Summary
The induction of pluripotency or trans-differentiation of one cell type to another can be accomplished with cell lineage-specific transcription factors. Here we report that repression of a single RNA binding protein PTB, which occurs during normal brain development via the action of miR-124, is sufficient to induce trans-differentiation of fibroblasts into functional neurons. Besides its traditional role in regulated splicing, we show that PTB has a previously undocumented function in the regulation of microRNA functions, suppressing or enhancing microRNA targeting by competitive binding on target mRNA or altering local RNA secondary structure. A key event during neuronal induction is the relief of PTB-mediated blockage of microRNA action on multiple components of the REST complex, thereby de-repressing a large array of neuronal genes, including miR-124 and multiple neuronal-specific transcription factors, in non-neuronal cells. This converts a negative feedback loop to a positive one to elicit cellular reprogramming to the neuronal lineage.
Introduction
Feedback and feed-forward circuits are key regulatory strategies for maintaining gene expression programs in specific cell types, and factors that alter the homeostatic balance of these circuits can induce enduring program switches in cell differentiation and development (Zernicka-Goetz et al., 2009). Many regulatory pathways use such mechanisms to control the output of biological responses through gene networks in transcription (Amit et al., 2009), RNA metabolism (Coutinho-Mansfield et al., 2007; Lareau et al., 2007), signal transduction (Carracedo and Pandolfi, 2008), macromolecular synthesis and degradation (Auld and Silver, 2006).
MicroRNA have emerged as key mediators in various modes of feedback and feed-forward regulation (Leung and Sharp, 2010). MicroRNA are expressed in all higher eukaryotes, ~800 in humans, and a recent estimate suggests that up to 60% of human genes may be subjected to microRNA control (Bartel, 2009). In many cases, homeostasis is achieved by feedback controls in which specific transcription factors or RNA binding proteins regulate the expression and biogenesis of microRNAs, which in turn suppress the expression of their regulators.
Neuronal differentiation is a well-studied paradigm as a consequence of transcription reprogramming (Li and Jin, 2010). Recent studies show that a set of neuronal-specific transcription factors is sufficient to trans-differentiate fibroblasts into functional neurons (Caiazzo et al., 2011; Kim et al., 2011; Pang et al., 2011; Qiang et al., 2011; Vierbuchen et al., 2010; Yang et al., 2011). Several neuronal-specific microRNA, such as miR-9/9* and miR-124, also play key roles in this process (Yoo et al., 2011). miR-124 is a well known regulator of the transcription silencing complex built on REST, which represses a large array of neuronal-specific genes in non-neuronal cells; this includes miR-124 itself, thus forming an auto-regulatory loop during neuronal differentiation (Ballas et al., 2005; Conaco et al., 2006). Forced expression of miR-124 can drive differentiation of neural progenitor cells to neurons (Cheng et al., 2009) and C2C12 cells to become neuronal-like cells (Watanabe et al., 2004).
Regulated RNA processing also plays a critical role in neuronal differentiation. The polypyrimidine tract binding protein PTB and its homolog nPTB undergo a programmed switch during neuronal differentiation (Boutz et al., 2007; Makeyev et al., 2007; Zheng et al., 2012). miR-124 is able to modulate such switch by reducing PTB, thereby reprogramming an array of neuronal-specific alternative splicing events and forced expression of PTB is able to block miR-124 induced neuronal differentiation (Makeyev et al., 2007). However, it has been unclear whether the PTB/nPTB switch is sufficient to initiate neuronal differentiation, and if so, which specific PTB/nPTB-regulated splicing events contribute to such cell fate switch.
We previously reported global PTB-RNA interactions in the human genome (Xue et al., 2009). In PTB-depleted cells, we unexpectedly observed conversion of diverse cell types into neuronal-like cells. In addition to induced alternative splicing events, we found an extensive involvement of PTB in the regulation of microRNA targeting either through direct competition or induced switch of local RNA secondary structure. A key event is the activation of the miR-124/REST loop in which PTB not only serves as a target, but also acts as a potent regulator. Consequently, regulated PTB expression induces massive reprogramming at both the splicing and microRNA levels to drive the cell fate decision towards the neuronal lineage.
Results
PTB down-regulation switches multiple cell types to neuronal-like cells
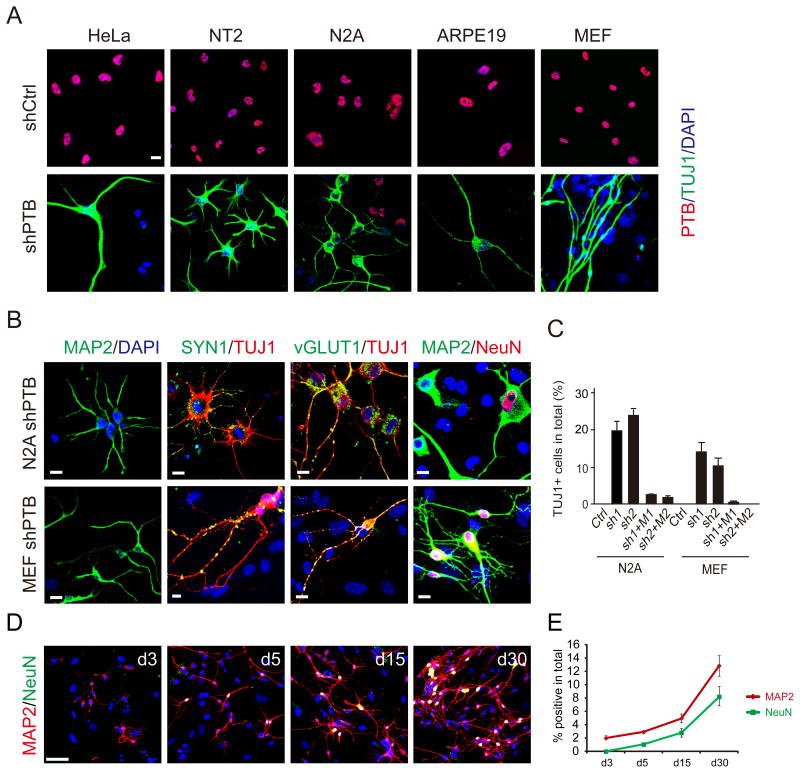
We attempted to use specific shRNAs to stably knock down PTB in order to systematically analyze PTB-regulated splicing. As expected, shPTB induced nPTB expression in HeLa cells (Figure S1A). We noted a slow growth phenotype of shPTB-treated cells, which was also seen by others (He et al., 2007). Strikingly, many PTB-depleted HeLa cells exhibited neurite outgrowth and further analysis revealed the expression of several neuron markers, including class III β-tubulin (known as Tuj1) and MAP2 (Figure 1A and Figure S1A), suggesting that PTB knockdown converted highly transformed HeLa cells to neuronal-like cells.
Figure 1. Differentiation of diverse cell types into neuronal-like cells in response to PTB knockdown.
(A) Induction of neuronal morphology and the expression of the neuronal marker Tuj1 in multiple cell types in response to depletion of PTB. Scale bar: 20 μm.
(B) Characterization of two cell types (N2A and MEF) with additional neural markers. Typical punctate staining is evident (yellow) with antibodies against Synapsin and vGLUT1. Scale bar: 20 μm.
(C) Quantification of induced neuronal-like cells derived from N2A and MEFs. The data were based on positive Tuj1 stained cells divided by initial plating cells in response to two separate shPTBs (sh1 and sh2). The effect could be efficiently rescued with the corresponding shRNA-resistant PTB expression units that contain mutations in the corresponding target sites (M1 and M2). Data are shown as mean ± SD.
(D) Time course analysis of neuronal induction on shPTB-treated MEFs after switching to N3 media. MAP2 and NeuN were stained at indicated time points. Scale bar: 60 μm
(E) Quantified temporal profile of PTB knockdown-induced neurons. Data shown as mean ± SD are based on 4 equivalent areas shown in D. See also Figure S1.
We extended this analysis to multiple cell types of diverse origin, including human embryonic carcinoma stem cells (NT2), mouse neural progenitor cells (N2A), human retinal epithelial cells (ARPE19), and primary mouse embryo fibroblasts (MEFs). Upon PTB knockdown (Figure S1B), all of these cells exhibited a neuronal-like morphology and showed strong Tuj1 staining (Figure 1A). Neuronal committed N2A and NT2 cells were potently induced to show typical neuronal morphology in ~5 days after PTB knockdown and develop more complex morphology after the cells were switched to N3 media containing a set of neural growth factors for 3 to 5 days. ARPE19 and MEFs took ~2 weeks to develop typical neuronal morphology in N3 media. Control shRNA treatment had no effect under these conditions.
