Abstract
Plant alkaloids have a rich chemical ecology that has been exploited for medicinal purposes for thousands of years. Despite being highly represented within today’s pharmacopoeia, relatively little is known about the biosynthesis, regulation and transport of these molecules. Understanding how nature synthesizes plant alkaloids will enhance our ability to overproduce—that is, to metabolically engineer—these medicinally useful compounds as well as new-to-nature compounds (with potentially improved bioactivity) derived from these natural scaffolds. Recent progress in the metabolic engineering of nitrogen-containing plant natural products—specifically the monoterpene indole alkaloids, the benzylisoquinoline alkaloids and the glucosinolates—was made possible through the characterization of various components in both native and engineered enzymatic pathways. The subsequent reconfiguration and tuning of these biological “parts” has enabled the production of selected products at increasingly higher titers.
Introduction
Plant natural products have been exploited for thousand of years. The alkaloids, particularly, have a long and storied narrative. This history is highlighted in the life of Cleopatra, who used alkaloid-containing extracts from belladonna (Italian for ‘beautiful woman’) to dilate her pupils so as to increase her beauty and thereby disarm her enemies [1]. Far from being confined to ancient chronicles, the alkaloids retain a palpable presence in today’s clinics. For example, optometrists still apply eye drops containing the alkaloid atropine, an active component of belladonna, to dilate the pupil during routine eye exams [1]. It is unsurprising that most alkaloids are bioactive given that the evolutionary process selects for products that confer an advantage to the producing organism. Despite the rich ethnopharmacological tradition and high usage of alkaloids in the modern era, relatively little is known about the biosynthesis, regulation and transport of these molecules. This lack of knowledge fundamentally impedes our ability to co-opt nature’s machinery in order to overproduce—that is, to metabolically engineer—these valuable compounds.
Notably, many drug screening efforts exclude plant natural products because of their high production costs and instead screen larger numbers of simpler molecules, which can be produced inexpensively and in fewer chemical steps [2]. Given the challenges of getting these molecules through the drug pipeline, we contend that natural products—including plant alkaloids—should be included in drug screens. Plant natural products have a high success rate as candidates and leads [3,4]. While the chemical syntheses of plant natural products, particularly the alkaloids, are dramatically improving [5], many syntheses are still too lengthy for commercial production or require industrially impractical separation steps. Therefore, alternative production platforms must be developed, evaluated and instituted. An increasing body of work enlists microbes as well as cell and tissue cultures to produce these valuable plant-derived products [4]. Biological systems have the potential to be scalable and selective, while simultaneously being more environmentally friendly and—importantly—less expensive than synthetic reactions [4].
In this Opinion, we highlight recent metabolic engineering efforts designed to improve production of selected plant-derived alkaloids. We focus on the monoterpene indole alkaloids (MIAs), the benzylisoquinoline alkaloids (BIAs) and the glucosinolates. Though not classically classified as alkaloids, the glucosinolates are nitrogen-containing compounds, that have been the subject of a compelling body of research that will inform the forward engineering of all plant natural products. These three classes of plant-derived nitrogen-containing natural products have been the subject of recent research efforts aimed at discovering and manipulating cellular activities, which include enzymatic function, metabolite transport and regulatory control. Ultimately this work may lead to biotechnologically useful enzymes and new drug candidates. Throughout this opinion, we also highlight the challenges that arise in attempting to chart the underexplored landscape of plant biosynthesis and discuss the grand challenges that remain in the metabolic engineering of plant alkaloid products.
The Monoterpene Indole Alkaloids
Introduction
The monoterpene indole alkaloids (MIAs) have garnered interest over the past few decades in large part because of vinblastine 8 and vincristine 9, two potent and widely prescribed anti-cancer agents that are currently produced solely through harvest from the leaves of mature periwinkle plants (Catharanthus roseus) [6]. The concentrations of vinblastine and vincristine per gram of dry leaf material are approximately 0.01% and 0.003%, respectively, and are greatly dependent upon plant growth conditions [7]. Their low yields and lengthy production timeline have elicited intense efforts to engineer higher titers of these medicinally important MIAs.
The MIAs are encountered most commonly in the Apocynaceae, Loganiaceae and Rubiaceae families [8]. Most MIAs are built from the secoiridoid secologanin 3 and the indole-containing molecule tryptamine 2 (Figure 1a) [9]. Strictosidine synthase (STS) condenses these two molecules via a Pictet-Spengler condensation that forms strictosidine 4, which is believed to ultimately succumb to either of two chemical fates [9,10]. If the plant is not under herbivore attack, strictosidine 4 is deglucosylated and rearranged into the over 3000 MIAs found in nature [10]. Madagascar periwinkle has a subset of approximately 130 MIAs. Alternatively, if the plant is under herbivore attack, the strictosidine 4 pool (estimated to be approximately 10 mM in periwinkle leaf epidermal cells hormonally treated to mimic herbivore attack) can be directed to the nucleus for mass deglucosylation, leading to a reactive dialdehyde species capable of cross-linking proteins [10]. This mechanism has been dubbed the strictosidine nuclear “bomb” in reference to the “mustard oil bomb” mechanism of glucosinolate biosynthesis (see below) [10]. Importantly, many of the MIA metabolites themselves have also been implicated in plant defense strategies [11].
Figure 1.
(a) The monoterpene indole alkaloid (MIA) pathway. TDC, tryptophan decarboxylase; STS, strictosidine synthase; Glc, glucose. (b) Introduction of halogenation into the MIA pathway. RebH and PyrH are both flavin-dependent halogenases from actinomycetes species; STSvm, strictosidine synthase Val214Met mutant. (c) Reengineering of halogenase to preferentially chlorinate tryptamine 2 over the natural substrate tryptophan 1.
Obtaining the building blocks for metabolic engineering efforts – A case study on the discovery of P450s involved in MIA biosynthesis
The enzymatic pathways leading to the MIAs have not yet been fully elucidated in any organism. These uncharacterized biochemical steps may utilize novel chemistries or possess informative and interesting specificities that enable the enzymes to be employed in various synthetic metabolic pathway designs [12,13]. Notably, many plants—including MIA producers—are predicted to contain a high percentage of cytochromes P450 (P450s). Some estimates place P450s at approximately 1% of representative plant genomes [14], over 5-fold higher than the proportion of P450s found in the human genome [15]. By using molecular oxygen to tailor hydrocarbon skeletons, P450s facilitate a panel of difficult chemical transformations and are, consequently, utilized in many alkaloid biosynthetic pathways [13]. P450s have also been successfully engineered for biotechnological purposes [15,16]. Various technologies, such as nanodiscs [17], and N-terminus reengineering efforts [16] have improved the expression of membrane-bound P450s, making this class of enzymes accessible to a full suite of biochemical and biophysical characterization techniques.
