Abstract
Congenital hyperinsulinism/hyperammonemia (HI/HA) syndrome is caused by an activation mutation of glutamate dehydrogenase 1 (GDH1), a mitochondrial enzyme responsible for the reversible interconversion between glutamate and α-ketoglutarate. The syndrome presents clinically with hyperammonemia, significant episodic hypoglycemia, seizures, and a frequent incidences of developmental and learning defects. Clinical research has implicated that although some of the developmental and neurological defects may be attributed to hypoglycemia, some characteristics cannot be ascribed to low glucose and as hyperammonemia is generally mild and asymptomatic, there exists the possibility that altered GDH1 activity within the brain leads to some clinical changes. GDH1 is allosterically regulated by many factors, and has been shown to be inhibited by the ADP-ribosyltransferase sirtuin 4 (SIRT4), a mitochondrially localized sirtuin. Here we show that SIRT4 is localized to mitochondria within the brain. SIRT4 is highly expressed in glial cells, specifically astrocytes, in the postnatal brain and in radial glia during embryogenesis. Furthermore, SIRT4 protein decreases in expression during development. We show that factors known to allosterically regulate GDH1 alter gliogenesis in CTX8 cells, a novel radial glial cell line. We find that SIRT4 and GDH1 overexpression play antagonistic roles in regulating gliogenesis and that a mutant variant of GDH1 found in HI/HA patients accelerates the development of glia from cultured radial glia cells.
Keywords: glutamate dehydrogenase, sirtuin 4, gliogenesis, radial glia, CTX8 cell line
INTRODUCTION
Glutamate Dehydrogenase (GDH) catalyzes the reversible interconversion between glutamate and α-Ketoglutarate (αKG), using NADP+ as a cofactor. Found in all living organisms, GDH exists in vivo as a homohexamer, with each subunit ranging in size from approximately 450 amino acids in prokaryotes to about 500 in eukaryotes (505 in humans). GDH provides a source of αKG for the Krebs cycle, thereby indirectly regulating the amount of NADH available for the electron transport chain. Mammalian GDH resides within the inner mitochondrial matrix of all cells and is composed of 2 homotrimers packing directly on top of each other, with the bases contacting one another (Stanley, 2009). Interestingly, although many publications have suggested a significant role for GDH1 in the brain, actual data related to its expression and function are sparse. Both GDH1 and its human non-intronic variant GLUD2 are expressed in the human brain (Shashidharan et al., 1994; Spanaki et al., 2010). Initially, experiments to determine the exact cell types in which GDH1 is expressed were largely inconclusive and highly contested (Nicklas, 1984); however, it has become clear that GDH1 is expressed in glia (Aoki et al., 1987; Dutuit et al., 2000; Schmitt and Kugler, 1999), specifically in regions where high expression of glutamatergic neurons occur, including the cortex (Aoki et al., 1987).
An inhibitor of GDH, sirtuin 4 (SIRT4), is one of seven mammalian sirtuins that make up a family of class III histone deacetylases. Exogenously expressed SIRT4 localizes to mitochondria (Haigis et al., 2006; Michishita et al., 2005) in pancreatic β cells. Overexpression of SIRT4 represses GDH activity and limits the metabolism of glutamate and glutamine to generate ATP (Haigis et al., 2006). Furthermore, a knock-out mouse was generated by removing exons 1-3 of the SIRT4 gene, and this mouse exhibited approximately 2-fold increase in GDH activity in liver lysates, confirming not only the knock-out of SIRT4, but also the role of SIRT4 in regulating GDH in vivo (Haigis et al., 2006). Mechanistically, SIRT4, unlike the other class III HDACs, does not display deacetylase activity but instead ADP-ribosylates GDH (Haigis et al., 2006).
Studies on congenital hyperinsulinism/hyperammonemia (HI/HA) syndrome have elucidated the role of GDH1, and potentially SIRT4, in the brain since mutations of GDH1 appear to result in neurodevelopmental defects seen in patients with this syndrome. HI/HA syndrome is the second most common form of hyperinsulinism, occurring in approximately 1:30,000 live births. Patients with this disorder are characterized by hypoglycemia (transient or chronic) that can occur spontaneously or after meals, particularly those rich with protein, along with persistently elevated ammonia levels. HI/HA syndrome typically presents by age 2, with a mean initial diagnosis age from 4 to 11 months, (MacMullen et al., 2001; Stanley et al., 2000), although the disease is thought to be present at birth. Infants are born with normal birth weights and do not present with any significant obvious morphologic abnormalities (de Lonlay et al., 2002a; MacMullen et al., 2001), although they can develop some physical characteristics common to all children with hyperinsulinism later in life (de Lonlay et al., 2002b). The most prominent neurological feature of HI/HA syndrome is seizures. HI/HA patients exhibit petit mal or absence type seizures (Raizen et al., 2005) manifested by loss of normal cognition, staring, and lack of generalized shaking or twitching, with any present twitching being limited to eyelids and lips. The underlying mechanisms involved in the neurological defects observed in patients with HI/HA syndrome remain unclear, since changes in glucose and ammonia levels cannot fully explain developmental defects and seizure activity.
Given that mutations of GDH1 within the brain likely contribute to neurodevelopmental defects of HI/HA syndrome and that GDH1 is negatively regulated by SIRT4, we decided to examine the functional role of GDH1 and SIRT4 within the CNS, particularly in astroglial development. Here we show that GDH1 and SIRT4 are localized to mitochondria within the brain, expressed at high levels in astrocytes in the postnatal brain and radial glia in embryonic tissues, and expression decreases during development. Using cultures of CTX8 cells, a novel radial glial cell line (Li et al., 2012), we further show that factors known to allosterically regulate GDH1 alter gliogenesis and that SIRT4 and GDH1 overexpression play antagonistic roles in regulating gliogenesis. Importantly a mutant variant of GDH1 found in HI/HA patients promotes glial development of cultured CTX8 radial glia cells. Thus, our studies elucidate a novel role for GDH1 and SIRT4 in the regulation of radial glial differentiation.
MATERIALS AND METHODS
Reagents
GAPDH, GFAP, MAP2, nestin, and vimentin antibodies were from Millipore. GDH1 and SIRT4 (Western blotting) antibodies were from US Bio and SIRT4 antibody (immunostaining) was from Abcam.
Primary culture of cortical neurons
Neuronal cultures were prepared from cortices of rat embryos at 18 days gestation as our laboratory has described previously for hippocampi (Charych et al., 2006; Firestein et al., 1999; Kwon et al., 2011; Sweet et al., 2011; Zhang et al., 2008). Briefly, the cortices were dissociated, and cells were plated on poly-D-lysine-coated glass coverslips (12 mm diameter) at a density of 1800 cells/mm2 or 1 × 106 cells on 35mm culture dishes. Cultures were maintained in Neurobasal media (Gibco) supplemented with B27 (Gibco), penicillin/streptomycin, and Glutamax (Gibco). Cells were grown for the indicated number of days in vitro (DIV) and used for specific experiments as indicated below.
Culturing of CTX8 cells
CTX8 cells were grown as neurospheres in suspension in plastic dishes. CTX8 cells, were maintained in +FGF media (96.8% DMEM/F12, 20% B-27, 10%Pen/Strep, 10 ng/ml bFGF, 2μg/ml heparin), and were typically passaged once every 2-3 days. bFGF was refreshed daily to ensure adequate bFGF levels to keep cells in an undifferentiated state. Cells were passaged a maximum of three times. Dissociation and passaging of CTX8 cells was performed by collecting spheres into a conical tube and briefly centrifuging at 1000 × g for 30 seconds. The cell pellet was trypsinized quickly using 0.25% trypsin, followed by the addition of an equal volume of 25 mg/ml trypsin inhibitor to stop the trypsinization process. Cells were centrifuged at 1000 × g for 5 minutes, and cells were dissociated into +FGF media. Cells were seeded onto Matrigel-coated plates for experiments.
Isolation of mitochondria, cytosol, and nuclei from rat brain
A whole rat brain was washed twice with 2 volumes of Extraction Buffer (10 mM HEPES, pH 7.5, 200 mM mannitol, 70 mM sucrose, and 1 mM EGTA). The brain was cut into small slices and homogenized using a pestle and glass homogenizer on ice with 10 volumes of Extraction Buffer containing 2 mg/ml BSA. The homogenate was centrifuged at 600 × g for 5 min at 4°C to pellet the nuclei. The supernatant was centrifuged at 11,000 × g for 10 min at 4°C to yield the cytosolic supernatant and a pelleted mitochondria fraction. The nuclei pellet was resuspended in 10 volumes of Extraction Buffer and centrifuged at 600 × g for 5 min at 4°C to yield purified nuclear extract. The mitochondria were resuspended in 10 volumes of Extraction Buffer and centrifuged at 11,000 × g for 10 min at 4°C to collect purified mitochondria. The cytosol was centrifuged at 11,000 × g for 10 min at 4°C to yield purified cytosol.