We further characterized two of these cell lines (N2A and MEFs) by examining additional neural markers, including Synapsin 1 (SYN1), vGLUT1 and NeuN (Figure 1B). SYN1 and vGLUT1 showed a typical punctate staining pattern on Tuj1-positive cells, but not on undifferentiated cells in the same field (Figure 1B). We also detected strong staining of GABA channel receptors on these derived neurons (data not shown). As previously described (Vierbuchen et al., 2010), immunostaining and RT-PCR analyses ruled out potential contamination of our starting MEFs with neural crest cells (Figure S1C and S1D).
Both N2A cells and MEFs were efficiently converted by two distinct shRNAs against PTB to neuronal-like cells (Figure S1E). Importantly, the effect of specific shPTB molecules could each be rescued with the shRNA-resistant PTB expression unit that carries synonymous mutations (M1 or M2) in their targeting sites, thus ruling out potential off-target effects (Figure 1C). Time-course analysis demonstrated that PTB knockdown progressively converted MEFs to neuronal-like cells with complex morphology (Figure 1D and 1E). These data strongly suggest that PTB down-regulation potently induced these cells to differentiate (in the case of N2A cells) or trans-differentiate (in the case of MEFs) into neurons.
MEF-derived neurons are functional with synaptic activities
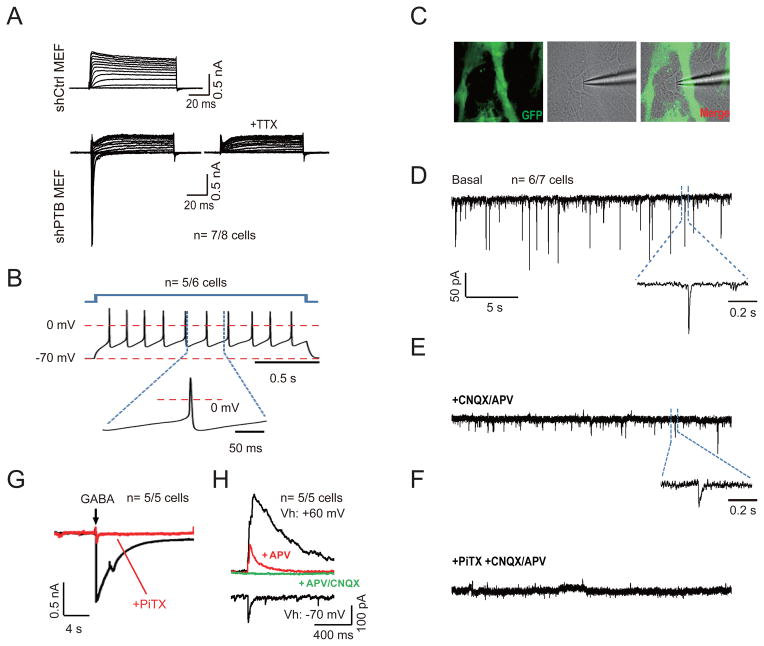
To determine the functionality of differentiated cells, we patch-clamped both shPTB-induced neurons from N2A cells and MEFs. We observed that 11 out of 12 N2A cell-derived neurons exhibited fast inward Na+ currents and action potential upon membrane depolarization (Figure S2A) and that 7 out of 8 shPTB-induced MEFs showed a similar response, which could be blocked by the sodium channel inhibitor TTX (Figure 2A). Both of these induced cell types showed depolarization-induced Ca++ influx (Figure S2B and S2C). We next determined whether MEF-derived neurons are fully functional in the presence of primary astrocytes, which is known to be essential for trans-differentiated MEFs to become synaptically competent (Vierbuchen et al., 2010). After co-culture for a week with freshly isolated astrocytes free of contaminating neurons from the brain of a GFP-transgenic rat, we detected repetitive action potentials of varying frequencies driven by current pulse in 5 out of 6 MEFs-derived neurons (Figure 2B). Importantly, we recorded synaptic activities on 6 out of 7 such neurons examined (Figure 2C and 2D).
Figure 2. Synaptic activities on neurons derived from shPTB-induced MEFs.
(A) Representative traces of whole-cell currents on control shRNA-treated (top) and shPTB-treated (bottom) MEFs. Only shPTB-treated MEFs exhibited fast inward sodium currents, which could be blocked by 1 μM sodium channel inhibitor TTX.
(B) Representative trace of action potentials in response to step current injections on shPTB-induced neurons after co-culturing with rat glial cells.
(C) Image of an shPTB-induced neuron co-cultured with GFP-marked rat glial cells. Recording electrode was patched on the shPTB-induced neuron (middle and right).
(D to F) Representative traces of spontaneous postsynaptic currents on shPTB-induced neurons (D). The cell was held at −70mV, revealing events of various amplitudes and frequencies. The insert shows a representative trace of synaptic response. Glutamategic synaptic currents were blocked with 20 μM CNQX plus 50 μM APV (E). The insert highlights the remaining GABA current. GABA currents were blocked with 50 μM PiTX (F).
(G) Induction of GABA currents by focal application of 1 mM GABA, which could be blocked by PiTX (red).
(H) Representative trace of synaptic currents recorded on shPTB-induced neurons. Vh: holding potential. AMPA-R mediated EPSC was recorded at −70mV. Blockage of Mg++ to NMDA-R was relieved at +60mV, revealing both AMPA and NMDA EPSCs, which could be sequentially blocked with 50 μM APV (antagonist of NMDA-type glutamate receptors) and 20 μM CNQX (antagonist of AMPA receptors).
The number of cells that show the representative response against total cells examined is indicated in each panel. See also Figure S2.
The detected postsynaptic currents likely reflect both glutamatergic and GABAergic responses, because CNQX+APV (antagonists of glutamatergic channel receptors) and Picrotoxin (PiTX, antagonist of GABAA channel receptors) could sequentially block the expected signals (Figure 2E and 2F). We further recorded GABA-induced, PiTX-sensitive currents upon focal application of GABA (Figure 2G). In the presence of PiTX, we detected AMPA receptors-mediated excitatory postsynaptic currents (EPSC) with fast kinetics when holding the neuron at −70mV with an external solution containing 2 mM Mg++ (Figure 2H), which is known to inhibit NMDA EPSC with slow kinetics (Nowak et al., 1984). By holding the neuron at +60mV to relieve the inhibitory effect of Mg++, we detected both NMDA and AMPA EPSCs, which could be progressively blocked by the NMDA channel inhibitor APV and the AMPA channel antagonist CNQX (Figure 2H). These data demonstrated that shPTB had trans-differentiated MEFs into functional neurons.
PTB regulates the expression of many neuronal genes in non-neuronal cells
Because of the induced neuronal morphology and the availability of the genome-wide PTB-RNA interaction map on HeLa cells (Xue et al., 2009), we initially took this cell type as a surrogate model to understand shPTB-induced cellular reprogramming. We identified by RNA-seq a large number of up- or down-regulated genes induced by shPTB (Figure S3A) and we further confirmed a panel of these events by RT-qPCR (Figure S3B). Gene Ontology (GO) analysis showed that many such altered genes were linked to neuronal functions (Figure S3C). These observations indicate that PTB is extensively involved in the regulation of neuronal genes in non-neuronal cells.