Given the high sequence similarity among P450s, identifying a P450 that facilitates a specific biochemical reaction within a biosynthetic pathway remains a challenge. This has greatly slowed the discovery and characterization of new P450s within the plant kingdom. However, Giddings et al. recently used co-expression analysis to identify P450s with expression profiles similar to known MIA biosynthetic genes [12]. By functionally assaying these candidates in Saccharomyces cerevisiae, Giddings et al. discovered one P450 (CYP71BJ1) that hydroxylated the 19 position of either lochnericine or tabersonine 7, an intermediate that is positioned at a metabolic branch point [12]. Hydroxylation of tabersonine 7 at the 16 position commits the intermediate to vindoline and vinblastine 8 biosynthesis, whereas hydroxylation at the 19 position commits the molecule to 19-O-acetylhörhammericine formation [12].
Engineering “unnatural” natural products
Plant alkaloids often require modification to improve their pharmacological properties for human consumption. Halogenation, particularly fluorination and chlorination, is a pervasive modification in successful pharmaceutical candidates [18,19]. Halogens often confer the potency of a drug, alter its pharmacokinetics or function as site-specific handles for subsequent modification [18–20]. Halogens can be introduced into MIA pathways by a number of methods. One particular example employed mutasynthesis, a process whereby natural biosynthesis is first blocked by genetic silencing of the natural precursor, and is then rescued by feeding with structural analogs of the precursor [21]. In this case, in conjunction with the RNAi-mediated knockdown of tryptophan decarboxylase (TDC), unnatural tryptamine 2 analogs were added to a chemically “silent”—non-alkaloid producing—background and fluorinated MIA analogs were observed [21]. In a separate engineering strategy, strictosidine synthase (STS)—the enzyme situated at the first committed step of MIA biosynthesis—was engineered to accept an expanded range of halogenated tryptamine 2 precursors [22]; the utility of this enzyme was demonstrated in planta by feeding previously unaccepted unnatural precursors to C. roseus hairy roots [23].
Finally, Runguphan et al. interfaced RebH and PyrH—two tryptophan halogenases isolated from soil-dwelling actinomycetes species—with the MIA metabolism of periwinkle to produce halogenated natural products de novo [24••] (Figure 1b). However, the RebH- and RebF-overexpressing lines also displayed a brown and slow growth morphology [24••]. Runguphan et al. hypothesized that this morphology was the result of the accumulation of 7-chlorotryptan 1a, an analog of primary metabolite L-tryptophan 1 that is somewhat structurally similar to 4-chloroindole-3-acetic acid, an auxin known to be involved in regulating plant growth [24••].
To circumnavigate this problem, Glenn et al. employed structure-guided protein design to engineer a halogenase that preferentially chlorinated tryptamine 2, a more direct MIA precursor (Figure 1c) [25]. Microgram per gram fresh weight quantities of 12-chloro-19,20-dihydroakuamicine 5a were observed with this strategy, but neither 7-chlorotryptophan 1a nor 7-chlorotryptamine 2a accumulated in planta, indicating the chlorinated precursor was being effectively shuttled into MIA metabolism [25]. Engineering halogenation into MIA metabolism highlights an important need to interface specialized metabolism with primary carbon and nitrogen metabolism.
The Benzylisoquinoline Alkaloids
Introduction
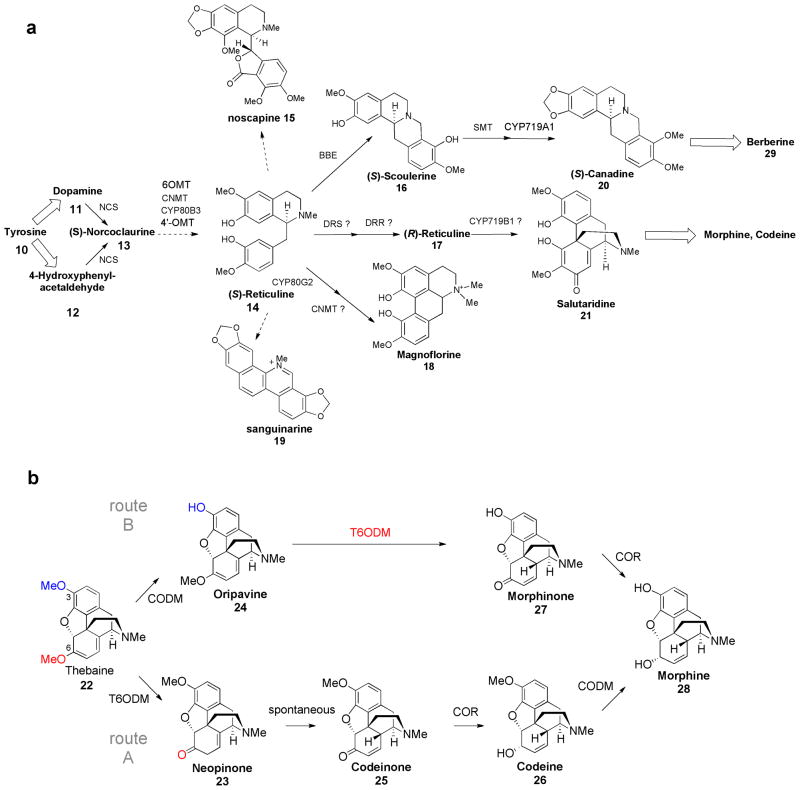
Benzylisoquinoline alkaloids (BIA) are found mainly in the Papaveraceae, Ranunculaceae, Berberidaceae and Menispermaceae plant families. Approximately 2500 BIAs have been isolated to date [26]. This class of compounds has been used throughout human history and contains pharmaceuticals that are still widely used today, including the narcotic and analgesic morphine 28, the cough suppressant codeine 26, the muscle relaxant papaverine, and the anti-microbial agents sanguinarine 19 and berberine 29. All known BIAs, like the MIAs, are derived from a single intermediate, which, for this class of compounds, is norcoclaurine 13. Norcoclaurine synthase (NCS) catalyzes the condensation between dopamine 11 and 4-hydroxyphenylacetaldehyde 12 to yield the central intermediate norcoclaurine 13. Notably, the biosynthetic pathways of several benzylisoquinoline alkaloids—morphine 28, sanguinarine 19 and berberine 29—have been fully elucidated at the genetic level, which has enabled sophisticated metabolic engineering approaches. The application of metabolic engineering strategies for BIAs has focused predominantly on improving the yields of specific alkaloid compounds that exhibit medicinal value.