Western blotting
Dissociated cell cultures were placed on ice and lysed in ice cold 1X RIPA buffer (50 mM TrisHCl, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP40, 1 mM PMSF), after a 1 hr extraction, samples were centrifuged at 15,000 × g for 15 minutes to remove insoluble debris. Brain samples were homogenized in 5 × volume/weight of RIPA buffer, protein extracted for 1 hr, and centrifuged at 15,000 × g for 15 minutes to remove insoluble debris. Proteins were resolved by SDS-PAGE and immunoblotted for the indicated proteins.
Altered glucose formulations
Due to the fact that DMEM/F12 does not come glucose free, glucose free DMEM (Life Technologies cat# 11966) was substituted instead of DMEM/F12, to allow for custom addition of glucose. No significant change in cell growth or morphology was observed when the cells were placed in DMEM instead of DMEM/F12; however, 50 μM L-glutamic acid was added to keep glutamate concentrations the same between DMEM and DMEM/F12. Clinically relevant glucose concentrations in the DMEM media to mimic as closely as possible the glucose concentrations in the rat brain were as follows: low glucose, 0.3 mM; normal glucose, 1.5 mM; high glucose, 4.5 mM.
Glutamate dehydrogenase (GDH) assay
This assay was followed verbatim according to the manufacturer’s protocol (Biovision cat #K729-100). For obtaining tissue samples, either 50 mg whole rat brain or cell pellet from a 60 mm tissue culture dish of confluent CTX8 cells was collected and homogenized in assay buffer. Of this homogenate, 5 μl was used for the assay. To obtain accurate measurements of GDH activity, each sample was tested using two separate conditions. One condition used the provided 2 M glutamate, the other used the equivalent volume of plain assay buffer. As almost all tissue, especially brain, will contain baseline NADH activity, the samples without glutamate will elucidate background NADH production, and the ones with glutamate can be compared to the one with GDH stimulation and background, with the difference being GDH only activity.
Glucose Assay
Similar to the GDH Assay, this test was followed verbatim according to the company protocol (Biovision cat #K606-100). One ml media supernatant was collected from confluent CTX8 cell cultures and briefly spun at 5000 × g for 5 min to remove cellular debris. Supernatant was then collected in a clean tube and placed on ice. Remaining media was stored at −20°C.
Immunocytochemistry of dissociated cells
CTX8 cells cultured in dishes were fixed with phosphate buffered saline (PBS) containing 4% paraformaldehyde (PFA) and 4% sucrose for 20 minutes at RT. Blocking was performed using PBS containing 0.2%Triton X-100 (PBSTX) and 5% Bovine Serum Albumin (BSA) for 30 min at RT. Cells were incubated with primary antibodies diluted in PBSTX containing 1% BSA for 4-6 hours at RT. Cells were washed and incubated with appropriate secondary antibodies in PBSTX containing 1% BSA for 45 minutes at RT. Coverslips were affixed to glass slides using Fluoromount antifade agent (Southern Biotechnology).
Immunocytochemistry of brain sections
Animals were perfused with 0.9% saline solution followed by 4% paraformaldegyde in PBS, brains were isolated and cryoprotected in 30% sucrose solution, and sectioned at 40 μm. If antigen retrieval was required (only for GDH1 labeling), sections were incubated in standard sodium citrate buffer and placed in a water bath at 95-100°C for 30 min. Sections were allowed to cool to RT. Blocking, primary, and secondary antibody labeling and all washing was performed as described in dissociated cell protocol above.
Subcloning
The mOrange-N1 vector was obtained from Dr. James Zheng’s lab at Emory University. For the rat SIRT4 construct, gene specific primers for Rattus norvegicus Sirt4 were designed: 5′ primer: 5′-ATGAGGGGGT TGATTTTCAG GCCGACAAGG-3′, 3′ primer: 5′-CTGTGGGTCTATTAAGGGCAGCAGC-3′. RT-PCR was performed using adult rat RNA library as a template. The rat SIRT4 cDNA was then ligated into the mOrange vector between the NheI and SalI restriction sites, respectively. For the RatSIRT4-hrGFPII-pCAG construct, hrGFPII was amplified from the pGE vector using PCR and used to replace mOrange in the mOrange-N1 vector at the SalI and NotI sites. Primers for this PCR were: 5′ primer: 5′-CCCC GTCGACATGGTTGGGATAAGGCTGGATT-3′. 3′ primer: 5′-CCCCGCGGCCGC TTACACCCACTCGTGCAGGCTG-3′. Once this cloning was completed, rat SIRT4 was inserted between NheI and SalI sites in the newly formed hrGFPII-N1 vector. Once the ratSIRT4-hrGFPII plasmid was created, ratSIRT4-hrGFPII was inserted into the pCAG vector since the CMV promoter does not allow sufficient expression in CTX8 cells. This subcloning used NotI restriction sites on both sides using the 5′ primer for ratSIRT4-Orange-pCAG vector and the 3′ primer for the original insertion of hrGFPII into the mOrange vector. For the RatGDH-mOrange plasmid, an adult rat RNA library was used a to amplify rat GDH. Primers were designed to be directly inserted into the mOrange vector, between the NheI and SalI sites as follows: 5′ primer: 5′-CCCCGCTAGC CGCCACCATGTACCGCCGTCTGGGCGAAGTGCTGCTAC-3′. 3′ primer: 5 ′-CCCCGTCGACGGCTGTGAAGGTCACGCCAGCCTCATTGTACACC-3′. For the RatGDH-mOrange-pCAG plasmid, essentially identical protocols were used to insert ratGDH-mOrange into the pCAG as was used to insert ratSIRT4-mOrange into pCAG. NotI restriction sites were used on both ends of the insert using 5′ primer: 5′-CCCCGCGGCCGCCGCCACCATGTACCGCCGTCTGGGCGAAGTGCTGCTAC-3′ and 3′ primer: Same used as for ratSIRT4-Orange-pCAG. For the H454Y-rGDH-Orange-pCAG plasmid, site-directed mutagenesis was performed using Agilent’s Site Directed Mutagenesis kit, and followed exactly according to the manufacturer’s protocols. Primers used were as follows: 5′ primer o r: 5 ′-CTGAGAAAGACATCGTGTACTCTGGCTTGGCCTAC-3′ and 3′ primer: REV Primer: 5′-GTAGGCCAAGCCAGAGTACACGATGTCTTTCTCAG-3′. Sequencing was then performed to confirm the correct mutation.
Transfection of CTX8 cells
To transfect CTX8 cells, we used the Rat Hippocampal Amaxa nucleofector kit (Lonza cat# VPG-1003). Neurospheres were collected and pelleted. 100 μl premixed Amaxa solution was added to pellet and gently agitated to redissolve pellet. Then indicated DNA was added (less than 5 μl), and the solution was gently shaken again a few times. The cells were electroporated using the G-013 code. 500 μl of pre-warmed culture media was immediately added to the cuvette and total contents were removed and diluted to appropriate concentration in culture media and placed in the tissue culture incubator. After 15-20 minutes, cells were plated on the appropriate substrate.
Image analysis
ImageJ was used for analysis of images. For analysis of co-expression, the co-localization plugin of ImageJ was utilized. After running the plugin, the images were converted to binary threshold images of equivalent threshold parameter. This parameter was defined as pixel intensity at least 5-fold higher than background intensity. The area of overlap was then calculated using the ImageJ area measure utility for each image, and a calculation of the percentage of co-localization was performed. For analysis of cell type marker expression, cell borders were first defined by pixels at least 5-fold higher than background intensity. Images of markers were then overlayed with the images of cells in a manner similar to that of co-localization analysis. Only cells with co-localization of markers that were within the cell border, followed the contours of the cell, and had at least 50% co-localization were considered positive for each marker. This minimized the possibility that coincidental co-localization would be considered positive. A background image that was acquired using identical camera settings but a blank slide was subtracted from the pixel intensity of all images. Prior to acquisition and analysis, all images were coded to blind the experimenter to condition.
Statistical analysis
Unless noted, significance was determined using one way ANOVA followed by Tukey’s HSD with p<0.05 considered statistically significant.
RESULTS
Expression of GDH and SIRT4 in the rodent brain
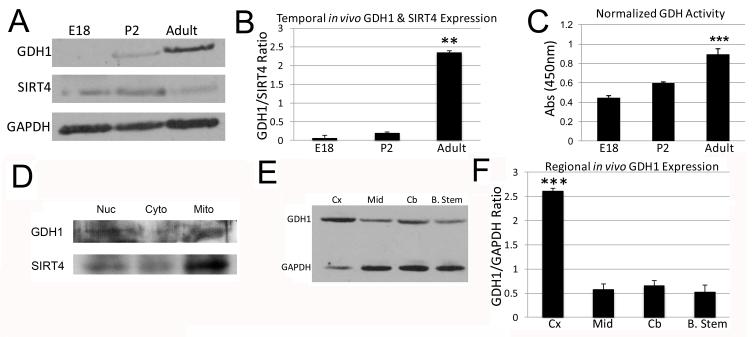
It has been previously reported that GDH protein expression increases in the hippocampus during postnatal rodent development (Kugler and Schleyer, 2004). To investigate embryonic expression of GDH, we performed Western blotting on equal volumes of whole brain lysate from embryonic day 18 (E18), postnatal day 2 (P2), and adult rat brain. Membranes were probed with antibodies recognizing GDH1 and SIRT4, and we observed a steady increase in GDH1 expression through development, with an associated increase in SIRT4 from E18 to P2 and then decreased expression by adulthood (Figure 1A). Ratios of SIRT4 to GAPDH, GDH1 to GAPDH, and GDH1 and SIRT4 expression were plotted, demonstrating a steady increase in the ratio of GDH1 expression as compared to SIRT4, progressively increasing into adulthood (Figure 1B). Since increases in the ratio of protein expression of GDH1 to its inhibitor SIRT4 do not necessarily equate with increased GDH1 activity, we utilized an in vitro assay to determine GDH1 activity. We found that in addition to an increase in protein expression, GDH activity also increases with development (Figure 1C). Furthermore, both GDH1 and SIRT4 localize to mitochondria (Figure 1D).