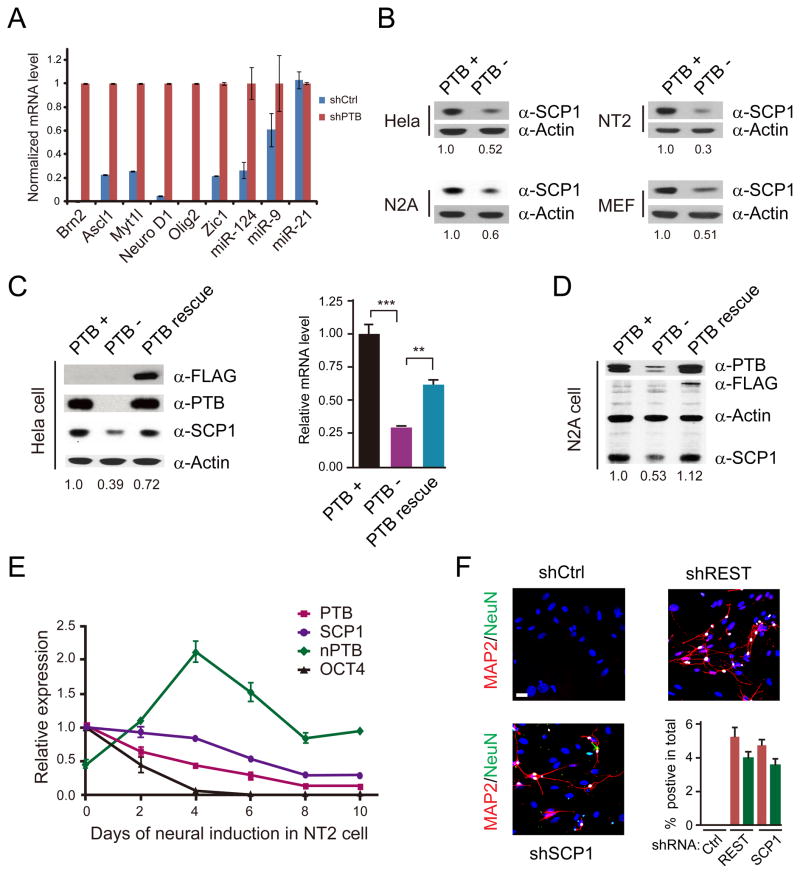
We noted the induction of Brn2 and Myt1l, which correspond to 2 out of 5 key transcription factors previously shown to be sufficient to induce trans-differentiation of fibroblasts into neurons (Vierbuchen et al., 2010). Because HeLa cells have a severely re-arranged genome, we performed a focused analysis on MEFs by RT-qPCR (Figure 3A), detecting the induction of all five critical transcription factors (Ascl1, Brn2, Myt1l, Zic1, and Olig2) as well as NeuroD1 known to enhance neurogenesis in human fibroblasts (Pang et al., 2011). We also observed the induction of miR-124 and miR-9 (Figure 3A), which have been shown to synergize with neuronal-specific transcription factors in promoting neurogenesis (Yoo et al., 2011). These data explain the compatible functionality of shPTB-induced neurons to that converted by a set of lineage-specific transcription factors.
Figure 3. De-repression of neuronal-specific genes in response to PTB knockdown.
(A) RT-qPCR analysis of a panel of transcription factors and microRNAs in shPTB-treated MEFs. Data are normalized against Actin; miR-21 served as a negative control.
(B) Down-regulation of SCP1 in multiple cell types determined by Western blotting.
(C and D) Rescue of SCP1 expression in PTB knockdown cells by an shRNA-resistant PTB in HeLa (C) and N2A (D) cells.
(E) Time course analysis of neural induction by retinoic acid (RA) on NT2 cells analyzed by RT-qPCR. Oct4 was analyzed as a control. Data are shown as mean ± SD.
(F) Induction of neuronal differentiation on MEFs with shRNA against SCP1 or REST. The induction efficiency was calculated based on the number of cells with positive MAP2 and NeuN staining divided by total plating cells. Data are shown as mean ± SD. See also Figure S3.
REST activity contributes a key part to the shPTB-induced neuronal program
The REST complex is known to repress a large set of neuronal genes in non-neuronal cells (Johnson et al., 2007). Interestingly, we noted that all induced transcription factors examined in Figure 3A contain significant REST ChIP-seq signals from the ENCODE data on C2C12 cells. We confirmed strong REST binding by ChIP-qPCR on most of these genes in MEFs, and as expected, REST knockdown induced the expression of these genes (Figure S3D and S3E). These data suggest that the function of the REST complex might be compromised in shPTB-treated MEFs.
To determine how the RSET complex was compromised, we examined the response of REST and REST co-factors to shPTB in HeLa cells. While REST expression was little affected from our RNA-seq analysis, we found that SCP1, a Pol II Ser5 phosphatase associated with the REST complex (Yeo et al., 2005), was significantly down regulated by shPTB in multiple cell types with induced neuronal morphology (Figure 3B). The effect of shPTB on SCP1 expression could be rescued on two of these cell types we examined (Figure 3C and 3D). During the course of retinoic acid-induced neural differentiation on NT2 cells, we observed that SCP1 expression was gradually reduced, which closely tracked PTB down-regulation and nPTB induction (Figure 3E). These findings suggest that PTB-regulated expression of the REST co-factor SCP1 may play a key role in neuronal differentiation under physiological conditions.
Recent studies suggest that REST is required for maintaining the population of neural stem cells (Gao et al., 2011) and genetic inactivation of REST does not efficiently turn fibroblasts into neurons, despite the induction of some neuronal genes (Aoki et al., 2012). However, a dominant negative SCP1 was able to efficiently drive neuronal differentiation on P19 cells (Yeo et al., 2005). We thus wished to directly test the contribution of SCP1 to shPTB-induced neurogenesis under our experimental conditions and we similarly tested REST for comparison. We found that both shSCP1 and shREST, but not control shRNA, were able to trigger neuronal differentiation on MEFs (Figure 3F). The neural induction efficiency by shSCP1 and shREST was similar, but lower than that induced by shPTB (compared Figure 1C and Figure 3F), indicating that other PTB-regulated events may additionally contribute to the induction of neurogenesis. The reason for efficient induction of neurogenesis with shPTB or shRNA against REST or a REST co-factor gene may be due to gradual switch of these cell lineage-specific regulators, which may mimic relevant developmental processes (see Discussion).
PTB-regulated splicing likely facilitates the development of the neural program
PTB is best known for its role in regulated splicing (Makeyev et al., 2007), which is consistent with our RNA-seq data from HeLa cells (Figure S3F). However, it has been unclear which altered splicing event(s) contributes to the development of the neuronal lineage in PTB-depleted cells. In light of the recent finding that the REST gene itself undergoes alternative splicing to produce a truncated, non-functional isoform (REST4) (Raj et al., 2011), we asked whether this splicing event might be subjected to PTB regulation. We detected some induction of the REST4 isoform in N2A cells, but not in other cell types we examined (Figure S3G).
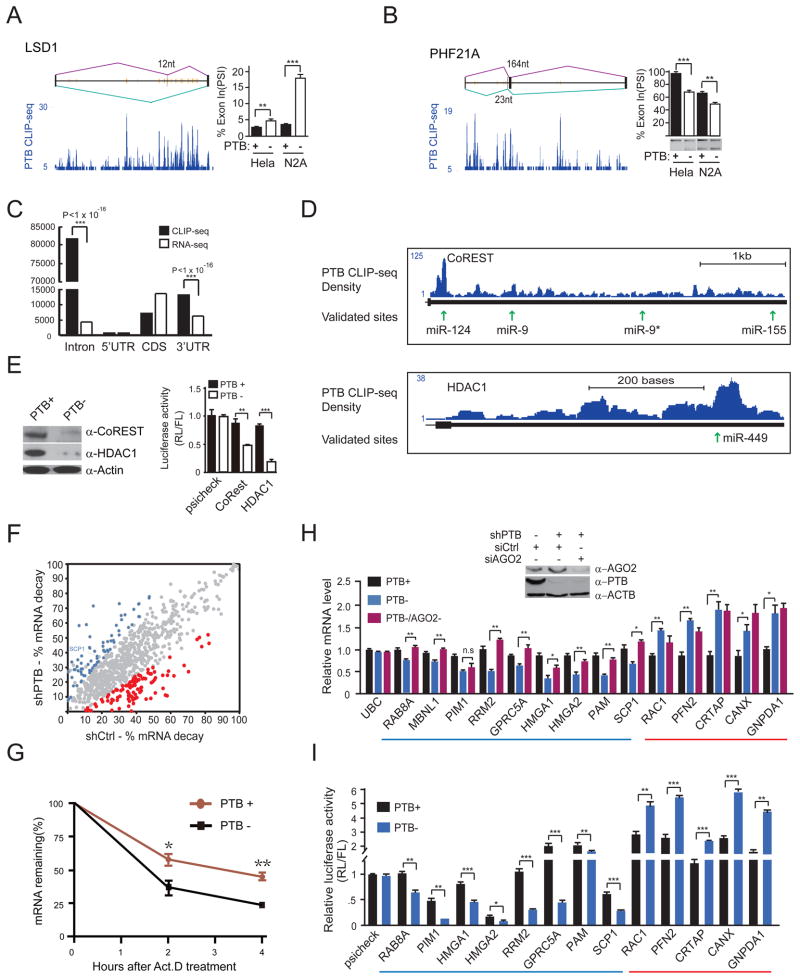
During the course of this investigation, we detected induced alternative splicing of two key genes, LSD1 (a histone lysine demethylase, a component of the REST complex) and PHF21A (a component of the histone deacetylase HDAC1 complex) upon PTB knockdown in HeLa and N2A cells (Figure 4A and 4B). This is consistent with multiple PTB binding events around the regulated exon in both cases from our published CLIP-seq data (Xue et al., 2009). Importantly, induced skipping of the alternative exon in LSD1 has recently been shown to affect neurite morphogenesis/maturation (Zibetti et al., 2010). Although it remains to be determined whether induced PHF21A splicing has any functional consequence, these findings suggest that some PTB-regulated splicing events may directly contribute to the neuronal phenotype observed in PTB down-regulated cells.