Enzyme discovery and engineering in BIA pathways
Several outstanding efforts in enzyme discovery have been reported for BIA biosynthetic pathways. In a recent effort, Hagel et al. characterized two O-demethylases that are involved in morphine biosynthesis, completing the characterization of the morphinan pathway (Figure 2b) [27•]. This work also clearly highlighted how co-expression analysis can be used to discover enzymes with unprecedented catalytic function. These enzymes offer the first examples of non-heme iron(II) oxoglutarate dioxygenases capable of catalyzing O-demethylation. Codeine-O-demethylase (CODM) regioselectively demethylates codeine 26 and thebaine 22 at the 3-position, while thebaine-6-O-demethylase (T6ODM) demethylates thebaine 22 and oripavine 24 at the 6-position. Swapping amino acid regions between the two demethylases resulted in a CODM mutant that selectively demethylates codeine (Figure 2b) [28]. This mutant—which effectively sidesteps oripavine 24 production by committing thebaine 22 to just one of two possible routes—could potentially impact titers of codeine 26 and morphine 28 in subsequent metabolic engineering efforts. Collectively, these studies highlight how characterizing individual pathways steps and understanding their specificity and selectivity can both inform and enable metabolic engineering efforts.
Figure 2.
(a) The benzylisoquinoline alkaloid (BIA) pathway. NCS, norcoclaurine synthase; 6-OMT, norcoclaurine 6-O-methyltransferase; CNMT, coclaurine-N-methyltransferase; 4′-OMT, 3′-hydroxy-N-methylcoclaurine-4′-O-methyltransferase; DRS, 1,2- dehydroreticuline synthase; DRR, 1,2-dehydroreticulene reductase; BBE, berberine bridge enzyme; SMT, scoulerine 9-O-methyltransferase; MAO, bacterial monoamine oxidase; CYP2D6, human cytochrome P450 enzyme; CYP80G2, CYP719B1 and CYP719A1, plant cytochrome P450 enzymes. (b) The morphinan alkaloid pathway. CODM, codeine O-demethylase; T6ODM, thebaine 6-O-demethylase; COR, codeinone reductase.
While transcript analysis has proven to be spectacularly successful in elucidating the demethylases of morphine 28 biosynthesis, Winzer et al. provide a rare example of gene clustering in a BIA pathway [29••]. The authors describe a 10-gene cluster in the poppy genome that putatively encodes the entire biosynthetic pathway of the BIA noscapine 15. This is the first gene cluster discovered for an alkaloid pathway, and it is the largest plant gene cluster discovered to date. The authors further successfully silenced six of the ten proposed genes using VIGS to validate their role in noscapine 15 biosynthesis [29••]. This study indicates that genomic data, in addition to expression data, can be used to decipher alkaloid pathways in plants.
Engineering in native hosts
In one of the earliest attempts to engineer BIA-producing plants, RNA interference (RNAi) was used to silence the expression of codeinone reductase (COR) in the opium poppy [30]. COR, the penultimate enzyme of morphine biosynthesis, converts codeinone 25 to codeine 26 (Figure 2b). While one might anticipate that the silencing of COR would lead to elevated levels of codeinone 25, the study found instead that COR-silenced plants accumulated reticuline 14—an intermediate seven steps upstream of codeinone 25—at the expense of morphine 28, codeine 26, oripavine 24 and thebaine 22. A feedback mechanism was proposed as an explanation for the elevated levels of reticuline 14, though testing this hypothesis has yielded conflicting results [30].
Other early attempts to improve the yields of BIA alkaloids include the over-expression of berberine bridge enzyme (BBE) in Eschscholzia californica root cultures. This effort resulted in elevated levels of downstream alkaloids and decreased levels of amino acids, though notably levels of tyrosine 10—the amino acid employed in BIA synthesis—were unaltered [31]. Conversely, the antisense suppression of BBE expression led to the effective silencing of BIA production and increased cellular amino acid levels, though, again, tyrosine 10 levels went largely unchanged (less than two-fold higher than in control lines) [32]. Nonetheless, these two studies highlight how perturbations in alkaloid metabolism can impact primary metabolism [31,32]. More recent studies suggest, however, that the RNAi suppression of BBE in E. californica leads to increased accumulations of (S)-reticuline 14 instead of various canonical amino acids [33]. These contradictory results are surprisingly common in the metabolic engineering of alkaloids in plants and cell cultures and provide us with the impetus to understand these pathways in greater detail, paying specific attention to their biochemical and molecular regulatory elements.
Reconstituting BIA biosynthesis into microbial systems
Many pathways in BIA biosynthesis are fully characterized, which opens the possibility of transplanting entire alkaloid pathways into microbial hosts. Though relatively difficult, the reconstitution of entire metabolic pathways into microbial hosts confers a number of advantages, including rapid biomass accumulation, facile purification and access to the host of tools available for workhorse organisms like E. coli and S. cerevisiae. A number of recent reports have successfully reconstituted portions of BIA pathways into S. cerevisiae, E. coli and combinations thereof in co-culture systems [34–36]. For example, Hawkins et al. were able to produce reticuline 14 as well as sanginarine/berberine-type and morphinan-type BIAs in yeast by overexpressing genes from mixed plant sources and human [35]. Notably, they were also able to tune enzyme expression levels through use of a glucocorticoid-inducible promoter and in situ promoter titration [35]. This level of tuning enables maximal pathway flux and minimal enzyme expression. The expression system is nominally taxed under these conditions, since valuable cellular resources are not used on the biosynthesis of supernumerary proteins and nucleic acids.
The Glucosinolates
Introduction
The glucosinolates are not classified as alkaloids, although, along with the alkaloids, these compounds are amino acid-derived, nitrogen-containing small molecules of plant origin. The glucosinolates are included in this review because the recent and creative metabolic engineering studies performed on this class of compounds will undoubtedly inform the forward engineering of all plant natural products, including the alkaloids.
Glucosinolates are sulfur- and nitrogen-containing compounds that are derived from glucose and various amino acids (Figure 3a) [37]. They are found in cruciferous vegetables (the Brassicaceae plant family) and have been shown to possess a range of bioactivities [37]. The glucosinolates occupy an essential space in the chemical ecology of their host organisms by attracting specialist crucifer pollinators and insects and deterring predatory herbivores [38]. Specifically, crucifers employ myrosinases (hydrolases) to cleave the glucose moiety of glucosinolates in response to predation and herbivory (Figure 3b) [39]. The myrosinases and glucosinolates are physically separated, coming into contact only upon disruption of the plant tissue (Figure 3b) [39]. Upon hydrolysis, the resultant unstable aglycone intermediate spontaneously rearranges into the corresponding isothiocyanate via a Lossen-type rearrangement [39]. The three known types of specifier proteins, Thiocyanate-Forming Proteins (TFPs), Nitrile-Forming Proteins (NFPs) and Epithiospecifier Proteins (ESPs)—which can be found in planta or in various specialist insects—can redirect glucosinolate hydrolysis from isothiocynate products toward thiocyanate, simple nitrile and epithionitrile products, respectively (Figure 3b) [39]. Notably, many specifiers can direct glucosinolate hydrolysis to more than one product [39]. Early workers on this plant defense and pollination system dubbed it “The Mustard Oil Bomb” [38]. To date, over 120 glucosinolates have been identified [40].