Figure 1. Western blot analysis of SIRT4 and GDH1 in the rodent brain.
A: Western blot of GDH1 (~56 kDa) and SIRT4 (~38 kDa) at Embryonic day 18 (E18), Postnatal day 2 (P2) and Adult. The blot was then reprobed with GAPDH (~38 kDa) to ensure that equal amounts of protein were loaded. B: Graphical representation of the normalized ratio of band intensity as a function of GDH1/SIRT4 for each given age of the blot in A. ***p<0.001. Adult ratio was significantly different than both E18 and P2. C: Changes in GDH1 activity using an in vitro GDH Assay Kit, as measured by absorbance at 450 nm in whole brain extracts at the indicated ages. ***p<0.001. Adult activity is significantly different than E18. P2 activity does not differ from that of E18 but is different than that of adult (p<0.01). D: Western blot of nuclear, cytosolic, and mitochondrial fractions probed for GDH1 and SIRT4. E: Western blot of GDH1 in adult rat brain samples in the specified regions; cerebral cortex (Cx), striatal cortex (Mid), cerebellum (Cb) and brain stem (B. Stem). The blot was reprobed with GAPDH to ensure that equal amounts of protein were loaded. F: Graphical representation of the normalized ratio of band intensity as a function of GDH1/GAPDH for each given region of the blot in E. ***p<0.001. There is a significant difference between Cx and Mid, Cx and Cb, and Cx and B. Stem.
Since our Western blot analysis used whole brain lysate, we asked whether there were differences in GDH1 expression within various adult rat brain regions. Equal volumes of samples were obtained from cerebral cortex, striatal cortex (midbrain), cerebellum and brain stem (approximately at the level of the medulla), and probed for GDH1 protein (Figure 1E). GDH1 levels were compared to GAPDH levels of the same sample, and quantified using densitometry. We found an approximate 4-fold increase in GDH1 protein levels in cortex compared to the other brain regions examined (Figure 1F), predicting a significant role for GDH within the cerebral cortex. It should be noted that SIRT4 expression within adult rat brain was too low to be detected when we separated out brain regions, so only GDH1 was examined.
Since GDH1 is most highly expressed in the adult cortex, we then asked whether expression of GDH1 and SIRT4 in the cortex is homogenous throughout all cell types, or as previous publications have implicated, strongly biased towards glial expression (Aoki et al., 1987; Kugler and Schleyer, 2004; Schmitt and Kugler, 1999). Although many specific cell types exist within the cortical layers, we analyzed three broad categories of cells: radial glia and directly related cells (ependymal cells, B and C cells), true glia (astrocytes, oligodendrocytes, microglia), and neurons. We cultured the three cell types, attempting to keep them as homogenous as culturing methods would permit. The CTX8 radial glial cell line, which possesses the capability to differentiate into either glia or neurons, can be kept in an “undifferentiated” radial glial fate (Li et al., 2012). Cultures of E18 cortical neurons treated with AraC represented pure neurons. Finally, astrocyte cultures obtained from E15 cortices served as our glial sample. We found that while there is little to no expression of GDH1 in radial glial and neuronal cultures, significant expression occurs in astrocyte cultures (data not shown). There was low expression of approximately equal amounts of SIRT4 in purified CTX8 cells and neurons, while expression of SIRT4 was undetectable in astrocyte cultures (data not shown). In contrast, GDH1 is highly expressed in astrocytes, with little detection in CTX8 cells and neurons. These findings help support previous data showing a predominantly glial, specifically astrocytic, expression of GDH1 and show that SIRT4 is expressed at low levels in the three cell types examined.
In vitro immunofluorescence expression of GDH1 and SIRT4 within the CNS
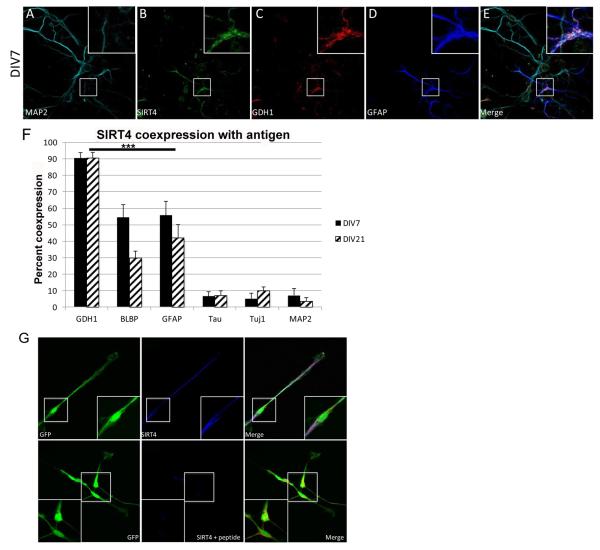
To perform an analysis of GDH1 and SIRT4 expression in specific cell types, we cultured cortical cells from rat embryos at embryonic day 18 and examined by immunocytochemical staining at various times in culture. Cells were fixed at DIV7 or DIV21 and labeled with antibodies against MAP2 (neurons), GFAP (astroglia), and SIRT4 or GDH1, and imaged using confocal microscopy (Figure 2A-E). SIRT4 and GDH1 are expressed in GFAP+ cells but are not expressed in MAP2+ cells. Since MAP2 only labels dendrites and does not label smaller neurites and axons, additional immunocytochemistry was performed with antibodies against tau and Tuj1, two neuronal projection markers (Menezes and Luskin, 1994; Tytell et al., 1984), and the Brain Lipid Binding Protein (BLBP), a marker for immature and mature glial cells (Malatesta et al., 2008). SIRT4 and GDH1 were co-expressed to the greatest degree, about 90%, followed by co-expression with BLBP and GFAP (Figure 2F). Co-expression of SIRT4 and GDH1 with the neuronal markers tau, Tuj1, and MAP2 showed a percentage that was not significantly greater than that of DAPI (data not shown), and therefore could be classified as coincidence rather than what could be inferred as actual physiologic co-expression. Furthermore, pre-absorption with SIRT4 antibody eliminated SIRT4 immunostaining, confirming specificity of the antibody (Figure 2G). These results support our biochemical findings of a predominantly glial expression pattern of both SIRT4 and GDH1 and allowed us to establish the basis for investigating the in vivo expression of these two proteins.
Figure 2. Expression of SIRT4 and GDH1 in dissociated cortical cultures.
A-D: Expression of the indicated protein in mixed neuronal/glial co-cultures from E18 cortical cultures, shown at DIV7, 60X magnification. E: Merged image of A-D. Boxed area in center indicates additional 3.4X magnified area at top right of each image (2A-E and 2G). F: Quantitation of percentage of co-expression of SIRT4 with indicated protein at DIV7 and DIV21. ***p< 0.001. GDH1, BLBP and GFAP are significantly different from tau, Tuj1 and DAPI staining. BLBP does not differ from GFAP. ***p < 0.001. Co-expression with GDH1, BLBP and GFAP is significantly different than co-expression with tau, Tuj1 and DAPI. G: Expression of GFP in CTX cells at DIV2. SIRT4 expression without and with SIRT4 specific antigenic peptide preincubation.