Figure 4. PTB-regulated splicing and RNA stability.
(A and B) PTB-regulated alternative splicing of LSD1 and PHF21A. The CLIP-seq mapped PTB binding events (blue) are shown along with deduced PTB binding peaks (orange lines) on each gene model. PTB knockdown induced alternative splicing was determined by RT-qPCR in the case of LSD1 and by semi-quantitative RT-PCR in the case of PHF21A.
(C) Relative enrichment of PTB binding in intronic and 3′UTR regions. Significant enrichment of PTB binding events is indicated by the p-values in each case.
(D) PTB binding on two REST component genes, showing that multiple PTB binding peaks overlap with validated targeting sites by miR-124 and miR-9.
(E) Reduced CoREST and HDAC1 proteins (left) and diminished reporter activities (right) in PTB-depleted HeLa cells.
(F) Genome-wide analysis of PTB-regulated RNA stability. The calculated decay rate was compared in the presence (shCtrl-treated) or absence (shPTB-treated) of PTB. Genes with increased and decreased decay are highlighted in red and blue, respectively, based on triplicated RNA-seq data (p<0.05).
(G) Accelerated SCP1 mRNA decay detected by RT-qPCR in PTB-depleted HeLa cells.
(H) The effect of knocking down PTB (PTB−) or both PTB and Ago2 (PTB−/Ago2−) on the expression of a panel of genes that show PTB and Ago2 binding events in their 3′UTRs. A gene (UBC) without binding evidence for PTB and Ago2 severed as a negative control.
(I) Re-capture of PTB-dependent regulation with the 3′UTR of individual genes analyzed in H. Note that the MBNL1 gene was not included in this analysis because its 3′UTR is too long to clone.
Data in individual panels are shown as mean ± SD. **p<0.01; ***p<0.001. See also Figure S4.
PTB is involved in the RNA stability control of key neuronal genes
Because many PTB-affected genes could not be explained by induced splicing, we searched for other potential mechanisms. PTB has been reported to regulate RNA stability in multiple cases through C/U-rich sequences in the 3′UTR, but the mechanism has remained elusive (Knoch et al., 2004; Kosinski et al., 2003; Pautz et al., 2006; Porter et al., 2008; Tillmar and Welsh, 2002; Woo et al., 2009). By examining the PTB-RNA map (Xue et al., 2009), we noted extensive PTB binding events in the 3′UTR of all of those reported genes (Figure S4A). Globally, PTB binding on both intronic regions and 3′UTRs are more prevalent than 5′UTRs and exons compared to the RNA-seq signals in these regions (Figure 4C). However, PTB binding alone does not seem to be sufficient to regulate RNA stability, as we showed by using an MS2-based tethering assay (Figure S4B-4D). We noted that all of those previously mapped PTB binding sites localize closely with predicted microRNA targeting sites (Figure S4A), raising an intriguing possibility that PTB may regulate RNA stability via functional interplay with microRNA.
Multiple PTB binding peaks are evident in the 3′UTR of CoREST and HDAC1 (Figure 4D), both of which have been implicated in neurogenesis as key components of the REST complex (Dovey et al., 2010; Hsieh et al., 2004). These mapped PTB binding sites are coincident with three previously validated targeting sites by miR-124, miR-9 and miR-449 (Baudet et al., 2012; Packer et al., 2008; Selbach et al., 2008). Indeed, PTB knockdown in HeLa cells dramatically reduced the expression of both CoREST and HDAC1 at the protein level and diminished the luciferase activity of the reporters containing the 3′UTR of these genes (Figure 4E). These data strongly suggest that PTB down-regulation caused dismantling of multiple components of the REST complex, which likely contribute in a collective fashion to the induction of neuronal-specific genes in non-neural cells.
PTB regulates RNA stability in conjunction with microRNA
From this point, we used HeLa cells to understand the mechanism underlying PTB-regulated gene expression mainly because of the experimental manipulability of the cell type, although it is important to emphasize that caution must be taken when extrapolating deduced molecular mechanism from one cell type to another. To determine how extensively PTB is involved in RNA stability control, we performed RNA-seq on mock-depleted and PTB-depleted cells before (T0) or after blocking transcription with Actinomycin D for 4 hours (T4). This allowed us to calculate mRNA decay [(T0–T4)/T0 ×100%] and determine how such decay might be influenced by PTB for each expressed gene in the human genome. We identified a total of 142 genes that showed significantly increased (red dots in Figure 4F) or decreased (blue dots in Figure 4F) decay (p<0.05) in response to PTB knockdown. Interestingly, SCP1 is among these genes, which was further confirmed by RT-qPCR (Figure 4G).
We next selected a panel of PTB-bound genes to determine whether these PTB-regulated events were dependent on the microRNA machinery (Figure 4H). We found that many PTB down-regulated genes (blue underlined in Figure 4H) lost the response to PTB knockdown when Ago2 was inactivated. We found no or little effect on several PTB up-regulated genes after Ago2 RNAi (red underlined in Figure 4H), consistent with the possibility that microRNA no longer acted on these genes in PTB-depleted cells. To determine whether the 3′UTR of PTB-regulated genes might mediate the response to PTB knockdown, we constructed a series of luciferase reporters containing the 3′UTR of these genes, finding that the reporters re-captured PTB-dependent suppression or enhancement (Figure 4I). These data illustrate that PTB is involved in the regulation of RNA stability and/or translational control in conjunction with the action of microRNA on the 3′UTR of many genes.
The 3′UTR of SCP1 contains multiple microRNA targeting sites
We used SCP1 as a model to investigate the functional interplay between PTB and microRNA. We compiled PTB and Ago2 CLIP-seq signals (see below in Figure 7) in the 3′UTR of this gene before and after PTB knockdown in order to select relevant regions for functional analysis. We detected one lost and at least six gained Ago2 binding events in the 3′UTR of SCP1 in response to PTB knockdown (Figure 5A). In the F1 region, for example, we noted three PTB binding sites that weakly overlap with the mapped Ago2 binding sites before PTB knockdown, and in response to PTB knockdown, one of these Ago2 CLIP peaks was significantly enhanced, which is near the predicted target site for miR-124. The predicted microRNA targeting sites in the F1 region are each flanked by a PTB binding consensus motif (Figure 5A, lower panel). Multiple mapped PTB and Ago2 binding events also show various degrees of overlap with the predicted targeting sites for miR-124 and miR-96 in the F2 and F3 regions.
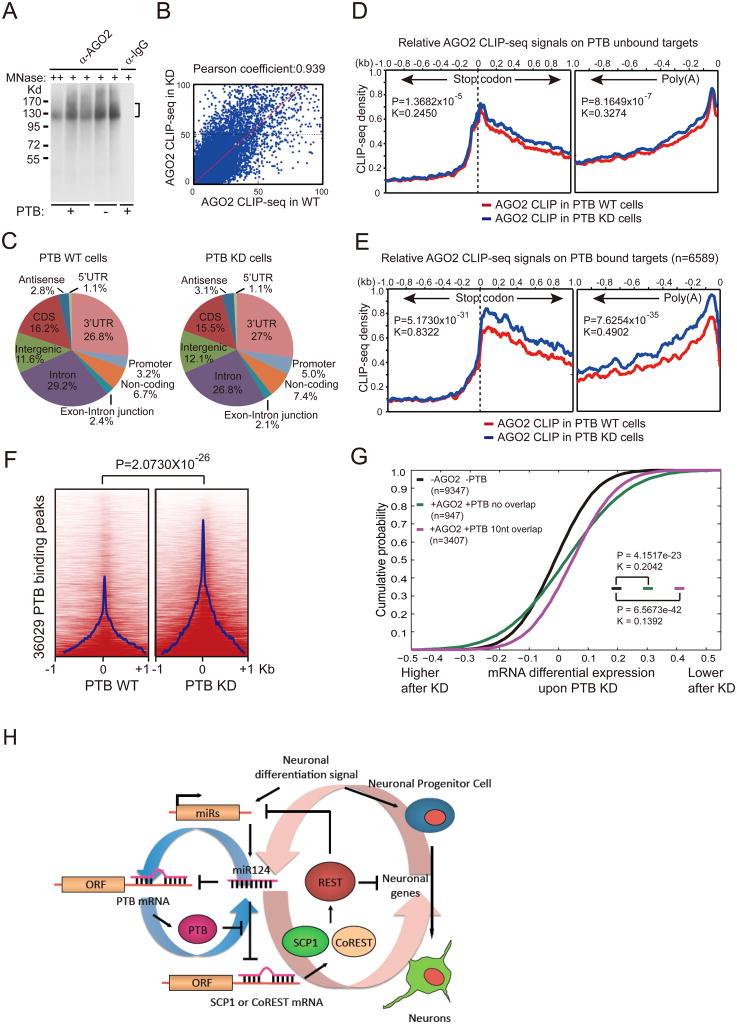
Figure 7. Global analysis of Ago2 binding in response to PTB knockdown.