Figure 3.
(a) The glucosinolate (GLS) pathway. GSH, glutathione. (b) Glucosinolate hydrolysis to form epithionitrile, nitrile, thiocyanate and isothiocyanate.
Improving yield in non-native hosts
The reconstitution of entire metabolic pathways into heterologous plant hosts requires the use of efficient and facile “gene stacking” methodologies. A spectacularly successful example is the engineering of benzylglucosinolate biosynthesis into Nicotiana benthamiana. Benzylglucosinolate was reconstituted in N. benthamiana using a transient expression system. In this study, Geu-Flores et al. identified a γ-glutamyl peptidase bottleneck, suggesting that reduced sulfur is incorporated into glucosinolates via glutathione conjugation (Figure 3a) [37]. The co-expression of this peptidase augmented the yield of benzylglucosinolate 5.7-fold, indicating how consideration of primary metabolite resources can impact natural product yield [37]. In a separate metabolite analysis, MØdlrup et al. monitored the accumulation of desulfobenzylglucosinolate, the penultimate product in the benzylglucosinolate pathway [41]. Directing sulfur from primary to secondary metabolism through the co-expression of adenosine 5-phosphosulfate kinase—which provides the 3′-phosphoadenosine-5′-phosphosulfate (PAPS) co-substrate necessary for the final step of benzylglucosinolate biosynthesis (Figure 3a)—in the N. benthamiana expression system alleviated the subsequent bottleneck and increased the benzylglucosinolate yield by 16-fold [41]. In yeast, Mikkelsen et al. were able to reconstitute the biosynthesis of indolylglucosinolate [42]. This example was a proof-of-concept study for a technology that enables the stacking of large numbers of genes, a requirement for total pathway reconstitution.
Notably, the benzylglucosinolate biosynthetic pathway has also been stably transformed into Nicotiana tabacum, another non-cruciferous plant, which does not normally produce glucosinolates [43•]. This reengineered plant has been shown to attract the diamondback moth (Plutella xylostella) and encourage oviposition (the deposition of eggs), highlighting its potential utility as a dead-end trap crop to deter predatory insects and prevent billion-dollar damages to cruciferous crops worldwide [43•].
Future Directions and Conclusions - Alkaloids and Beyond
Historically, altering metabolic pathways in plants to achieve a given end has been difficult. Metabolic engineering in plants is still in its infancy and has largely been confined to single-gene expression or silencing events in the background of endogenous plant cell metabolism. The complexity of the plant host’s metabolism has been shown, in many cases, to effectively mute the engineering effort or lead to unpredictable results (Table 1). However, in recent years, a wealth of new approaches has expanded the capabilities of multi-gene pathway expression in both plants and microbes and has highlighted our increasing ability to engineer the production of plant natural products in both plants and heterologous systems. The increase in available and reliable sequencing and expression data enables the (relatively) facile discovery of gene, transporter and regulatory elements, the identification of which is often a prerequisite for multi-step metabolic engineering efforts. The three case studies in this review (MIAs, BIAs and glucosinolates) exemplify the challenges and progress in metabolically engineering plant-derived natural products. While we have made a special effort to highlight the advantages and pitfalls of individual techniques and efforts throughout this Opinion, a number of grand challenges for plant metabolic engineering remain to be tackled in the coming years.
Table 1.
Examples of recent alkaloid engineering studies and their outcomes.
| Class | Engineering Strategy | System | Observations | Reference |
|---|---|---|---|---|
| MIAs | Overexpression of TDC | C. roseus crown gall | Did not significantly alter alkaloid yield | [53] |
| MIAs | Overexpression of transcription factor CrWRKY1 | C. roseus hairy root cultures | 3-fold increase in serpentine 10-fold increase in ajmalicine 2-fold decrease in catheranthine |
[54] |
| MIAs | Overexpression of feedback insensitive anthranilate synthase alpha subunit | C. roseus hairy root cultures | 300-fold increase in tryptophan 10-fold increase in tryptamine 1.8-fold in lochnericine |
[55] |
| MIAs | Overexpression of alpha or alpha and beta subunits of anthranilate synthase and feeding of 10-deoxy-D-xylulose, loganin and secologanin | C. roseus hairy root cultures | 2.3-fold increase in horhammericine 1.5-fold increase in cathenamine 1.3-fold increase in catharanthine 1.8-fold increase in ajmalicine 2.1-fold increase in lochnericine 4.5-fold increase in tabersonine |
[56] |
| MIAs | Methyl jasmonate elicitation Fed loganin and/or tryptamine |
C. roseus hairy root tissue cultures | 3.2-fold increase in strictosidine 8.8-fold increase in ajmalicine 8.4-fold increase in tabersonine Substrate feeding did not increase yield in elicited cultures |
[57] |
| MIAs | Feeding with unnatural tryptamine precursors | C. roseus tissue cultures | observed ug/ gram fresh weight quantities of many modified MIAs | [58] |
| MIAs | Mutasynthesis | Fed tryptamine isomers to TDCi-silenced periwinkle hairy roots | observed ug/ gram fresh weight quantites of many modified MIAs Achieved “alkaloid free” background |
[21] |
| MIAs | Substrate feeding to roots harboring STS with expanded substrate specificity | C. roseus hairy root cultures | observed ug/gram fresh weight quantities of previously metabolically inaccessible modified products | [23] |
| MIAs | Overexpression of RebH/RebF and PyrH/RebF in planta | C. roseus hairy root cultures | observed the de novo production of 12-chloro-19,20-dihydroakuammicine (25 ug/ gram fresh weight) Observed accumulation of 7-chlorotryptophan (50 ug/g fresh weight) |
[24] |
| MIAs | Overexpression of RebF and reengineered RebH in planta | C. roseus hairy root cultures | Alleviated TDC bottleneck (no accumulation of 7-chlorotryptophan) observed the de novo production of 12-chloro-19,20-dihydroakuammicine (2.7 ug/ gram fresh weight) over-expression of TDC led to slow-growing hairy roots |
[25] |
| BIAs | heterologous expression system for production of S-reticuline and downstream alkaloids | Artificial pathway in yeast | Produced S-reticuline in yields up to 150 mg/L Demonstrated production of (S)-tetrahydroberberine (≪5 mg/ L) Demonstrated production of (S)-scoulerine Demonstrated production of (S) tetrahydrocolumbarmine (60 mg/L) |
[35] |
| BIAs | heterlogous expression system for production of S-reticuline | Artificial pathway in E. coli | Produced (S)-reticuline in yields up to 40 mg/L | [36] |
| BIAs | Suprression of codeinone reductase expression | RNAi in Opium poppy | Accumulation of (S)-reticuline (7 metabolic steps upstream) | [30] |
| BIAs | Overexpression of BBE in E. califonica | E. californica root cultures | 5.8-fold increase of total downstream alkaloids 3.2-fold decrease in leucine concentration 2.4-fold decrease in threonine concentration 2.1-fold decrease in valine concentration Tyrosine levels unaltered |
[31] |