In vivo immunofluorescence expression of GDH1 and SIRT4 within the CNS
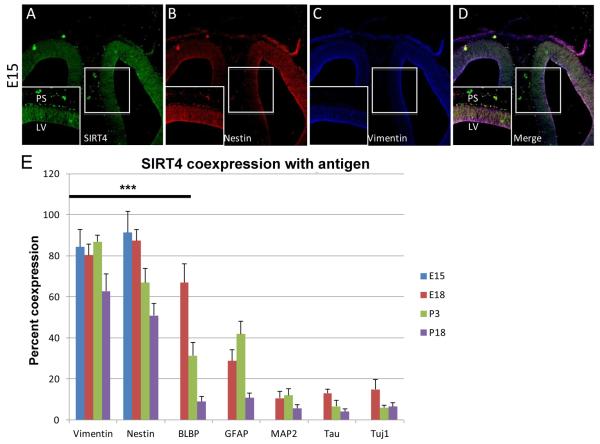
Our biochemical data showed that while GDH1 expression was low in the embryonic brain, SIRT4 expression was relatively high. During embryonic time points, when development of the rat cerebral cortex occurs, astrocytes have not yet fully developed, and the predominant cell types are migratory neurons and radial glia as radial glia are present in the rat brain from about E14-E15 (Malatesta et al., 2008). We analyzed brains from rats at E15 as a starting point for immunohistochemistry. We also analyzed brains from rats at E18, P3 and P18. Since we could not find a reliable GDH1 antibody that worked in in vivo sections, only co-expression of SIRT4 with cell-type specific markers was examined. SIRT4 is co-expressed with vimentin and nestin, and we found a high degree of co-expression, furthermore, we saw that a general decrease in co-expression occurs throughout cortical development (Figure 3E). When co-expression with the other cell type-specific markers was examined, we observed a pattern similar to that of dissociated cultures, with BLBP and GFAP having greater co-expression than any of the neuronal makers, MAP2, tau or Tuj1 (Figure 3E). At E18, only BLBP+ cells showed expression of SIRT4 above baseline, and by P3 and P18 only vimentin+ and nestin+ cells showed expression of SIRT4 above baseline (Figure 3E). Interestingly, we observed a relatively filamentous localization of SIRT4 in cells that were likely radial glia (Figure 3A), which looks similar to the expression pattern of nestin and vimentin at E15 (Figure 3B and 3C). The merged image highlights the co-localization of SIRT4 with nestin and vimentin (Figure 3D). As our findings and that of previous publications have shown that SIRT4 is mitochondrially localized (Haigis et al., 2006), we were concerned that perhaps the localization was non-specific because mitochondria are known to undergo a fission-fusion phenomena and have been shown to form filament-like structures in various cell types and under certain disease conditions (Chen and Chan, 2009; Hom and Sheu, 2009).
Figure 3. Expression of SIRT4 in cortical brain sections from rat.
A-C: Expression of indicated protein in E15 coronal cortical sections, 10X magnification. D: Merged image of A-C. Boxed area in center indicates additional 1.8X magnified area at bottom left of each image. PS= Pial Surface, LV= Lateral Ventricle. E: Quantitation of percentage of colocalization of SIRT4 with indicated protein at E15, E18, P3 and P18. ***p<0.001. At E18, there is significant co-expression of SIRT4 with vimentin and nestin and significant co-expression of SIRT4 with vimentin, nestin, and BLBP, but not GFAP, tau, Tuj1, and MAP2. At P3, there is significant co-expression of SIRT4 with vimentin and nestin, but not BLBP, GFAP, tau, Tuj1 and MAP2. At P18, there is significant co-expression of SIRT4 with vimentin and nestin, but not BLBP, GFAP, tau, Tuj1, and MAP2.
To address the concern of the unexpected SIRT4 expression pattern in vivo and distinguish between non-specific antibody labeling, we labeled E15 brain sections with the SIRT4 antibody with and without a SIRT4 specific antibody blocking peptide. The peptide consists of the same last 13 residues (of human SIRT4) that comprise the antibody’s antigen binding site, so when used in conjunction with the SIRT4 antibody, specific antibody binding should be eliminated and only non-specific labeling should remain. Results showed that when the SIRT4 blocking peptide was used in conjunction with the SIRT4 antibody, a significant decrease in SIRT4 labeling along cortical fibers occurred (data not shown). Sections were additionally labeled with vimentin antibody to ensure that the peptide did not eliminate all antibody binding. Fluorescence of vimentin remained essentially unchanged (data not shown).
Characterization of SIRT4 and GDH1 expression in CTX8 cells
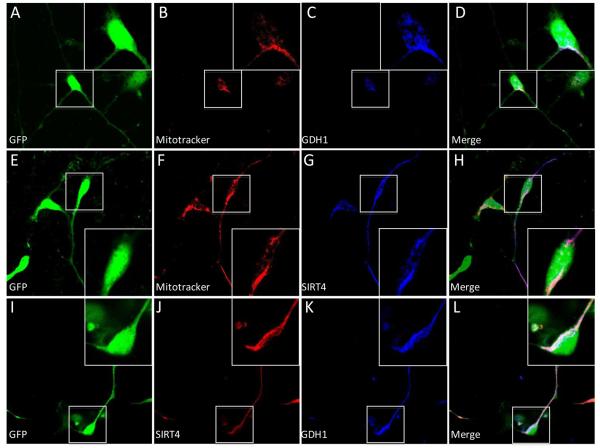
To determine the role of GDH1 and SIRT4 in neuron/astrocyte fate, we used CTX8 cells as a model. These GFP-positive cells were isolated from E14.5 cortices of rats expressing GFP under control of the β-actin promoter and were selected for their radial glial characteristics, which can be maintained in medium containing basic fibroblast growth factor (Li et al., 2012). CTX8 cells can also differentiate into both neurons and astroglia. To determine whether these cells express endogenous SIRT4 and GDH1 and where these proteins are localized, we labeled live cells with Mitotracker-Orange, which incorporates and is fluorescent only in the mitochondrial membrane and is retained post fixation. After fixation we also co-immunostained cells for either SIRT4 or GDH1 (Figure 4). Merged images show that both SIRT4 and GDH1 localized almost exclusively with Mitotracker and with each other, suggesting a mitochondrial localization and high degree of co-localization of the two proteins in cultured radial glial cells Figure 4D, H, and L).
Figure 4. Expression of SIRT4 and GDH1 in CTX8 cells.
A, E, I: GFP expression in DIV2 CTX8 cells. B, F: Mitotracker labeling of the CTX8 cells in A and E, respectively. C: Expression of GDH1 in cell A. G: Expression of SIRT4 in cell E. J, K: Expression of indicated protein in cell I. D, H, L: Merged image of A-C, E-G, I-K, respectively. Boxed area indicates additional 1.5X magnified area at respective corner.
Characterization of CTX8 radial glial cell line
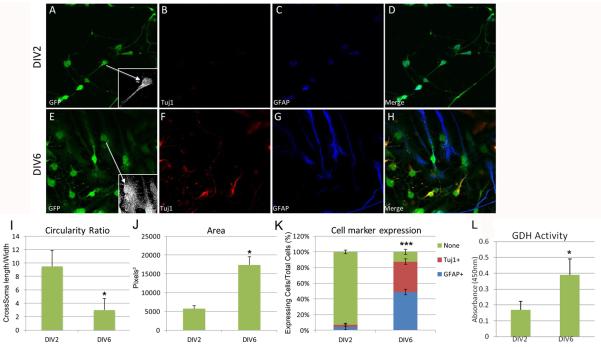
CTX8 cells can differentiate into both Tuj1+ and GFAP+ cells (Li et al., 2012). As such, we further characterized CTX8 cells before using them to alter GDH1 or SIRT4 levels. Radial glia, when cultured in vitro, are difficult to characterize by either morphology or cell-specific markers alone (Pollard and Conti, 2007). They contain long, thin projections (normally numbering 2), with one projection being longer and thinner than the other and a small oval-shaped soma. Cell-specific markers, such as vimentin, nestin, BLBP, RC2, and Pax6, can be used to identify radial glia; however, some of these markers (BLBP, vimentin and nestin) persist past the radial glial state into immature and even mature glial cells (Rakic, 2007). Additionally, the Excitatory Amino Acid Transporter 1 (EAAT1), a known marker for differentiating astrocytes, cannot reliably be used for identifying mature astrocytes, as radial glia (and CTX8 cells) express the marker (data not shown). Although in primates GFAP is expressed in both radial glia and astrocytes, it is only expressed in rodent astrocytes. Therefore, for our purposes of classifying CTX8 cell progeny, any cells that had filamentous GFAP expression were considered astrocytes. Likewise, Tuj1 is not expressed in radial glia, so any cells that expressed filamentous Tuj1 were considered neurons. We did not observe any CTX8 cells that express both GFAP and Tuj1, and cells that did not express either marker were placed in a separate category and considered radial glia not yet destined for either fate by DIV6 or that would not develop into either neurons or astrocytes. We observed typical radial glial morphology of the CTX8 cells at DIV2 and development into both neurons and astrocytes by DIV6 (Figure 5). The merged images highlight the mutual exclusivity of GFAP and Tuj1 expression within each cell at DIV 2 and DIV 6 (Figure 5D and 5H, respectively). An approximate 3-fold decrease in the circularity ratio of the cells occurred from DIV2 to DIV6 under normal culture conditions (Figure 5I). Cell area increased approximately 3.5 fold over the development of CTX8 cells, with large variability due to the differences in area of cells destined to become astrocytes, which are significantly larger than those becoming neurons (Figure 5J). Finally, when we quantified the difference in number of Tuj1+ and GFAP+ cells from DIV2 to DIV6, we found a significant increase from 3% to 40% Tuj1+ and 4% to 50% GFAP+ cells, and a large decrease in those cells that did not express either protein, from 92% at DIV2 to 10% at DIV6 (Figure 5K). We also measured GDH activity and found that there was a significant increase in GDH activity from DIV2 to DIV6 (Figure 5L). Collectively, these measures of changes in cell morphology and protein expression/activity gives us an adequate method of determining the changes that CTX8 cells undergo throughout development in culture, and as subsequent experiments would be aimed at examining how GDH1 activity may influence the formation of astrocytes from radial glia, we used the quantitation of differences in Tuj1 and GFAP expression as our measure of astrocyte formation.