(A) CLIP signals detected with anti-Ago2 before and after PTB knockdown. No signal was detected with IgG control.
(B) Comparison between the two Ago2 CLIP-seq datasets in 1kb windows across the human genome before and after PTB depletion.
(C) Genomic distribution of Ago2 binding events before (left) and after (right) PTB knockdown, showing prevalent Ago2 binding in the 3′UTR region.
(D and E) Ago2 binding in the 3′UTR of PTB unbound (D) and bound (E) targets before (red) and after (blue) PTB knockdown. Dramatic differences were detected on PTB bound targets (n=6589) in E, which compares to much less responses on a similar number of randomly selected PTB unbound targets in D. Statistical significance was determined for the differences on both the stop codon and poly (A) sides by two-tailed Kolmogorov-Smirnov test with both the p- and k-values shown in the insert.
(F) Induction of significant Ago2 binding on and near the PTB binding sites. The p-value for the differences is indicated on the top.
(G) Functional correlation between PTB/microRNA interplay and gene expression. Genes with induced and repressed expression are plotted in a cumulative fashion. Statistical significance was determined by KS-test.
(H)Model for the PTB-regulated miR124-REST loop. See also Figure S7.
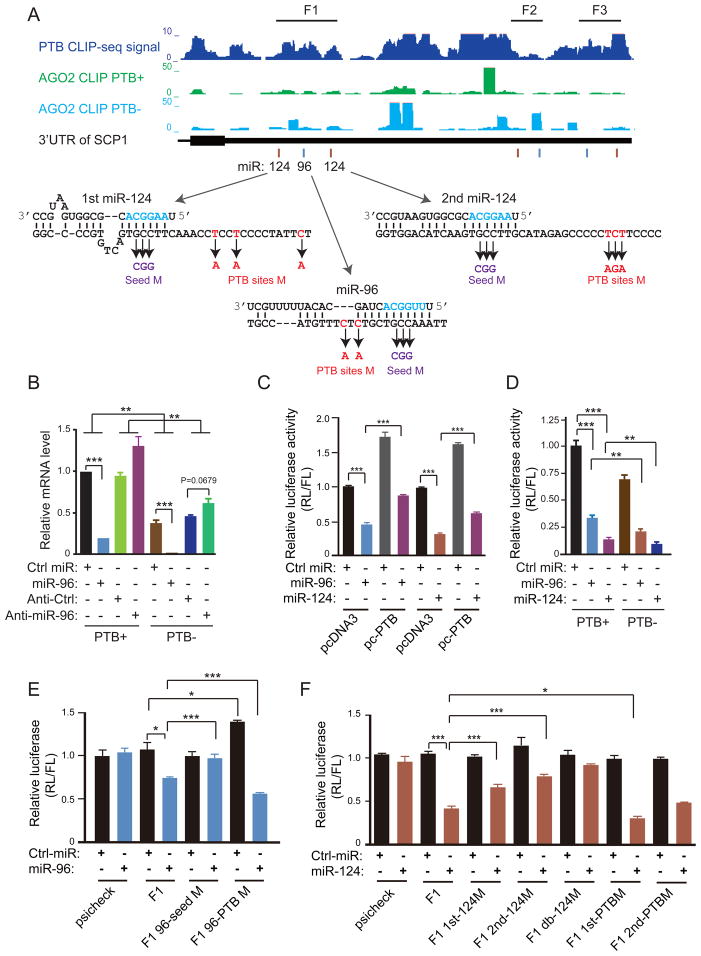
Figure 5. PTB competition with microRNA targeting in the 3′UTR of SCP1.
(A) The mapped PTB binding events in the 3′UTR of the SCP1 gene (top). Above the gene model also show the mapped Ago2 binding density before (green) and after (cyan) PTB knockdown in HeLa cells. Below the gene model indicate multiple predicted microRNA target sites for miR-124 (brown lines) and miR-96 (cyan lines). Arrow-highlighted are deduced base-paired regions between the mRNA and individual microRNAs. Also illustrated are the mutations in the 3′UTR of the SCP1 gene that correspond to the sequence on the microRNA targeting sites in the seed region (violet) or on the PTB binding site (red) in each case.
(B) The effects on the endogenous SCP1 mRNA by overexpressed miR-96 and its antagomir before and after PTB knockdown.
(C) Blockage of the effect of overexpressed miR-96 and miR-124 by PTB overexpression on the luciferase reporter containing the F1 fragment from the SCP13′UTR.
(D) Enhanced effect of overexpressed miR-96 and miR-124 in response to PTB knockdown on the luciferase reporter containing the F1 fragment from the SCP13′UTR.
(E) The requirement for the seed region in the miR-96 target site to respond to overexpressed miR-96. While the mutations in the PTB binding site impaired miR-96 targeting (compared lanes 3 and 7), the mutants enhanced the overall effect of miR-96 on the luciferase reporter (compare lanes 3/4 and lanes 7/8).
(F) Contribution of individual miR-124 target sites in the SCP1 F1 region to microRNA-mediated down-regulation of the luciferase activity. The mutations in the seed region of miR-124 targeting sites progressively reduced the response to overexpressed miR-124 (compare lanes 3 to 10). The mutations in the PTB binding site near the first miR-124 targeting sites enhanced miR-124 mediated down-regulation (compare lanes 4 and 12). The statistical significance in comparing different groups was determined by paired t-test.
Data in individual panels are shown as mean ± SD. **p<0.01; ***p<0.001. See also Figure S5.
Previous studies showed that forced miR-124 expression could switch the gene expression profile towards that of brain in HeLa cells (Lim et al., 2005). Relevant to the present study, miR-124 has also been shown to subject to regulation by SCP1 during neurogenesis in vivo (Visvanathan et al., 2007). Collectively, these observations suggest an important pathway for neuronal differentiation that involves the functional interplay between miR-124, PTB and SCP1/REST.
PTB directly competes with microRNA targeting on the 3′UTRof SCP1
Perturbation experiments confirmed the role of PTB in the regulation of microRNA function. For example, overexpression of miR-96 suppressed SCP1 expression and PTB knockdown enhanced the effect, whereas miR-96 antagomir showed the opposite response (Figure 5B). A non-targeting miR-339 (labeled as Ctrl miR) served as a negative control. We could recapitulate these effects with a luciferase reporter containing the entire 3′UTR of the SCP1 gene (Figure S5A). We then analyzed individual segments (F1 to F3) in the 3′UTR of SCP1, finding that overexpression of either miR-96 or miR-124 could suppress the activity of the reporter containing the F1 fragment (Figure 5C). PTB overexpression antagonized, but PTB knockdown enhanced, the effect of both microRNAs (Figure 5C and 5D). We made a similar observation on the luciferase reporter containing the F2 (Figure S5B) or F3 (Figure S5C) fragment.
To determine the sequence requirement for both microRNA- and PTB-mediated actions, we carried out mutational analysis in the seed region of individual microRNA target sites and on the nearby PTB binding sites (Figure 5A). We found that the mutant (GCC to CGG) in the miR-96 seed region no longer responded to the overexpression of this microRNA (Figure 5E). The mutations (double C-to-A) in the nearby PTB binding site enhanced the effect of the microRNA, even though these mutations impaired miR-96 targeting to some degree, thus causing an increase in the reporter activity in control microRNA-treated cells (Figure 5E). Similarly, the mutations (GCC to CGG) in each of the miR-124 targeting sites attenuated and the double mutation abolished the response to transfected miR-124 (Figure 5F). In comparison, at least one of the mutations in nearby PTB binding sites (the triple A mutant in the first miR-124 targeting site shown in Figure 5A) enhanced the effect of miR-124 (compare lanes 4 and 12 in Figure 5F). Together, these data demonstrated that PTB directly competes with microRNA on multiple targeting sites in the 3′UTR of the SCP1 gene.