| BIAs | Suppression of BBE expression | E. californica root cultures | Elevated levels of (S)-reticuline to 310 ug/g cell fresh weight (S)-reticuline levels at 6 mg/20 mL in the media |
[33] |
| BIAs | Systematic knockdown of morphinan pathway enzymes through VIGS | Opium poppy seedlings | Suppression of SalSyn, SalR, T6ODM, CODM resulted in concomitant increase of direct precursors Suppression of SalAT resulted in the accumulation of salutiridine (not direct precursor) Suppression of COR resultined in increases of (S)- reticuline (not direct precursor) |
[59] |
| GlucosinolatesBenzylglucosinolate pathway reconstitution | Transient expression in N. benthamiana Coexpression of gamma-glutamyl peptidase |
Produced benzylglucosinolate in yields up to 0.57 nmol/ mg freshweight leaf tissue coexpression of peptidase raised yield 5.7-fold | [37] | |
| GlucosinolatesBenzylglucosinolate pathway reconstitution | Transient expression in N. benthamiana coexpression of adenosine 5-phosphosulfate kinase yeast | Production of benzylglucosinolate in yields up to 1.8 nmol/ mg freshweight expression of kinase elevated yield by 16-fold | [41] | |
| GlucosinolatesIndolylglucosinolate pathway reconstitution | Reach indolylglucosinolate titers as high as 1.07 mg/ ml product excreted into the media | [42] | ||
| GlucosinolatesBenzylglucosinolate pathway reconstitution | Stable expression in N. tabacum | Benzylglucosinolate titers as high as 0.5 nmol/mg fresh weight Increase oviposiiton on non-cruciferous plants | [43] | |
Effective mining strategies
Effective mining strategies, such as those employed by Giddings et al. [12], Hagel et al. [27•], Winzer et al. [29••] and Liscombe et al. [44], are required to sift through the mounting data of the sequencing age. Hanson et al. (this issue) provides a more comprehensive review of effective mining strategies and phylogenetic analyses. Traditionally, plant enzyme discovery methods have relied heavily upon time-intensive reverse genetics. Bioinformatic techniques that engage co-expression analyses and comparative metabolite profiling to limit the gene space to be investigated are improving the discovery process. Moreover, a suite of new silencing tools, including VIGS [7], RNAi [23] and the IL-60 system [45], can provide rapid insight into the physiological function of plant enzymes.
Metabolic engineering in native versus non-native hosts
Many efforts aimed at improving the yield of alkaloids in native hosts have focused on feeding precursors and over-expressing transcription factors or enzymes positioned at metabolic bottlenecks. While these efforts often result in modest improvements to yield (Table 1), many are often accompanied by adverse morphological effects that may significantly stunt the growth of plant and tissue cultures, highlighting the tight regulation of metabolic processes within highly organized plant cells and tissues. In native systems especially, the slow growth morphologies may result from the depletion of cellular resources used in synthesizing a surfeit of transcripts and enzymes or from the accumulation of intermediates that negatively impact growth and development. Engineering in native hosts or heterologous plant species is attractive because not having to build the starting substrates and supply the co-factors greatly simplifies engineering efforts. However, with this strategy, maintaining the balance between primary metabolism and the engineered metabolism—a feat that will likely improve growth morphologies—is complicated precisely because endogenous primary metabolite pools are expropriated for the overproduction of selected metabolites. Also, despite the advantage of minimal gene stacking, the often uncharacterized and unanticipated complex metabolism and regulatory elements of native systems can lead to engineering outcomes that are particularly difficult to control and predict. The industrial scale production of plant natural products will likely require more comprehensive engineering efforts than single-gene over-expression or silencing events in the context of native plant hosts. Engineering in faster-growing and “chemically silent” heterologous hosts may increase biomass accumulation and simplify purification.
Controlling metabolic flux through new expression constructs, scaffolds and tunable regulatory elements
The intricate relationship between primary metabolism (i.e. glycolysis, the TCA cycle) and the native or heterologous secondary metabolism (i.e. isoprenoid and alkaloid pathways) must be considered. In plants, Park et al. alluded to this interplay by demonstrating that BBE expression levels vastly affect amino acid levels [31,32]. As metabolic engineering strategies in plants become more sophisticated, we should also begin to consider flux analyses, taking into account that natural product pathways are evolutionarily optimized to channel intermediates toward product through a highly choreographed system of protein-protein interactions, localization and regulation [46]. The overall goal is to maximally channel metabolic resources to the desired products without over taxing the host system.
Co-localization through scaffolding is a proven way to channel metabolites in E. coli. These systems attempt to mock natural megasynthases, which efficiently shuttle metabolites between active sites. Essentially, scaffolding increases the local metabolite and enzyme concentrations and effectively lowers the Km of the substrate. These systems are widely modular and are known to improve titers, alleviate metabolic bottlenecks and reduce metabolic loads by preventing carbon from exiting the pathway. Under conditions of low enzyme expression (decreased metabolic load), Dueber et al. successfully achieved a 77-fold enhancement in mevalonate production by building a scaffold based on the protein-protein interactions of GBD, SH3 and PDZ domains and their cognate ligands [47]. They built the scaffold on hydroxymethylglutaryl-CoA reductase, the enzymatic bottleneck of mevalonate production [47]. Notably, these scaffolding systems require that the enzyme at the metabolic bottleneck, the subsequent enzyme and the substrate be co-localized [47], underscoring why they may be untenable for some highly compartmentalized systems. Nonetheless, the prospect of engineering metabolons into plants is exciting.
The effective metabolic engineering of plant natural products will inevitably require advanced, but easy-to-use, gene stacking techniques. Traditionally, multi-gene expression in plants has been plagued with inadvertent silencing events, the incomplete incorporation of all genes and lengthy and technically challenging procedures [48]. A number of new technologies, however, are being developed to assemble and transplant large fragments of DNA easily [48]. Golden Gate cloning and USER fusion have been used to clone multiple pathway elements [48,49]. Additionally, synthetic plant chromosomes and the universal expression and silencing IL-60 platform both have the demonstrated capability of introducing multiple plant pathway elements into plants [48,50]. For example, under the transformation-free IL-60 platform, Mozes-Koch et al. expressed an entire bacterial operon in tomato and produced pyrollinitrin, which they observed after only two days [50•].