Figure 5. Characterization of CTX8 radial glial cells.
A, E: Typical morphology of CTX8 cells at DIV2 and DIV6, respectively. Inset highlights morphological characteristics. B, F: Tuj1 expression at DIV2 and DIV6, respectively. Note that there is no visible Tuj1 expression at DIV2. C, G: GFAP expression at DIV2 and DIV6, respectively. Note the mild, centralized nuclear expression of GFAP at DIV2 and filamentous expression at DIV6. Only filamentous expression of GFAP is quantified as GFAP expression. D, H: Merged images of A-C and E-G, respectively. I: Quantitation of the changes in mean circularity ratio as defined by the quotient of cell length divided by width passing through the center of the soma. J: Quantitation of the changes cell area. K: Quantitation of changes in expression of Tuj1 and GFAP in DIV2 and DIV6 CTX8 cells. Expression of “none” is defined as a cell with no visible expression of either Tuj1 or GFAP. L: GDH activity assay performed on extracts from CTX cells at DIV2 and DIV6. GDH activity increases over time. *p<0.05 and ***p <0.001 by t-test for all panels.
Alterations in gliogenesis in CTX8 cells with changes in culture conditions
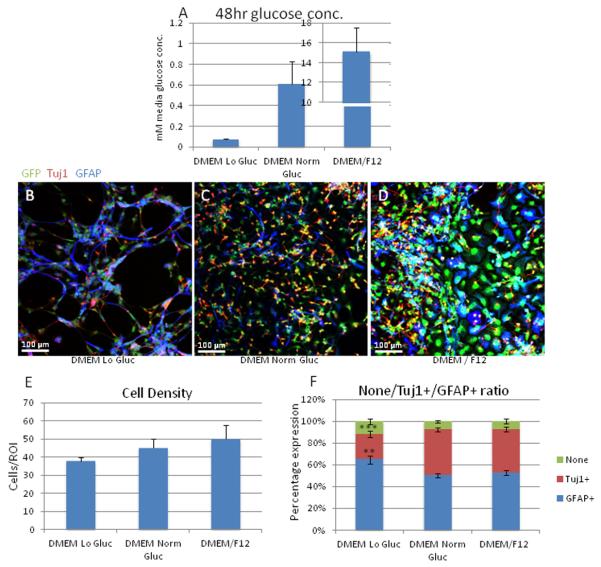
Once we established that CTX8 cells possess the ability to form astrocyte-like cells and that they express GDH1, we asked whether factors related to the allosteric regulation of GDH1 could affect the formation of astrocytes from CTX8 radial glia. We considered factors which have been shown to alter GDH1 activity such as glucose, glutamate and glutamine, alpha-ketoglutarate and EGCG concentrations. We also treated cells with trans-PDC, an EAAT1-5 blocker, as a potential blocker of glutamate uptake, to assess effects on astrocyte formation. We first set out to alter the glucose concentration in CTX8 media, and thus, created media conditions that more accurately represent low glucose conditions that occur in the brain during clinically significant hypoglycemic events. Although previous publications have induced GDH activity by complete withdrawal of glucose, we chose to simulate glucose concentrations similar to those experienced in the brain, less than 0.3 mM, during clinically significant hypoglycemia (Canabal et al., 2007). We preferred this low glucose condition to a complete lack of glucose which is unlikely to occur in vivo. Since standard DMEM/F12 contains a glucose concentration of 17.5 mM, which is approximately 3.5 fold higher than mean plasma rat (and human) glucose, which averages about 5 mM (Gain et al., 1981), and approximately 17 fold higher than what occurs within the rat brain, which averages about 1 mM and ranges between 0.5 mM to 1.5 mM (Song and Routh, 2006), we used a glucose-deficient DMEM and supplemented it with the appropriate amount of D-glucose to create a “normal glucose DMEM” (glucose concentration of 1 mM) and a “low glucose DMEM” (glucose concentration of 0.25 mM, as would be present in the brain during symptomatic hypoglycemia (Canabal et al., 2007). We also cultured cells in standard DMEM/F12 as a comparison to determine what affect the change from DMEM/F12 to “normal glucose DMEM” would have on CTX8 cell differentiation. To reduce variability, all of our DMEM-only media was supplemented with an additional 50 μM L-glutamic acid to mimic the glutamate concentration of DMEM/F12 as DMEM contains no glutamate. Since changes in glutamate concentration could potentially alter GDH1 activity and therefore the GFAP/Tuj1 ratio, this supplementation was necessary to reduce the factors that could alter the GFAP/Tuj1 ratio and potentially mask or exaggerate changes caused by the altered glucose concentrations. To ensure that the amount of glucose consumed by the cultured cells did not completely deplete media glucose concentrations, accidentally creating unanticipated hypoglycemic conditions in the normal glucose concentration media, we performed a glucose assay. After 48 hours in altered glucose conditions, concentrations were still within originally set limits (Figure 6A). To ensure that our media conditions contain high glucose levels (DMEM/F12) and normal glucose levels (DMEM, which has significantly lower glucose than DMEM/F12), we measured glucose levels of CTX8 cells after 48hours in DMEM/F12 and found that glucose levels remained considerably high (15 mM versus original 17.5 mM). To ensure no further decrease in glucose concentration, we changed the media every 2 days until DIV6, at which time we fixed and immunostained cells to quantify changes in GFAP/Tuj1 ratio (Figure 6B-D). We found no significant change in total cell number occurred with increasing glucose concentration (Figure 6E). In contrast, the percentage of GFAP+ cells significantly increased when cells were incubated in low glucose conditions, and the percentage of Tuj1+ cells significantly decreased (Figure 6F). These results suggest that extracellular glucose concentration may play a role in development of astrocytes as well as neurons, and that decreases in the availability of glucose lead to the preferential formation of glia at the expense of neurons.
Figure 6. Alteration in GFAP/Tuj1 ratio with altered media glucose levels.
A: Quantitation of glucose levels in media after 48 hours of media change to ensure that glucose concentration remained within intended ranges. DMEM Lo Gluc was 0.3 mM at time 0 and decreased to 0.08 mM at 48 hrs, DMEM Norm Gluc was 1 mM at time 0 and decreased to 0.6mM at 48 hrs, and DMEM/F12 was 17 mM at time 0 and decreased to 15m M at 48hrs. B-D: Images of CTX8 cells immunostained for GFAP and Tuj1 in indicated glucose conditions. E: Quantitation of cell density in each glucose condition. While no significant differences were observed, there was a trend toward more cells per area as the glucose concentration increased. F: Quantitation of changes in GFAP/Tuj1 ratio with changes in glucose levels in media at DIV6. No significant difference was seen in the number of cells that did not express either protein. There is significant decrease in the number of Tuj1+ cells (***p<0.001) in the Lo glucose condition compared to both the Norm glucose and DMEM/F12. A significant increase in the number of GFAP+ cells was seen (**p< 0.01) in the Lo glucose condition compared to both the Norm glucose and DMEM/F12.