PTB can also boost microRNA action on specific genes
Our RNA-seq experiments and luciferase-based assays revealed both up- and down-regulated genes in response to PTB knockdown. While many up-regulated genes likely resulted from de-repression, we detected multiple examples of up-regulated genes in PTB knockdown cells that appear to depend on their 3′UTRs (Figure 4I). Such effect might be due to PTB-regulated switch of polyadenylation from the distal to proximal site, thereby shortening the 3′UTR in some genes that reduce microRNA targeting potentials. We tested and ruled out this possibility by measuring RNA-seq tags at the 3′ end of each expressed gene in response to PTB knockdown (Figure S5D and S5E).
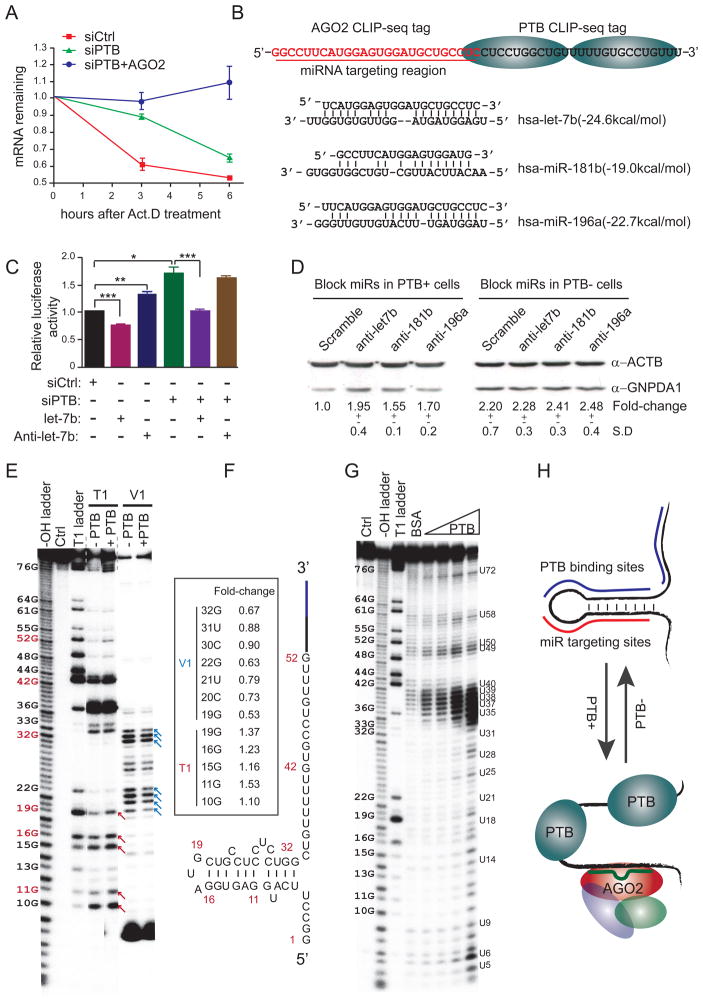
To understand how PTB knockdown could induce gene expression, we took GNPDA1 as a model, which was up regulated by PTB via its 3′UTR (Figure 4I). We validated that PTB knockdown enhanced the stability of the endogenous GNPDA1 transcript (Figure 6A). We noted that the CLIP-seq mapped PTB binding events are coincident with two stretches of C/U-rich sequences on the 3′UTR of the GNPDA1 gene (Figure 6B). We confirmed high affinity PTB binding on this element by gel mobility shift (Figure S6A). Importantly, the PTB binding sites are immediately downstream of the mapped Ago2 binding sites that contain potential targeting sites for several microRNA, including Let-7b, miR-181b, and miR-196a (Figure 6B). As expected, Let-7b overexpression suppressed the expression of the luciferase reporter containing this region, while anti-Let-7b showed the opposite effect (Figure 6C). The reporter activity could be further enhanced by PTB knockdown in HeLa (Figure 6C) and NT2 cells (Figure S6B). We also showed that both anti-Let-7b and anti-miR-181b enhanced GNPDA1 protein expression in a PTB-dependent manner (Figure 6D). These data demonstrated that microRNAs act more effectively on GNPDA1 in the presence of PTB.
Figure 6. Enhanced microRNA targeting by modulatingRNA secondary structure.
(A) Stabilization of the GNPDA1 transcript in response to PTB and/or Ago2 knockdown in the presence of the transcription inhibitor Act.D.
(B) Potential microRNA targeting sites near the mapped PTB binding site in the 3′UTR of GNPDA1.
(C) Overexpressed Let-7b suppressed and antagomir Let-7b enhanced the expression of the luciferase reporter containing the 3′UTR of GNPDA1 (lanes 1 to 3). PTB knockdown enhanced the luciferase activity (compared between lanes 1 and 4). Overexpression of Let-7b still suppressed the luciferase activity, but anti-Let-7b no longer showed the effect in PTB knockdown cells.
(D) Antagomir Let-7b, miR-196a and miR-181b increased GNPDA1 protein in the presence, but not absence, of PTB in transfected HeLa cells. The protein levels were quantified with the SD shown in the bottom.
(E and F) Mapping the secondary structure in the 3′UTR of GNPDA1. Individual G residues were labeled on the left with red indicating several key positions in the deduced secondary structure (E), as modeled (F). Red and blue arrows respectively indicate PTB enhanced and suppressed cleavages in the deduced stem-loop region. Quantified fold-changes at key positions are indicated in the box inserted in panel F.
(G and H) Increased single-strandness of RNA in the presence of increasing amounts of PTB detected by in-line probing (G). A proposed model indicates PTB-mediated opening of the stem-loop that facilitates microRNA targeting (H).
Data in A, C, and D are shown as mean ± SD. *p<0.05; **p<0.01; ***p<0.001. See also Figure S6.
PTB facilitates microRNA action by changing local RNA secondary structure
To uncover the mechanism for PTB-dependent enhancement of microRNA action, we determined the secondary structure in the 3′UTR of GNPDA1 gene using RNase T1 to cut single-stranded RNA after the nucleotide G, and RNase V1 to cleave double-stranded RNA (Figure 6E). This analysis suggests a stem-loop between 6U and 33G (Figure 6F), which appears to be undertaking a dynamic switch between the single- and double-stranded states, as evidenced by T1 and V1 cleaved products in the same stem region. In the presence of PTB, we reproducibly detected enhanced single-strandness of the stem-loop, as indicated by increased T1 cleavage from 10G to 19G (red arrows in Figure 6E) and concurrent decreased V1 cleavage from 19G to 32G (blue arrows in Figure 6E), which were quantified on a modeled RNA structure (boxed in Figure 6F).
We substantiated the increase of single-strandness by in-line probing, an approach widely used to detect riboswitches, which measures spontaneous RNA cleavage in solution with strong preference for U-rich residues (Regulski and Breaker, 2008). With increasing amounts of PTB, we found that the entire region gradually opened up, as indicated by enhanced cleavage on nearly all residues from 10G to 33G and the flanking U-rich PTB binding sites from 34C to 40U (Figure 6G). Thus, PTB appears to induce the exposure of the microRNA target site through binding to multiple pyrimidine-rich regions, including that directly involved in base-paring with microRNA (Figure 6H). In principle, such modulation of RNA secondary structure by PTB or other RNA binding proteins may enhance or shield microRNA target sites in adjacent regions, thus affecting RNA stability in both directions.
PTB globally regulates microRNA-mRNA interactions in the human genome
To assess the global impact of PTB on both positive and negative modulation of microRNA targeting, we conducted CLIP-seq mapping of Ago2 before and after PTB knockdown in HeLa cells. As previously described (Chi et al., 2009), we detected Ago2-RNA crosslinking adducts IPed with anti-Ago2 above the position of the Ago2 protein on SDS-PAGE (Figure 7A). We obtained ~23 million uniquely mapped CLIP-seq tags and identified 4314 and 4087 genes that contain at least one Ago2 peak in their 3′UTRs before and after PTB knockdown, respectively. Comparison of these datasets suggests that PTB knockdown generally enhances Ago2 binding in the human genome (Figure 7B). Ago2 binding events were significantly enriched in the 3′UTR of protein-coding genes in both mock-treated and shPTB-treated cells (Figure 7C), especially near the stop codon and the poly(A) site (Figure 7D and 7E).