A number of RNA-based silencing systems, including RNAi, have also been engineered and applied to medicinal plants [21]. Notably, RNAi, which provides a permanent pheno- or chemotype, has been employed to block shunt pathways and channel metabolic resources toward a desired product, enhancing our ability to engineer in multiple dimensions [21].
Lastly, promoter libraries, engineered untranslated regions (i.e. 5′ untranslated regions and intergenic regions), genetic circuit designs and biosensor regulators have been tremendously helpful in microbial engineering. Applying these design principles to the metabolic engineering of plants may greatly enrich our efforts to produce valuable and chemically diverse alkaloids. Notably, a variety of constitutive and inducible plant promoters and expression systems are now widely available. Synthetic RNA elements, ribosome binding site elements and a combination of different strength promoters strategically placed in front of stacked pathway genes could theoretically enable tunable protein expression [35,51]. These elements could potentially limit the expression of toxic activities or the accumulation of toxic metabolites until the stationary phase (or an appropriate stage) of growth, thereby absolving the system of unsustainable metabolic burden.
Localization and transport – Engineering in multiple dimensions
Many alkaloid biosynthetic pathways are highly compartmentalized at both the inter- and intracellular levels. For example, at least three cell types are required for the biosynthesis of many MIAs [6]. The forward engineering of plant natural products will require sifting through the increasing amount of available sequencing and expression data and untangling the complexity of the plant cell and different tissue types. In addition to the linear design and channeling of metabolic pathways, the successful metabolic engineering of plant natural products will require engineering in the “third dimension,” namely at the level of localization and cell type [52].
Combinatorial biosynthesis in Plants – Mixing and matching pathways and engineering new enzyme specificities
De novo combinatorial biosynthesis in plant systems has gone largely underexplored. Most of the few efforts to engineer unnatural natural products have utilized precursor feeding or mutasynthesis-based approaches, which can be costly and time intensive. The de novo biosynthesis of unnatural natural products will require that constituent enzymes have reengineered or broad specificity. Notably, directed evolution has been successfully used to alter enzyme specificity. However, enzyme engineering efforts are greatly enhanced if the protein structure is known and the mechanism is well understood. Then, the enzyme can be subjected to structure-guided techniques, such as site-directed mutagenesis and domain swapping, which create smaller protein libraries enriched with functional mutants.
As we seek further to convert plants and microbes into the chemical factories to meet our medicinal needs, we should remember that, although many plant natural products are bioactive and serve as important lead compounds, they often require modification before making it to the clinic. Therefore, the forward engineering of “unnatural” or “new-to-nature” natural products must also be a grand challenge if plant natural products are to be shuttled from the annals of human tradition into the drug development programs and clinics of tomorrow.
Highlights.
Recent metabolic engineering efforts for plant alkaloids
Characterizing, reconfiguring and fine-tuning metabolic “parts” improves titers
Additional strategies are necessary to produce “unnatural” natural products
Acknowledgments
The laboratory of S.E.O. is funded in part by NIH (GM074820), BBSRC (BB/J004561/1), and the John Innes Foundation. W.S.G. gratefully acknowledges a National Science Foundation Predoctoral Fellowship. S.E.O. receives salary support through a synergy initiative between the John Innes Centre and the University of East Anglia.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shibamoto T, Bjeldanes LF. Introduction to Food Toxicology. Academic Press (Elsevier); 2009. [Google Scholar]
- 2.Lipinski C, Hopkins A. Navigating chemical space for biology and medicine. Nature. 2004;432:855–861. doi: 10.1038/nature03193. [DOI] [PubMed] [Google Scholar]
- 3.Rosén J, Gottfries J, Muresan S, Backlund A, Oprea LA. Novel chemical space exploration via natural products. J Med Chem. 2009;52:1953–1962. doi: 10.1021/jm801514w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leonard E, Runguphan W, O’Connor SE, Prather KJ. Opportunities in metabolic engineering to facilitate scalable alkaloid production. Nat Chem Biol. 2009;5:292–300. doi: 10.1038/nchembio.160. [DOI] [PubMed] [Google Scholar]
- 5.Martin DBC, Nguyen LQ, Vanderwal CD. Syntheses of strychnine, norfluorocurarine, dehydrodesacetylretuline, and valparicine enabled by intramolecular cycloadditions of Zincke aldehydes. J Org Chem. 2012;77:17–46. doi: 10.1021/jo2020246. [DOI] [PubMed] [Google Scholar]
- 6.Murata J, Roepke J, Gordon H, De Luca V. The leaf epidermome of Catharanthus roseus reveals its biochemical specialization. Plant Cell. 2008;20:524–542. doi: 10.1105/tpc.107.056630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liscombe DK, O’Connor SE. A virus-induced gene silencing approach to understanding alkaloid metabolism in Catharanthus roseus. Phytochemistry. 2011;72:1969–1977. doi: 10.1016/j.phytochem.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seigler DS. Plant Secondary Metabolism. Kluwer Academic Publishers; 1998. [Google Scholar]
- 9.O’Connor SE, Maresh JJ. Chemistry and biology of monoterpene indole alkaloid biosynthesis. Nat Prod Rep. 2006;23:532–547. doi: 10.1039/b512615k. [DOI] [PubMed] [Google Scholar]
- 10.Guirimand G, Courdavault V, Lanoue A, Mahroug S, Guihur A, Blanc N, Giglioli-Guivarc’h N, St-Pierre B, Burlaut V. Strictosidine activation in Apocynaceae: towards a “nuclear time bomb”? BMC Plant Biology. 2010:10. doi: 10.1186/1471-2229-10-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dewick PM. Medicinal Natural Products A Biosynthetic Approach. 2. John Wiley & Sons; 2001. [Google Scholar]