Alterations in gliogenesis in CTX8 cells occurs when GDH1 activity is altered
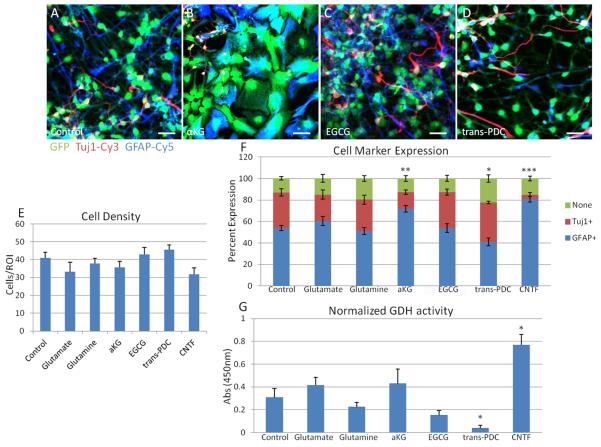
Once we established that CTX8 cells possess the ability to form astrocyte-like cells and that they express GDH1, we asked whether factors related to the allosteric regulation of GDH1 can affect the formation of astrocytes from CTX8 radial glia. We added either glutamate or α-KG, the two direct substrates and products of GDH1, to the medium to determine whether they can modify gliogenesis. Glutamine, which can be converted into glutamate by glutaminase, was added to the media to regulate GDH1 via increased mitochondrial glutamate concentrations (Smith and Stanley, 2008). Lastly, we incubated the cells with the green tea polyphenol, EGCG, a known inhibitor of GDH activity at a previously determined concentration of 1 μM (Li et al., 2006) or with L-trans-2,4-PDC (Blitzblau et al., 1996; Matsui et al., 1999), an inhibitor of EAAT1-5, which has been shown to inhibit glutamate uptake in glial cells (Dugan et al., 1995). As a positive control to ensure that our paradigm accurately represented the changes that occurred in culture, we treated cells with ciliary neurotrophic factor (CNTF), a known activator of the JAK-STAT pathway, which in turn can function as an activator of astrocytes (Escartin et al., 2006; Escartin et al., 2007; Wu et al., 2006). We then allowed CTX8 cells to differentiate spontaneously for 6 days in DMEM/F12 media without bFGF, in the presence of the given specific factor, followed by fixation and immunostaining for GFAP and Tuj1 (Figure 7A-D), and quantified as described above. No significant changes in overall cell density were observed in any condition (Figure 7E). CNTF yielded a significantly greater percentage of GFAP+ cells, about 81%, with an associated significant decrease in Tuj1+ cells. The presence of glutamate did skew towards a greater percentage of GFAP+ cells, although this change was not statistically significant, while the addition of glutamine had no effect. Incubation with α-KG did, however, result in significantly more GFAP+ cells and less Tuj1+ cells (Figure 7F), while EGCG had no effect (Figure 7F). Interestingly, treatment of cells with L-trans-2,4-PDC resulted in a significant decrease in the number of GFAP+ cells without affecting the Tuj1+ number and yielded more CTX8 cells that did not express either marker (Figure 7F). These data suggest a shift in the formation of GFAP+ cells to those cells maintaining a radial glial state. To determine whether these changes in GFAP expression had any correlation to GDH1 activity, we measured GDH1 activity in each condition. Although there was an apparent decrease in GDH1 activity with glutamate and α-KG treatment, they were not statistically significant (Figure 7G). Only treatment with CNTF produced a significant increase in GDH activity (Figure 7G). Interestingly, however, treatment with trans-PDC caused a significant decrease in GDH1 activity (Figure 7G), suggesting that the blocking glutamate uptake in CTX8 cells may be related to the activity of GDH1.
Figure 7. Analysis of GDH activity and differentiation of treated CTX8 cells.
Differentiation analysis of treated CTX8 cells. A-D: Merged images of representative DIV6 CTX8 cells with various treatments (A, Control; B, α-ketoglutarate; C, EGCG; D, trans-PDC). Scale bar represents 20 μM. E: Quantitation of cell density for each specific treatment to appreciate any changes in cell population. No significant changes in cell density with treatment occurred. F: Quantitation of changes in expression of Tuj1 and GFAP in DIV6 CTX8 cells with indicated treatment. Expression of “none” is defined as a cell with no visible expression of either tuj1 or GFAP. Compared to control conditions, there is a significant increase in the number of cells expressing neither Tuj1 or GFAP only with PDC treatment. Compared to control conditions, there is a significant decrease in the number of cells expressing Tuj1 with aKG and CNTF treatment. Compared to control conditions, there is a significant increase in the number of cells expressing GFAP with aKG and CNTF treatment, and a significant decrease in the number of cells expressing GFAP with PDC treatment. *p < 0.05 ** p < 0.01, ***p < 0.001. G: Quantitation of Normalized GDH activity using a GDH assay kit, as measured by absorbance at 450nm, from lysate of DIV6 CTX8 cells with indicated treatments. One way ANOVA followed by Dunnett’s showed that there was a significant decrease in GDH activity with trans-PDC treatment compared to control, * p <0.05, and a significant increase in GDH activity with CNTF treatment, * p <0.05.
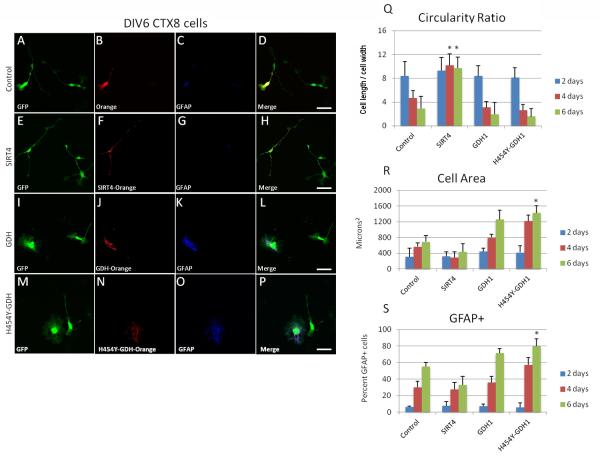
Overexpression of GDH1, H454Y-GDH1 and SIRT4 in CTX8 cells
To more thoroughly establish the role that GDH1 plays in the development of glia, we created overexpression vectors of GDH1, its overactive mutant variant H454Y-GDH1 that is found in patients with HI/HA (Fang et al., 2002; MacMullen et al., 2001; Stanley et al., 1998), and SIRT4 to represent a potential GDH1 inhibitor. We transfected the CTX8 cells at the time of plating, and cells were allowed to express the protein of interest for 2, 4, or 6 days. Cells were then fixed and immunostained for GFAP and three measurements were analyzed: circularity ratio, total cell area and presence of filamentous GFAP expression. When examined for circularity ratio, there were no significant differences at 2 days of expression in all conditions, and cells exhibited a high ratio, signifying long processes. However at 4 and 6 days of expression, cells became more circular except for cells overexpressing SIRT4, which maintained long thin processes and a typical radial glial morphology over the course of the experiment and had a significantly higher circularity ratio at DIV4 and 6 than cells in all other conditions (Figure 8A-H and 8Q). These results suggest that SIRT4 plays a role in maintaining cells with a radial glial morphology. Analysis of cell area revealed no difference at 2 days post transfection of any plasmid; however, there was a steady increase in cell area at 4 and 6 days in cells overexpressing GDH1 and H454Y-GDH1, with a significant increase in cell area in cells overexpressing H454Y-GDH1 at DIV6 (Figure 8I-P and 8R), and a trend towards decreased cell area in cells overexpressing SIRT4 (Figure 8E-H and 8R), although this change was not statistically significant. As CTX8 cell area generally increases with days in culture, it was intriguing to find that SIRT4 and GDH1 play antagonistic roles in regulating cell size (Figure 8R). We observed a significant increase in percentage of cells that expressed filamentous GFAP at 6 days in culture in cells overexpressing H454Y-GDH (Figure 8S) although we did not see a change in GFAP expression in cells overexpressing SIRT4 (Figure 8S). These findings of GFAP expression support the hypothesis that the activity of GDH1 affects the formation of glia from radial glia, and our morphological findings support the idea that SIRT4 and GDH1 play antagonistic roles in the regulation of gliogenesis in CTX8 cells.
Figure 8. Overexpression of SIRT4 and GDH1 in CTX8 cells.
A, E, I, M: GFP expression in CTX8 cells at DIV6. B, F, J, N: Expression of indicated protein in A, E, I, M, respectively. C, G, K, O: GFAP expression in CTX8 cells of the respective row. D, H, L, P: Merged image of A-C, E-G, I-K and M-O, respectively. Q: Quantitation of circularity ratio as defined by the ratio of cross somal length divided by width of indicated cells at DIV2, 4, and 6. There is a significant increase in the circularity ratio of cells overexpressing SIRT4 at DIV4 and 6 as compared to control, *p<0.05. R: Quantitation of cell area in μm2 of indicated cells at DIV2, 4, and 6. There is a significant increase in the cell area of cells overexpressing H454Y-GDH1 at DIV6 as compared to control, *p<0.05. S: Quantitation of percentage of GFAP+ cells at DIV2, 4 and 6. There is a significant increase in the percent of cells expressing GFAP in cells overexpressing H454Y-GDH1 at DIV6 as compared to control, *p<0.05.
DISCUSSION
The main finding of this study is that GDH1 and SIRT4 play opposing roles in the development of astroglia from radial glia. We have also shown for the first time that SIRT4 is expressed in the central nervous system during development and adulthood. Specifically, overexpression of GDH1 and a constitutively active mutant, H454Y-GDH found in patients with HI/HA, drive cells towards an astroglial phenotype. Pharmacological manipulation of GDH1 phenocopies this effect. Conversely, overexpression of SIRT4 drives cells away from an astroglial phenotype. This is the first study to show a role for these two proteins in gliogenesis. We also present a novel cell culture model for the study of differentiation of progenitor cells. CTX8 cells were isolated from E14.5 cortices and selected for their radial glial characteristics, which can be maintained in basic fibroblast growth factor (Li et al., 2011). In our study, we show that we can indeed use these cells to study factors that modulate gliogenesis.