We next compared the relationship between PTB and Ago2 occupancies in the 3′UTR of protein-coding genes in response to PTB knockdown. The Ago2 binding profiles were similar in the protein-coding side (upstream of the stop codon) on both wt and shPTB-treated cells, which provide important internal controls for our comparison. By separately analyzing PTB bound and unbound genes, we found that PTB depletion caused a dramatic increase in Ago2 binding in the 3′UTR of PTB bound targets, but had only a minor effect on PTB unbound targets (Figure 7D and 7E). These differences are highly statistically significant at the right side of the stop codon and the left side of the Poly(A) sites, as determined by two-tailed Kolmogorov-Smirnov test. This likely represents an underestimate of increased microRNA targeting events because the transcripts of many PTB bound genes were down regulated to various degrees in PTB knockdown cells. Global analysis further showed that PTB knockdown generally and significantly enhanced Ago2 binding on and around the mapped PTB binding sites (Figure 7F). In this analysis, we noted many altered Ago2 binding events around and away from the mapped PTB binding sites, suggesting that PTB binding may have both local and long-range effects on microRNA targeting.
PTB-regulated Ago2 binding functionally correlates to induced gene expression
To determine how such changes in Ago2 binding might be related to altered gene expression, we took a strategy recently used to analyze the interplay between HuR and microRNA (Mukherjee et al., 2011) to segregate expressed genes into 5 groups based on mapped PTB and Ago2 binding events in their 3′UTRs: (1) −Ago2, −PTB, (2) +Ago2, −PTB, (3) −Ago2, +PTB, (4) +Ago2, +PTB, but no overlap, and (5) +Ago2, +PTB with at least one overlapping binding event within 10 nt. This allowed us to compare gene expression changes in different groups in response to PTB knockdown by plotting genes in each group against induced transcript changes in a cumulative fashion (Figure 7G).
We found no significant differences between Groups 1–3, consistent with the lack of PTB and Ago2 actions on these genes. In comparison, relative to genes in Group 1 (black line), genes bound by both PTB and Ago2 but with little overlap (Group 4, green line) were linked to both repressed (right-shift at top) and enhanced gene expression (left-shift at bottom), consistent with changes in RNA secondary structure that caused increased or decreased microRNA targeting on different genes (Figure 7G). In contrast, genes bound by both PTB and Ago2 with extensive overlap (Group 5, purple line) mainly showed repressed expression (right-shift) as a result of enhanced microRNA targeting in the absence of PTB competition (Figure 7G). We obtained similar results in comparing genes in Group 2 (Figure S7A) and Group 3 (Figure S7B) with those in Group 4 and Group 5. These results demonstrate a large-scale involvement of PTB in regulated gene expression through its functional interplay with the microRNA machinery, which likely acts in synergy with regulated splicing to propel neurogenesis in mammalian cells.
Discussion
While the induction of pluripotency with a specific set of stem cell-specific transcription factors has been well accepted, the field has increasingly recognized the ability of lineage-specific transcription factors to induce differentiated cells to trans-differentiate into a different lineage without going back to the fully undifferentiated state. The classic example is MyoD-induced differentiation of fibroblasts to myotubes (Davis et al., 1987), and more recent examples include induced trans-differentiation of both embryonic and postnatal fibroblasts into neurons by Ascl1/Mash1 in combination with other enhancing factors (Caiazzo et al., 2011; Pang et al., 2011; Qiang et al., 2011; Vierbuchen et al., 2010). Interestingly, this pathway also involves specific microRNAs (Yoo et al., 2011), which are known to act on many critical target genes to induce neuronal differentiation (Li and Jin, 2010). We now report that the reduced expression of a single RNA binding PTB, which occurs during brain development, is able to potently induce differentiation or trans-differentiation of diverse cell types into neuronal-like cells or even functional neurons.
Our data highlights the contribution of specific regulated splicing during the induction of neuronal differentiation. More importantly, we discovered a PTB-regulated microRNA program responsible for dismantling of multiple components of REST. We provide further evidence that knockdown of SCP1 or the REST itself is sufficient to trigger trans-differentiation of MEFs into neuronal-like cells. The REST complex is well known for its role in suppressing many neuronal genes, including miR-124, in non-neuronal cells, while miR-124 and other neuronal specific microRNAs target various REST components, including SCP1 and CoREST. This creates a potent regulatory loop (Figure 7H). However, this loop is inefficient, at least in the initial phase of neuronal induction, unless PTB is first down regulated by miR-124. Thus, PTB not only serves as a key target of miR-124, but also functions as a negative regulator for this and other microRNAs to act on their target genes. This represents an interesting regulatory paradigm where the large auto-regulatory loop consisting of miR-124 and components of the REST complex is controlled by another feed-forward loop that involves PTB.
Strikingly, PTB down-regulation induced the expression of all critical transcription factors previously shown to be sufficient to induce trans-differentiation of MEFs into functional neurons. Our data provide a mechanism for the induction of these transcription factors because all of these transcription factors appear to be direct REST targets. The puzzle is why genetic ablation of REST or HADC1 impaired self-renewal of neural stem cells, thus preventing unintended neurogenesis in various cellular and animal models (Dovey et al., 2010; Gao et al., 2011; Lee et al., 2002). While the cellular context undoubtedly contributes to such restriction of neurogenesis in vivo, it is possible that PTB knockdown may mimic a gradual and sequential switch of a series of events during normal developmental processes by preventing abrupt induction of gene expression that may cause cell death before differentiation. We note that the PTB-regulated RNA program takes place in cells containing induced nPTB and our preliminary results indicate that simultaneous knockdown of PTB and nPTB greatly compromised the development of neuronal morphology. This may indeed represent critical sequential events during normal brain development (Zheng et al., 2012).
Mechanistically, our study joined PTB to a growing list of RNA binding proteins, including HuR, Dnd1, CRD-BP, and PUM1, that have been implicated in modulating microRNA targeting in mammalian cells (van Kouwenhove et al., 2011). In comparison with previous studies where specific RNA binding proteins appear to either positively or negatively regulate microRNA targeting, we found that PTB can function in both ways, competing with microRNA targeting on some genes, but promoting microRNA targeting on the others. These two modes of regulation may simultaneously occur on different locations in the same genes, and thus, the net effect of positive and negative regulation may dictate the final functional outcome. These working principles may be generally applicable to many other RNA binding proteins involved in the regulation of microRNA-mRNA interactions. Our global analysis of Ago2 binding in response to PTB knockdown also suggests that PTB binding may have some long-range effects on microRNA targeting in addition to local events. This may result from potential PTB-mediated RNA looping, as proposed earlier (Oberstrass et al., 2005), the action of other induced microRNAs, or synergy with other RNA binding proteins, all of which represent interesting regulatory paradigms to be investigated in future studies.
Experimental Procedures
Cell culture, RNAi, immunocytochemistry and electrophysiological analysis
Cell culture conditions, treatments with siRNA and shRNA, and immunocytochemistry are detailed in the supplemental experimental procedures. Glial cells were isolated from GFP-transgenic rat brain (Hakamata et al., 2001) and single cell patch clamp recordings were performed using an Axopatch 200B amplifier and pClamp 10.0 software (HEKA Elektronik, Lambrecht/Pfalz, Germany), as described in the supplemental information.
RT-qPCR, Western blotting, and luciferase assays
qPCR was performed with Fast Start universal SYBR green master mix using gene specific primers listed in Table S1. Luciferase activity was measured 24 hrs post-transfection. Western blotting analysis was conducted with various specific antibodies as detailed in the supplemental procedure.
RNA-seq, CLIP-seq, and probing of RNA secondary structure
RNA-seq and CLIP-seq was performed as previously described (Xue et al., 2009). Normalized Ago2 tags are plotted relative to the stop codon at the 3′ end of genes as described (Chi et al., 2009). Two-sided Kolmogorov-Smirnov statistics (in the R package, http://cran.r-project.org/) was used to determine the significance of the shift in pair-wise comparison. RNA foot-printing by RNase T1 and V1 was according to the manual from Ambion. The in-line probing assay was as previously described (Regulski and Breaker, 2008), which is also detailed in the supplemental information.
Supplementary Material
Highlights.