- 12.Giddings L-A, Liscombe DK, Hamilton JP, Childs KL, DellaPenna D, Buell CR, O’Connor SE. A stereoselective hydroxylation step of alkaloid biosynthesis by a unique cytochrome P450 in Catharanthus roseus. J Biol Chem. 2011;286:16751–16757. doi: 10.1074/jbc.M111.225383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizutani M, Sato F. Unusual P450 reactions in plant secondary metabolism. Arch Biochem Biophys. 2011;507:194–203. doi: 10.1016/j.abb.2010.09.026. [DOI] [PubMed] [Google Scholar]
- 14.Mizutani M, Ohta D. Diversification of P450 genes during land plant evolution. Annu Rev Plant Biol. 2010;61:291–315. doi: 10.1146/annurev-arplant-042809-112305. [DOI] [PubMed] [Google Scholar]
- 15.Sawayama AM, Chen MMY, Kulanthaivel P, Kuo M-S, Hemmerle H, Arnold FH. A panel of cytochrome P450 BM3 variants to produce drug metabolites and diversify lead compounds. Chem Eur J. 2009;15:11723–11729. doi: 10.1002/chem.200900643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leonard E, Koffas MAG. Engineering of artificial plant cytochrome P450 enzymes for synthesis of isoflavones by Escherichia coli. Appl Environ Microbiol. 2007;73:7246–7251. doi: 10.1128/AEM.01411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denisov I, Sligar SG. Cytochromes P450 in nanodiscs. Biochemica et Biophysica. 2011;1814:223–229. doi: 10.1016/j.bbapap.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neumann CS, Fujimori DG, Walsh CT. Halogenation strategies in natural product biosynthesis. Chem Biol. 2008;15:99–109. doi: 10.1016/j.chembiol.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 19.Herrera-Rodriguez LN, Khan F, Robins KT, Meyer H-P. Perspective on biotechnological halogenation Part I: Halogenated products and enzymatic halogenation. Chemistry Today. 2011;29:31–33. [Google Scholar]
- 20.Blasiak LC, Drennan CL. Structural perspective on enzymatic halogenation. Acc Chem Res. 2009;42:147–155. doi: 10.1021/ar800088r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Runguphan W, Maresh JJ, O’Connor SE. Silencing of tryptamine biosynthesis for production of nonnatural alkaloids in plant cultures. Proc Natl Acad Sci U S A. 2009;106:13673–13678. doi: 10.1073/pnas.0903393106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernahardt P, McCoy E, O’Connor SE. Rapid identification of enzyme variants for reengineered alkaloid biosynthesis in periwinkle. Chem Biol. 2007;14:888–897. doi: 10.1016/j.chembiol.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Runguphan W, O’Connor SE. Metabolic reprogramming of periwinkle plant culture. Nat Chem Biol. 2009;5:151–153. doi: 10.1038/nchembio.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Runguphan W, Qu X, O’Connnor SE. Integrating carbon-halogen bond formation into medicinal plant metabolism. Nature. 2010;18:461–464. doi: 10.1038/nature09524. The authors produce halogenated monoterpene indole alkaloids de novo by interfacing two tryptophan halogenases with the alkaloid metabolism of Madagascar periwinkle (Catharanthus roseus). This is the first example of de novo combinatorial biosynthesis in a medicinal plant. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glenn WS, Nims E, O’Connor SE. Reengineering a tryptophan halogenase to chlorinate a direct alkaloid precursor. J Am Chem Soc. 2011;133:19346–19349. doi: 10.1021/ja2089348. [DOI] [PubMed] [Google Scholar]
- 26.Facchini P, De Luca V. Opium poppy and Madagascar periwinkle: model non-model systems to investigate alkaloid biosynthesis in plants. Plant J. 2008;54:763–784. doi: 10.1111/j.1365-313X.2008.03438.x. [DOI] [PubMed] [Google Scholar]
- 27•.Hagel JM, Facchini PJ. Dioxygenases catalyze the O-demethylation steps of morphine biosynthesis in opium poppy. Nat Chem Biol. 2010;6:273–275. doi: 10.1038/nchembio.317. The authors characterize the first non-heme iron(II) oxoglutarate dioxygenases capable of catalyzing O-demethylation. The discovery of these enzymes completes the characterization of the morphinan pathway. [DOI] [PubMed] [Google Scholar]
- 28.Runguphan W, Glenn WS, O’Connor SE. Redesign of a dioxygenase in morphine biosynthesis. Chem Biol. 2012;19:674–678. doi: 10.1016/j.chembiol.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 29••.Winzer T, Gazda V, He Z, Kaminski F, Kern M, Larson TR, Li Y, Meade F, Teodor R, Vaistij FE, et al. A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science. 2012;336:1704–1708. doi: 10.1126/science.1220757. The authors describe a putative 10-gene cluster—the largest plant gene cluster discovered to date and the first discovered in an alkaloid pathway—in the poppy genome. The cluster is reported to be involved in the biosynthesis of the benzylisoquinoline noscapine. [DOI] [PubMed] [Google Scholar]
- 30.Allen RS, Milligate AG, Chilty JA, Thisleton J, Miller JA, Fist AJ, Gerlach WL, Lancin PJ. RNAi-mediated replacement of morphine with the nonnarcotic alkaloid reticuline in opium poppy. Nat Biotechnol. 2004;22:1559–66. doi: 10.1038/nbt1033. [DOI] [PubMed] [Google Scholar]
- 31.Park S-U, Yu M, Facchini PJ. Modulation of berberine bridge enzyme levels in transgenic root cultures of California poppy alters the accumulation of benzophenathridine alkaloids. Plant mol biol. 2003;51:153–164. doi: 10.1023/a:1021199311049. [DOI] [PubMed] [Google Scholar]
- 32.Park S-U, Facchini PJ. Antisense RNA-mediated suppression of benzophenathridine alkaloid biosynthesis in transgenic cell cultures of California poppy. Plant Physiol. 2002;128:696–706. doi: 10.1104/pp.010741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujii N, Inui T, Iwasa K, Morishige T, Sato F. Knockdown of berberine bridge enzyme by RNAi accumulates (S)-reticuline and activates a silent pathway in cultured California poppy cells. Transgenic Res. 2007;16:363–375. doi: 10.1007/s11248-006-9040-4. [DOI] [PubMed] [Google Scholar]
- 34.Minami H, Kim JS, Ikezawa N, Takemura T, Katayama T, Kumagai H, Sato F. Microbial production of plant benzylisoquinoline alkaloids. Proc Natl Acad Sci U S A. 2008;105:7393–7398. doi: 10.1073/pnas.0802981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hawkins KM, Smolke CD. Production of benzylisoquinoline alkaloids in Saccharomyces cerevisiae. Nat Chem Biol. 2008;4:564–573. doi: 10.1038/nchembio.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakagawa A, Minami H, Kim J-S, Koyangi T, Katayama T, Sato F, Kumagai H. A bacterial platform for fermentative production of plant alkaloids. Nat Commun. 2011:2. doi: 10.1038/ncomms1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geu-Flores F, Nielsen MT, Nafisi M, Møldrup ME, Olsen CE, Motawia MS, Halkier BA. Glucosinolate engineering identifies a gamma-glutamyl peptidase. Nat Chem Biol. 2009;5:575–577. doi: 10.1038/nchembio.185. [DOI] [PubMed] [Google Scholar]