Interestingly, the most studied sirtuin, SIRT1 has also been linked to determination of neuronal versus astroglial fate. SIRT1 null mice infrequently survive to adulthood with many dying in utero. Those that do survive postnatally are smaller, have underdeveloped brains and significant developmental defects (Teng et al., 2009). In neuroprogenitor cells, SIRT1 upregulation occurs in response to mild oxidative stress (Prozorovski 2008). This event results in an increase in progenitor cells developing towards an astrocyte versus neuronal fate. These findings were substantiated by addition of the SIRT1 activator Resveratrol and inhibition of SIRT1 by siRNA, both of which abolished the effect of mild oxidative stress (Prozorovski et al., 2008). In vivo knockdown of SIRT1 performed with in utero electroporation leads to an increase in cells differentiating towards a neuronal fate, a finding supported by induction of daily oxidative stress to mouse pups, which leads to an increase in SIRT1 levels and decrease in cells destined to a neuronal lineage (Prozorovski et al., 2008). Interestingly, we have found that SIRT4 increases the circularity ratio and decreases the cell area (Figure 8), which corresponds to keeping cells in a radial glial state. We do not believe that SIRT4 is sufficient in itself to drive radial glial towards a neuronal state as we did not see co-expression of SIRT4 and MAP2 or Tuj1 in vitro (Figure 2) or in vivo (Figure 3). Future studies will address whether SIRT4 acts as part of a pathway for neuronal differentiation, which may be inhibited by factors, such as TGF-β (Stipursky and Gomes, 2007). In contrast, activation of GDH results in rounder (closer to circularity index of 1), larger cells, which express GFAP, suggesting that GDH drives cells toward an astroglial fate.
SIRT4 is expressed outside of the brain, and it has been extensively studied for its role in liver, muscle, and calorie restriction. SIRT4 knockdown by RNAi in insulin secreting cell lines results in increased insulin secretion, and SIRT4 knock-out mice exhibit ~30% increased circulating insulin levels under normal feeding conditions, as well as an even greater increase under fasting conditions (Haigis et al., 2006). Amino-acid stimulated insulin secretion (AASIS), a known mechanism for stimulating insulin secretion (Cline et al., 2004; Kelly and Stanley, 2001), is also increased when leucine is added to SIRT4 RNAi expressing cells (Haigis et al., 2006). In addition, AASIS in SIRT4 knock-out mice under calorie restriction conditions show significantly increased insulin secretion. Taken together, these findings revealed a novel and important role for SIRT4 in regulating insulin secretion in vivo.
In line with this function for SIRT4, a dominant activating mutation of GDH1, H454Y-GDH, results in congenital hyperinsulinism/hyperammonemia (HI/HA) syndrome. The most prominent feature of HI/HA syndrome is seizures. Symptoms generally begin and end quickly. Furthermore, GDH protein levels are altered during the different phases of seizure in a rat model (Hammer et al., 2008). In a clinical study by Raizen et al (2005), fourteen patients with histories of HI/HA syndrome were examined for various neurological defects. Nine exhibited seizures, and among them only three had hypoglycemia at the time of seizure; the other six had normal blood sugars. Although hypoglycemia is a leading cause of pediatric seizures, aside from inborn developmental defects or trauma, the finding that only 1/3 of the patients in the Raizen (Raizen et al., 2005) study had hypoglycemia during seizure led to the inference that another yet undetermined mechanism for the seizure activity exists (Palladino and Stanley, 2010). Moreover, when general developmental aspects of children with HI/HA syndrome were studied in a group of French children, a significant portion were found to have some degree of developmental defects, ranging from mild to severe mental retardation (Bahi-Buisson et al., 2008). Based on our findings, we suggest that one of the mechanisms involved in the neurologocial defects observed in patients with HI/HA syndrome may be due to hyperactivity of GDH1 within the brain leading to an imbalance of glia and neurons in those with HI/HA syndrome. This imbalance may explain the seizures observed in patients with HI/HA syndrome.
ACKNOWLEDGEMENTS
We thank Drs. Hedong Li and Martin Grumet for providing CTX8 cells. This work was supported in part by NSF grant IBN-0919747 and March of Dimes Foundation Grant 1-FY08-464. D.K. was awarded an Anna B. and James Leathem Fellowship. C.L.R. and R.P.H. were supported by NIH R21 MH085088. C.L.R. was an NSF IGERT Trainee.
REFERENCES
- Aoki C, Milner TA, Sheu KF, Blass JP, Pickel VM. Regional distribution of astrocytes with intense immunoreactivity for glutamate dehydrogenase in rat brain: implications for neuron-glia interactions in glutamate transmission. J Neurosci. 1987;7:2214–2231. doi: 10.1523/JNEUROSCI.07-07-02214.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahi-Buisson N, Roze E, Dionisi C, Escande F, Valayannopoulos V, Feillet F, Heinrichs C, Chadefaux-Vekemans B, Dan B, de Lonlay P. Neurological aspects of hyperinsulinism-hyperammonaemia syndrome. Dev Med Child Neurol. 2008;50:945–949. doi: 10.1111/j.1469-8749.2008.03114.x. [DOI] [PubMed] [Google Scholar]
- Blitzblau R, Gupta S, Djali S, Robinson MB, Rosenberg PA. The glutamate transport inhibitor L-trans-pyrrolidine-2,4-dicarboxylate indirectly evokes NMDA receptor mediated neurotoxicity in rat cortical cultures. Eur J Neurosci. 1996;8:1840–1852. doi: 10.1111/j.1460-9568.1996.tb01328.x. [DOI] [PubMed] [Google Scholar]
- Canabal DD, Song Z, Potian JG, Beuve A, McArdle JJ, Routh VH. Glucose, insulin, and leptin signaling pathways modulate nitric oxide synthesis in glucose-inhibited neurons in the ventromedial hypothalamus. Am J Physiol Regul Integr Comp Physiol. 2007;292:R1418–1428. doi: 10.1152/ajpregu.00216.2006. [DOI] [PubMed] [Google Scholar]
- Charych EI, Akum BF, Goldberg JS, Jornsten RJ, Rongo C, Zheng JQ, Firestein BL. Activity-independent regulation of dendrite patterning by postsynaptic density protein PSD-95. J Neurosci. 2006;26:10164–10176. doi: 10.1523/JNEUROSCI.2379-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chan DC. Mitochondrial dynamics--fusion, fission, movement, and mitophagy--in neurodegenerative diseases. Hum Mol Genet. 2009;18:R169–176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline GW, Lepine RL, Papas KK, Kibbey RG, Shulman GI. 13C NMR isotopomer analysis of anaplerotic pathways in INS-1 cells. J Biol Chem. 2004;279:44370–44375. doi: 10.1074/jbc.M311842200. [DOI] [PubMed] [Google Scholar]
- de Lonlay P, Fournet JC, Touati G, Groos MS, Martin D, Sevin C, Delagne V, Mayaud C, Chigot V, Sempoux C, et al. Heterogeneity of persistent hyperinsulinaemic hypoglycaemia. A series of 175 cases. Eur J Pediatr. 2002a;161:37–48. doi: 10.1007/s004310100847. [DOI] [PubMed] [Google Scholar]
- de Lonlay P, Touati G, Robert JJ, Saudubray JM. Persistent hyperinsulinaemic hypoglycaemia. Semin Neonatol. 2002b;7:95–100. doi: 10.1053/siny.2001.0090. [DOI] [PubMed] [Google Scholar]
- Dugan LL, Bruno VM, Amagasu SM, Giffard RG. Glia modulate the response of murine cortical neurons to excitotoxicity: glia exacerbate AMPA neurotoxicity. J Neurosci. 1995;15:4545–4555. doi: 10.1523/JNEUROSCI.15-06-04545.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutuit M, Didier-Bazes M, Vergnes M, Mutin M, Conjard A, Akaoka H, Belin MF, Touret M. Specific alteration in the expression of glial fibrillary acidic protein, glutamate dehydrogenase, and glutamine synthetase in rats with genetic absence epilepsy. Glia. 2000;32:15–24. doi: 10.1002/1098-1136(200010)32:1<15::aid-glia20>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Escartin C, Brouillet E, Gubellini P, Trioulier Y, Jacquard C, Smadja C, Knott GW, Kerkerian-Le Goff L, Deglon N, Hantraye P, et al. Ciliary neurotrophic factor activates astrocytes, redistributes their glutamate transporters GLAST and GLT-1 to raft microdomains, and improves glutamate handling in vivo. J Neurosci. 2006;26:5978–5989. doi: 10.1523/JNEUROSCI.0302-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escartin C, Pierre K, Colin A, Brouillet E, Delzescaux T, Guillermier M, Dhenain M, Deglon N, Hantraye P, Pellerin L, et al. Activation of astrocytes by CNTF induces metabolic plasticity and increases resistance to metabolic insults. J Neurosci. 2007;27:7094–7104. doi: 10.1523/JNEUROSCI.0174-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Hsu BY, MacMullen CM, Poncz M, Smith TJ, Stanley CA. Expression, purification and characterization of human glutamate dehydrogenase (GDH) allosteric regulatory mutations. Biochem J. 2002;363:81–87. doi: 10.1042/0264-6021:3630081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein BL, Brenman JE, Aoki C, Sanchez-Perez AM, El-Husseini AE, Bredt DS. Cypin: a cytosolic regulator of PSD-95 postsynaptic targeting. Neuron. 1999;24:659–672. doi: 10.1016/s0896-6273(00)81120-4. [DOI] [PubMed] [Google Scholar]