Diminished PTB expression is sufficient to trans-differentiate fibroblasts to neurons
PTB plays a key role in a regulatory loop to suppress neuronal-specific genes
PTB regulates gene expression at both the splicing and RNA stability levels
PTB modulates microRNA targeting by competition or switching RNA structure
Acknowledgments
The authors are grateful to Drs. Gordon Gill, Dongxian Zhang, Wei Xiong for insightful advice during the course of this investigation and to Drs. Gordon Gill, Amy Pasquinelli, Bruce Hamilton, and Larry Goldstein for critical reading of the manuscript. We thank Drs. Douglas Black, Robert Darnell, Samuel Pfaff, Alysson Muotri and Geoff Rosenfeld for generous gifts of antibodies and Drs. Zhiping Pang and Marius Wernig for sharing detailed experimental protocols. This work was supported by the China 973 program (2011CB811300), a grant China NSFC International Corporation and Exchange program (B06018), and grants from NIH (GM049369, GM052872, HG004659).
Footnotes
Accession numbers
The RNA-seq and CLIP-seq data are available at the Gene Expression Omnibus under the accession number XXX and XXX.
Supplemental information includes supplemental experimental procedures, one table, and 7 figures along with legends, which can be found with this article online at XXX.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O, et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257–263. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki H, Hara A, Era T, Kunisada T, Yamada Y. Genetic ablation of Rest leads to in vitro-specific derepression of neuronal genes during neurogenesis. Development. 2012;139:667–677. doi: 10.1242/dev.072272. [DOI] [PubMed] [Google Scholar]
- Auld KL, Silver PA. Transcriptional regulation by the proteasome as a mechanism for cellular protein homeostasis. Cell Cycle. 2006;5:1503–1505. doi: 10.4161/cc.5.14.2979. [DOI] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudet ML, Zivraj KH, Abreu-Goodger C, Muldal A, Armisen J, Blenkiron C, Goldstein LD, Miska EA, Holt CE. miR-124 acts through CoREST to control onset of Sema3A sensitivity in navigating retinal growth cones. Nat Neurosci. 2012;15:29–38. doi: 10.1038/nn.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M, Jr, Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazzo M, Dell’anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, Sotnikova TD, Menegon A, Roncaglia P, Colciago G, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- Carracedo A, Pandolfi PP. The PTEN-PI3K pathway: of feedbacks and cross-talks. Oncogene. 2008;27:5527–5541. doi: 10.1038/onc.2008.247. [DOI] [PubMed] [Google Scholar]
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho-Mansfield GC, Xue Y, Zhang Y, Fu XD. PTB/nPTB switch: a post-transcriptional mechanism for programming neuronal differentiation. Genes Dev. 2007;21:1573–1577. doi: 10.1101/gad.1575607. [DOI] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Dovey OM, Foster CT, Cowley SM. Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc Natl Acad Sci U S A. 2010;107:8242–8247. doi: 10.1073/pnas.1000478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Ure K, Ding P, Nashaat M, Yuan L, Ma J, Hammer RE, Hsieh J. The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J Neurosci. 2011;31:9772–9786. doi: 10.1523/JNEUROSCI.1604-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakamata Y, Tahara K, Uchida H, Sakuma Y, Nakamura M, Kume A, Murakami T, Takahashi M, Takahashi R, Hirabayashi M, et al. Green fluorescent protein-transgenic rat: a tool for organ transplantation research. Biochem Biophys Res Commun. 2001;286:779–785. doi: 10.1006/bbrc.2001.5452. [DOI] [PubMed] [Google Scholar]
- He X, Pool M, Darcy KM, Lim SB, Auersperg N, Coon JS, Beck WT. Knockdown of polypyrimidine tract-binding protein suppresses ovarian tumor cell growth and invasiveness in vitro. Oncogene. 2007;26:4961–4968. doi: 10.1038/sj.onc.1210307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci U S A. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Kim J, Efe JA, Zhu S, Talantova M, Yuan X, Wang S, Lipton SA, Zhang K, Ding S. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci U S A. 2011;108:7838–7843. doi: 10.1073/pnas.1103113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoch KP, Bergert H, Borgonovo B, Saeger HD, Altkruger A, Verkade P, Solimena M. Polypyrimidine tract-binding protein promotes insulin secretory granule biogenesis. Nat Cell Biol. 2004;6:207–214. doi: 10.1038/ncb1099. [DOI] [PubMed] [Google Scholar]
- Kosinski PA, Laughlin J, Singh K, Covey LR. A complex containing polypyrimidine tract-binding protein is involved in regulating the stability of CD40 ligand (CD154) mRNA. J Immunol. 2003;170:979–988. doi: 10.4049/jimmunol.170.2.979. [DOI] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol Cell. 2010;40:205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Jin P. Roles of small regulatory RNAs in determining neuronal identity. Nat Rev Neurosci. 2010;11:329–338. doi: 10.1038/nrn2739. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene JD. Integrative Regulatory Mapping Indicates that the RNA-Binding Protein HuR Couples Pre-mRNA Processing and mRNA Stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak L, Bregestovski P, Ascher P, Herbet A, Prochiantz A. Magnesium gates glutamate-activated channels in mouse central neurones. Nature. 1984;307:462–465. doi: 10.1038/307462a0. [DOI] [PubMed] [Google Scholar]
- Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Sudhof TC, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;467:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautz A, Linker K, Hubrich T, Korhonen R, Altenhofer S, Kleinert H. The polypyrimidine tract-binding protein (PTB) is involved in the post-transcriptional regulation of human inducible nitric oxide synthase expression. J Biol Chem. 2006;281:32294–32302. doi: 10.1074/jbc.M603915200. [DOI] [PubMed] [Google Scholar]
- Porter JF, Vavassori S, Covey LR. A polypyrimidine tract-binding protein-dependent pathway of mRNA stability initiates with CpG activation of primary B cells. J Immunol. 2008;181:3336–3345. doi: 10.4049/jimmunol.181.5.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang L, Fujita R, Yamashita T, Angulo S, Rhinn H, Rhee D, Doege C, Chau L, Aubry L, Vanti WB, et al. Directed conversion of Alzheimer’s disease patient skin fibroblasts into functional neurons. Cell. 2011;146:359–371. doi: 10.1016/j.cell.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Raj B, O’Hanlon D, Vessey JP, Pan Q, Ray D, Buckley NJ, Miller FD, Blencowe BJ. Cross-Regulation between an Alternative Splicing Activator and a Transcription Repressor Controls Neurogenesis. Mol Cell. 2011;43:843–850. doi: 10.1016/j.molcel.2011.08.014. [DOI] [PubMed] [Google Scholar]
- Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Methods Mol Biol. 2008;419:53–67. doi: 10.1007/978-1-59745-033-1_4. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Tillmar L, Welsh N. Hypoxia may increase rat insulin mRNA levels by promoting binding of the polypyrimidine tract-binding protein (PTB) to the pyrimidine-rich insulin mRNA 3′-untranslated region. Mol Med. 2002;8:263–272. [PMC free article] [PubMed] [Google Scholar]
- van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nature reviews Cancer. 2011;11:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Kameoka S, Gopalakrishnan V, Aldape KD, Pan ZZ, Lang FF, Majumder S. Conversion of myoblasts to physiologically active neuronal phenotype. Genes Dev. 2004;18:889–900. doi: 10.1101/gad.1179004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo KC, Kim TD, Lee KH, Kim DY, Kim W, Lee KY, Kim KT. Mouse period 2 mRNA circadian oscillation is modulated by PTB-mediated rhythmic mRNA degradation. Nucleic Acids Res. 2009;37:26–37. doi: 10.1093/nar/gkn893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Ng YH, Pang ZP, Sudhof TC, Wernig M. Induced neuronal cells: how to make and define a neuron. Cell Stem Cell. 2011;9:517–525. doi: 10.1016/j.stem.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo M, Lee SK, Lee B, Ruiz EC, Pfaff SL, Gill GN. Small CTD phosphatases function in silencing neuronal gene expression. Science. 2005;307:596–600. doi: 10.1126/science.1100801. [DOI] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernicka-Goetz M, Morris SA, Bruce AW. Making a firm decision: multifaceted regulation of cell fate in the early mouse embryo. Nat Rev Genet. 2009;10:467–477. doi: 10.1038/nrg2564. [DOI] [PubMed] [Google Scholar]
- Zheng S, Gray EE, Chawla G, Porse BT, O’Dell TJ, Black DL. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat Neurosci. 2012;15:381–388. S381. doi: 10.1038/nn.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zibetti C, Adamo A, Binda C, Forneris F, Toffolo E, Verpelli C, Ginelli E, Mattevi A, Sala C, Battaglioli E. Alternative splicing of the histone demethylase LSD1/KDM1 contributes to the modulation of neurite morphogenesis in the mammalian nervous system. J Neurosci. 2010;30:2521–2532. doi: 10.1523/JNEUROSCI.5500-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.