- 38.Ratzka A, Vogel H, Kliebensten DJ, Mitchell-Olds T, Kroymann J. Disarming the mustard oil bomb. Proc Natl Acad Sci U S A. 2002;99:11223–11228. doi: 10.1073/pnas.172112899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wittstock U, Burow M. Tipping the scales--specifier proteins in glucosinolate hydrolysis. IUBMB Life. 2007;59:744–751. doi: 10.1080/15216540701736277. [DOI] [PubMed] [Google Scholar]
- 40.Mikkelsen MD, Buron LD, Salomonse B, Olsen CE, Hansen BG, MortenSen UH, Halkier BA. Microbial production of indolylglucosinolate through engineering of a multi-gene pathway in a versatile yeast expression platform. Metab Eng. 2012;14:104–111. doi: 10.1016/j.ymben.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 41.Møldrup ME, Geu-Flores F, Olsen CE, Halkier BA. Modulation of sulfur metabolism enables efficient glucosinolate engineering. BMC Biotechnol. 2011:11. doi: 10.1186/1472-6750-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mikkelsen MD, Buron LD, Salomonsen B, Olsen CE, Hansen BG, Mortensen UH, Halkier BA. Microbial production of indolylglucosinolate through engineering of a multi-gene pathway in a versatile yeast expression platform. Metab Eng. 2012;2:104–111. doi: 10.1016/j.ymben.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 43•.Møldrup ME, Geu-Flores F, de Vos M, Olsen CE, Sun J, Jander G, Halkier BA. Engineering of benzylglucosinolate in tobacco provides proof-of-concept for dead-end trap crops genetically modified to attract Plutella xylostella (diamondback moth) Plant Biotechnol J. 2012;10:435–442. doi: 10.1111/j.1467-7652.2011.00680.x. The authors reconstitute benzylglucosinolate biosynthesis in Nicotiana tabacum (tobacco) in a stable transformation. These engineered non-cruciferous plants encourage oviposition from the diamondback moth, which could prevent billions of dollars on damages to crucifers. [DOI] [PubMed] [Google Scholar]
- 44.Liscombe DK, Usera AR, O’Connor SE. Homolog of tocopherol C methyltransferases catalyzes N methylation in anticancer alkaloid biosynthesis. Proc Natl Acad Sci U S A. 2010;107:18793–18798. doi: 10.1073/pnas.1009003107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peretz Y, Mozes-Koch R, Akad F, Tanne E, Czosnek H, Sela I. A Universal expression/ silencing vector in plants. Plant Physiol. 2007;145:1251–1263. doi: 10.1104/pp.107.108217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weeks AM, Chang MCY. Constructing de novo biosynthetic pathways for chemical synthesis in living cells. Biochemistry. 2011;50:5404–5418. doi: 10.1021/bi200416g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KLJ, Keasling JD. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol. 2009;27:753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 48.Naqvi S, Farré G, Sanahuja G, Capell T, Zhu C, Christou P. Where more is better: multigene engineering in plants. Trends in Plant Science. 2009;15:48–56. doi: 10.1016/j.tplants.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 49.Nour-Eldin H, Geu-Flores F, Halkier BA. USER cloning and USER fusion: The Ideal Cloning Techniques for Small and Big Laboratories. In: Fett-Netto AG, editor. Plant Secondary Metabolism Engineering, Methods in Molecular Biology. Springer Science + Business Media, LLC; 2010. pp. 185–200. [DOI] [PubMed] [Google Scholar]
- 50•.Mozes-Koch R, Gover O, Tanne E, Peretz Y, Maori E, Chernin L, Sela I. Expression of an entire bacterial operon in plants. Plant Physiol. 2012;158:1883–1892. doi: 10.1104/pp.111.186197. The authors describe the expression of the pyrollinitrin operon in tomato. They observe pyrollinitrin formation after only two days. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang Al, Wolf JJ, Smolke CD. Synthetic RNA switches as a tool for temporal and spatial control over gene expression. Curr Opin Biotechnol. 2012;23:1–10. doi: 10.1016/j.copbio.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agapakis CM, Boyle PM, Silver PA. Natural strategies for the spatial optimization of metabolism in synthetic biology. Nat Chem Biol. 2012;8:527–535. doi: 10.1038/nchembio.975. [DOI] [PubMed] [Google Scholar]
- 53.Goddijin OJ, Pennings EJ, van der Helm P, Schilperoot RA, Verpoorte R, Hoge JH. Overexpression of a tryptophan decarboxylase cDNA in Catharanths roseus crown gall calluses results in increased tryptamine levels but not in increased terpenoid indole alkaloid production. Transgenic Res. 1995;4:315–323. doi: 10.1007/BF01972528. [DOI] [PubMed] [Google Scholar]
- 54.Suttipanta N, Pattanaik S, Kulshrestha M, Patra B, Singh SK, Yuan L. The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 2011;157:2081–2093. doi: 10.1104/pp.111.181834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hughes EH, Hong SB, Gibson SI, Shanks JV, San KY. Expression of a feedback-resistant anthranilate synthase in Catharanthus roseus hairy roots provides evidence for tight regulation of terpenoid indole alkaloid levels. Biotechnol Bioeng. 2004;86:718–727. doi: 10.1002/bit.20081. [DOI] [PubMed] [Google Scholar]
- 56.Peebles CA, Hong SB, Gibson SI, Shanks JV, San KY. Effects of terpenoid precursor feeding on Catharanthus roseus hairy roots over-expressing the alpha or the alpha and beta subunits of anthranilate synthase. Biotechnol Bioeng. 2006;93:534–540. doi: 10.1002/bit.20739. [DOI] [PubMed] [Google Scholar]
- 57.Goklany S, Loring RH, Glick J, Lee-Parsons CW. Assessing the limitations to terpenoid indole alkaloid biosynthesis in Catharanthus roseus hairy root cultures through gene expression profiling and precursor feeding. Biotechnol Prog. 2009;25:1289–1296. doi: 10.1002/btpr.204. [DOI] [PubMed] [Google Scholar]
- 58.McCoy E, O’Connor SE. Directed biosynthesis of alkaloid analogs in the medicinal plant Catharanthus roseus. J Am Chem Soc. 2006;128:14276–14277. doi: 10.1021/ja066787w. [DOI] [PubMed] [Google Scholar]
- 59.Wijekoon CP, Facchini PJ. Systematic knockdown of morphine pathway enzymes in opium poppy using virus-induced gene silencing. Plant J. 2012;69:1052–1063. doi: 10.1111/j.1365-313X.2011.04855.x. [DOI] [PubMed] [Google Scholar]