nucleocapsid protein p7 to a model RNA: elucidation of the structural determinants for function. J Mol Biol. 1999;287(1):59–75. doi: 10.1006/jmbi.1998.2521. [DOI] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423(6937):309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- Wacharapornin P, Lauhakirti D, Auewarakul P. The effect of capsid mutations on HIV-1 uncoating. Virology. 2007;358(1):48–54. doi: 10.1016/j.virol.2006.08.031. [DOI] [PubMed] [Google Scholar]
- Warrilow D, Harrich D. HIV-1 replication from after cell entry to the nuclear periphery. Curr HIV Res. 2007;5(3):293–299. doi: 10.2174/157016207780636579. [DOI] [PubMed] [Google Scholar]
- Warrilow D, Tachedjian G, Harrich D. Maturation of the reverse transcription complex: putting the jigsaw together. Rev Med Virol. 2009;19(6):324–337. doi: 10.1002/rmv.627. [DOI] [PubMed] [Google Scholar]
- Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460(7256):711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinderg JB, Matthews TJ, Cullen BR, Malim MH. Productive human immunodeficiency virus type 1 infection of nonproliferating human monocytes. J Exp Med. 1991;174(6):1477–1482. doi: 10.1084/jem.174.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker R, Hohenberg H, Tessmer U, Huckhagel C, Kräusslich HG. Biochemical and structural analysis of isolated mature cores of human immunodeficiency virus type 1. J Virol. 2000;74(3):1168–1177. doi: 10.1128/jvi.74.3.1168-1177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MC, Gorelick RJ, Musier-Forsyth K. Specific zinc-finger architecture required for HIV-1 nucleocapsid protein’s nucleic acid chaperone function. Proc Natl Acad Sci USA. 2002;99(13):8614–8619. doi: 10.1073/pnas.132128999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Henderson LE, Copeland TD, Gorelick RJ, Bosche WJ, Rein A, Levin JG. Human immunodeficiency virus 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J Virol. 1996;70(10):7132–7142. doi: 10.1128/jvi.70.10.7132-7142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander K, Sherman MP, Tessmer U, Bruns K, Wray V, Prechtel AT, Schubert E, Henklein P, Luban J, Neidleman J, Greene WC, Schubert U. Cyclophilin A interacts with HIV-1 Vpr and is required for its functional expression. J Biol Chem. 2003;278(44):43202–43213. doi: 10.1074/jbc.M305414200. [DOI] [PubMed] [Google Scholar]
- Zennou V, Petit C, Guetard D, Nerhbass U, Montagnier L, Charneau P. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell. 2000;101(2):172–185. doi: 10.1016/S0092-8674(00)80828-4. [DOI] [PubMed] [Google Scholar]
- Zhang H, Dornadula G, Pomerantz RJ. Endogenous reverse transcription of human immunodeficiency virus type 1 in physiological microenvironments: an important stage for viral infection of nondividing cells. J Virol. 1996;70(5):2809–2824. doi: 10.1128/jvi.70.5.2809-2824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang Y, Spicer TP, Abbott LZ, Abbott M, Poiesz BJ. Reverse transcription takes place within extracellular HIV-1 virions: potential biological significance. AIDS Res Hum Retroviruses. 1993;9(12):1287–1296. doi: 10.1089/aid.1993.9.1287. [DOI] [PubMed] [Google Scholar]
- Zhang S, Pointer D, Singer G, Feng Y, Park K, Zhao LJ. Direct binding to nucleic acids by Vpr of human immunodeficiency virus type 1. Gene. 1998;212(2):157–166. doi: 10.1016/s0378-1119(98)00178-4. [DOI] [PubMed] [Google Scholar]
- Zhao RY, Li G, Bukrinsky MI. Vpr-Host interactions during HIV-1 viral life cycle. J Neuroimmune Pharmacol. 2011;6(2):216–229. doi: 10.1007/s11481-011-9261-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Sokolskaja E, Jolly C, James W, Cowley SA, Fassati A. Transportin 3 promotes a nuclear maturation step required for efficient HIV-1 integration. PLoS Pathog. 2011;7(8):e1002194. doi: 10.1371/journal.ppat.1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K, Dobard C, Chow SA. Requirement for integrase during reverse transcription of human immunodeficiency virus type 1 and the effect of cysteine mutations of integrase on its interactions with reverse transcriptase. J Virol. 2004;78(10):5045–5055. doi: 10.1128/JVI.78.10.5045-5055.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]