- Gain KR, Malthus R, Watts C. Glucose homeostasis during the perinatal period in normal rats and rats with a glycogen storage disorder. J Clin Invest. 1981;67:1569–1573. doi: 10.1172/JCI110188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Hammer J, Alvestad S, Osen KK, Skare O, Sonnewald U, Ottersen OP. Expression of glutamine synthetase and glutamate dehydrogenase in the latent phase and chronic phase in the kainate model of temporal lobe epilepsy. Glia. 2008;56:856–868. doi: 10.1002/glia.20659. [DOI] [PubMed] [Google Scholar]
- Hom J, Sheu SS. Morphological dynamics of mitochondria--a special emphasis on cardiac muscle cells. J Mol Cell Cardiol. 2009;46:811–820. doi: 10.1016/j.yjmcc.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A, Stanley CA. Disorders of glutamate metabolism. Ment Retard Dev Disabil Res Rev. 2001;7:287–295. doi: 10.1002/mrdd.1040. [DOI] [PubMed] [Google Scholar]
- Kugler P, Schleyer V. Developmental expression of glutamate transporters and glutamate dehydrogenase in astrocytes of the postnatal rat hippocampus. Hippocampus. 2004;14:975–985. doi: 10.1002/hipo.20015. [DOI] [PubMed] [Google Scholar]
- Kwon M, Fernandez JR, Zegarek GF, Lo SB, Firestein BL. BDNF-promoted increases in proximal dendrites occurs via CREB-dependent transcriptional regulation of cypin. Journal of Neuroscience. 2011;31:9735–9745. doi: 10.1523/JNEUROSCI.6785-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Allen A, Kwagh J, Doliba NM, Qin W, Najafi H, Collins HW, Matschinsky FM, Stanley CA, Smith TJ. Green tea polyphenols modulate insulin secretion by inhibiting glutamate dehydrogenase. J Biol Chem. 2006;281:10214–10221. doi: 10.1074/jbc.M512792200. [DOI] [PubMed] [Google Scholar]
- Li H, Hader AT, Han YR, Wong JA, Babiarz J, Ricupero CL, Godfrey SB, Corradi JP, Fennell M, Hart RP, et al. Isolation of a novel rat neural progenitor clone that expresses Dlx family transcription factors and gives rise to functional GABAergic neurons in culture. Dev Neurobiol. 2012;72:805–820. doi: 10.1002/dneu.20977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMullen C, Fang J, Hsu BY, Kelly A, de Lonlay-Debeney P, Saudubray JM, Ganguly A, Smith TJ, Stanley CA. Hyperinsulinism/hyperammonemia syndrome in children with regulatory mutations in the inhibitory guanosine triphosphate-binding domain of glutamate dehydrogenase. J Clin Endocrinol Metab. 2001;86:1782–1787. doi: 10.1210/jcem.86.4.7414. [DOI] [PubMed] [Google Scholar]
- Malatesta P, Appolloni I, Calzolari F. Radial glia and neural stem cells. Cell Tissue Res. 2008;331:165–178. doi: 10.1007/s00441-007-0481-8. [DOI] [PubMed] [Google Scholar]
- Matsui K, Hosoi N, Tachibana M. Active role of glutamate uptake in the synaptic transmission from retinal nonspiking neurons. J Neurosci. 1999;19:6755–6766. doi: 10.1523/JNEUROSCI.19-16-06755.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes JR, Luskin MB. Expression of neuron-specific tubulin defines a novel population in the proliferative layers of the developing telencephalon. J Neurosci. 1994;14:5399–5416. doi: 10.1523/JNEUROSCI.14-09-05399.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas WJ. Amino acid metabolism in the central nervous system: role of glutamate dehydrogenase. Adv Neurol. 1984;41:245–253. [PubMed] [Google Scholar]
- Palladino AA, Stanley CA. The hyperinsulinism/hyperammonemia syndrome. Rev Endocr Metab Disord. 2010;11:171–178. doi: 10.1007/s11154-010-9146-0. [DOI] [PubMed] [Google Scholar]
- Pollard SM, Conti L. Investigating radial glia in vitro. Prog Neurobiol. 2007;83:53–67. doi: 10.1016/j.pneurobio.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Prozorovski T, Schulze-Topphoff U, Glumm R, Baumgart J, Schroter F, Ninnemann O, Siegert E, Bendix I, Brustle O, Nitsch R, et al. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat Cell Biol. 2008;10:385–394. doi: 10.1038/ncb1700. [DOI] [PubMed] [Google Scholar]
- Raizen DM, Brooks-Kayal A, Steinkrauss L, Tennekoon GI, Stanley CA, Kelly A. Central nervous system hyperexcitability associated with glutamate dehydrogenase gain of function mutations. J Pediatr. 2005;146:388–394. doi: 10.1016/j.jpeds.2004.10.040. [DOI] [PubMed] [Google Scholar]
- Rakic P. The radial edifice of cortical architecture: from neuronal silhouettes to genetic engineering. Brain Res Rev. 2007;55:204–219. doi: 10.1016/j.brainresrev.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt A, Kugler P. Cellular and regional expression of glutamate dehydrogenase in the rat nervous system: non-radioactive in situ hybridization and comparative immunocytochemistry. Neuroscience. 1999;92:293–308. doi: 10.1016/s0306-4522(98)00740-4. [DOI] [PubMed] [Google Scholar]
- Shashidharan P, Michaelidis TM, Robakis NK, Kresovali A, Papamatheakis J, Plaitakis A. Novel human glutamate dehydrogenase expressed in neural and testicular tissues and encoded by an X-linked intronless gene. J Biol Chem. 1994;269:16971–16976. [PubMed] [Google Scholar]
- Smith TJ, Stanley CA. Untangling the glutamate dehydrogenase allosteric nightmare. Trends Biochem Sci. 2008;33:557–564. doi: 10.1016/j.tibs.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Song Z, Routh VH. Recurrent hypoglycemia reduces the glucose sensitivity of glucose-inhibited neurons in the ventromedial hypothalamus nucleus. Am J Physiol Regul Integr Comp Physiol. 2006;291:R1283–1287. doi: 10.1152/ajpregu.00148.2006. [DOI] [PubMed] [Google Scholar]
- Spanaki C, Zaganas I, Kleopa KA, Plaitakis A. Human GLUD2 glutamate dehydrogenase is expressed in neural and testicular supporting cells. J Biol Chem. 2010;285:16748–16756. doi: 10.1074/jbc.M109.092999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley CA. Regulation of glutamate metabolism and insulin secretion by glutamate dehydrogenase in hypoglycemic children. Am J Clin Nutr. 2009;90:862S–866S. doi: 10.3945/ajcn.2009.27462AA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley CA, Fang J, Kutyna K, Hsu BY, Ming JE, Glaser B, Poncz M, HI/HA Contributing Investigators Molecular basis and characterization of the hyperinsulinism/hyperammonemia syndrome: predominance of mutations in exons 11 and 12 of the glutamate dehydrogenase gene. Diabetes. 2000;49:667–673. doi: 10.2337/diabetes.49.4.667. [DOI] [PubMed] [Google Scholar]
- Stanley CA, Lieu YK, Hsu BY, Burlina AB, Greenberg CR, Hopwood NJ, Perlman K, Rich BH, Zammarchi E, Poncz M. Hyperinsulinism and hyperammonemia in infants with regulatory mutations of the glutamate dehydrogenase gene. N Engl J Med. 1998;338:1352–1357. doi: 10.1056/NEJM199805073381904. [DOI] [PubMed] [Google Scholar]
- Stipursky J, Gomes FC. TGF-beta1/SMAD signaling induces astrocyte fate commitment in vitro: implications for radial glia development. Glia. 2007;55:1023–1033. doi: 10.1002/glia.20522. [DOI] [PubMed] [Google Scholar]
- Sweet ES, Previtera ML, Fernandez JR, Charych EI, Tseng CY, Kwon M, Starovoytov V, Zheng JQ, Firestein BL. PSD-95 alters microtubule dynamics via an association with EB3. J Neurosci. 2011;31:1038–1047. doi: 10.1523/JNEUROSCI.1205-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng FY, Hor CH, Tang BL. Emerging cues mediating astroglia lineage restriction of progenitor cells in the injured/diseased adult CNS. Differentiation. 2009;77:121–127. doi: 10.1016/j.diff.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Tytell M, Brady ST, Lasek RJ. Axonal transport of a subclass of tau proteins: evidence for the regional differentiation of microtubules in neurons. Proc Natl Acad Sci U S A. 1984;81:1570–1574. doi: 10.1073/pnas.81.5.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Liu RG, Zhou JP. Effect of ciliary neurotrophic factor on activation of astrocytes in vitro. Neurosci Bull. 2006;22:315–322. [PubMed] [Google Scholar]
- Zhang H, Das S, Li QZ, Dragatsis I, Repa J, Zeitlin S, Hajnoczky G, Bezprozvanny I. Elucidating a normal function of huntingtin by functional and microarray analysis of huntingtin-null mouse embryonic fibroblasts. BMC Neurosci. 2008;9:38. doi: 10.1186/1471-2202-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]