Abstract
Over the past decades, there has been growing recognition that light can provide a powerful stimulus for biological interrogation. Light-actuated tools allow manipulation of molecular events with ultra-fine spatial and fast temporal resolution, as light can be rapidly delivered and focused with sub-micrometer precision within cells. While light-actuated chemicals such as photolabile “caged” compounds have been in existence for decades, the use of genetically-encoded natural photoreceptors for optical control of biological processes has recently emerged as a powerful new approach with several advantages over traditional methods. Here we review recent advances using light to control basic cellular functions and discuss the engineering challenges that lie ahead for improving and expanding the ever-growing optogenetic toolkit.
Keywords: optogenetics, photoreceptors, optical dimerizers, light, inducible systems
Introduction
Living cells depend on precise spatial and temporal coordination of molecular events. In fields such as neurobiology and developmental biology, tools allowing inducible control over cellular processes have been indispensible for probing the inherent complexities of biological systems. Chemical-genetic strategies, in which small molecules are used to inducibly control cellular function on user-defined timescales, have provided a wealth of information for experimental biologists. While such strategies have been invaluable, chemical-genetic approaches generally do not provide spatial resolution and are not easily reversible. Photocaged and photolabile moieties attached to chemical groups overcome this problem as they provide spatial and temporal control over release of biologically active compounds, however such compounds can be expensive or difficult to obtain, are not typically photoreversible, are highly diffusive, and are difficult to deliver to cells in some cases.
Recently, there has been growing interest in using modified photoreceptor proteins as tools for conditional control, as such tools promise dose-dependent molecular manipulation with high spatiotemporal resolution in a genetically encoded system. The engineering and use of these tools has broadly been designated the field of optogenetics. Pioneering research with light-gated cation channels and anion pumps, such as Channelrhodopsin-2 and halorhodopsin, created the first widely-adopted optogenetic technologies (Boyden et al., 2005; Li et al., 2005; Han and Boyden, 2007; Zhang et al., 2007) These rhodopsin-based systems have revolutionized neurobiology as they provide a method for stimulating or silencing neural activity with cellular resolution and temporal precision. These innovative tools have been the subject of a number of reviews (Deisseroth, 2011; Yizhar et al., 2011; Zhang et al., 2011; Chow et al., 2012; Mei and Zhang, 2012) and will not be covered here.
One of the most recent advances in optogenetics is the development of a growing suite of light-controlled modules for interrogating cellular functions. These tools use natural or engineered photosensory proteins to control basic cellular functions, such as protein localization and protein-protein interactions. These new technologies allow activation of signaling cascades, transcription, and other cellular events with high spatial resolution and temporal precision using genetically-encoded proteins. In this review, we cover this second generation of optogenetic tools, emphasizing recent work engineering photoreceptor proteins for allosteric control and to control protein-protein interactions, and describing future engineering challenges faced by developers and adopters of these new technologies.
Biological photoreceptors: basic modules for optogenetic engineering
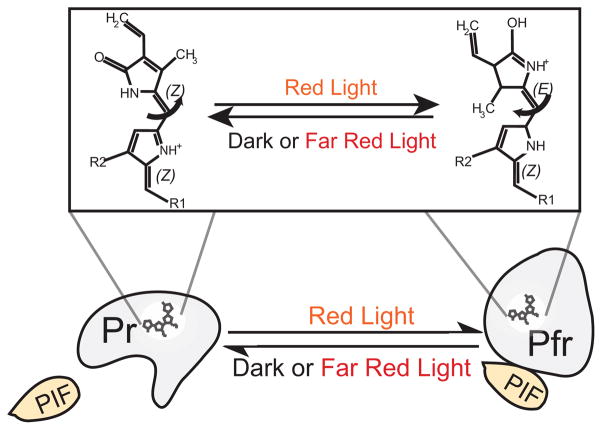
Photoreceptors proteins are used by organisms to sense and respond to optical cues in their environment. Generally, photoreceptors are stimulated to their photoexcited states via changes in a small molecule cofactor, termed a chromophore, that is bound tightly to the protein and is responsible for initial photon absorption. Photon capture causes a structural change in the chromophore, leading to a conformational change in the tightly associated photoreceptor protein. In the absence of additional photon stimuli, the stimulated photoreceptor relaxes over time to its original unexcited conformation, or ground state, in a process known as dark reversion. The rate at which the protein reverts from the photoexcited to ground state can vary on a time scale of seconds, minutes, or hours depending on the protein. As shown in Figure 1 with plant phytochrome B (phyB), the conformational switching of a photoreceptor protein between the photoexcited and ground states functions as a biological switch to alter signaling states in the protein.
Figure 1. Mechanism of phytochrome B photoactivation.
In dark or far-red light, phyB exists predominantly in a red light responsive ‘Pr’ form. Upon red light stimulation, the covalently bound chromophore phytochromobilin (top) undergoes a reversible Z-E isomerization, resulting in a conformational change in phyB to the Pfr signaling state. This state can be switched back to the Pr state by far-red light illumination. Light dependent effector proteins, such as PIF3 or PIF6, bind the photoreceptor preferentially in one conformational state.
Based on the associated chromophore, biological photoreceptors can be grouped in six different families: rhodopsins, xanthopsins, phytochromes, cryptochromes, LOV domain-containing and BLUF domain-containing (van der Horst and Hellingwerf, 2004; Moglich et al., 2010). In the first three families, photoswitching occurs via a cis-trans isomerization around a carbon double bond in the chromophore that takes place upon photon absorption. The latter three families contain flavin-based chromophores that engage in electron transfer or adduct formation upon photostimulation (Purcell and Crosson, 2008; Moglich et al., 2010). In addition to these families, at least one photoreceptor protein, the UV-B sensitive plant UVR8, does not bind an auxiliary chromophore but appears to perceive light by switching from a dimer to monomer state upon excitation of tryptophan residues contained within the protein (Christie et al., 2012a; Wu et al., 2012). In this review, we will provide an overview of photosignaling states of phytochromes, cryptochromes, and LOV domains, as these photoreceptors have been used most extensively for optical control.
Phytochromes
Classically, phytochromes are red/far-red light-sensing photoreceptor proteins found in higher plants, cyanobacteria, non-photosynthetic eubacteria, and some fungi (Davis et al., 1999; Rockwell et al., 2006). This superfamily also includes a class of related cyanobacteriochromes from cyanobacteria with diverse photocycles that respond to a broader range of light wavelengths (Ikeuchi and Ishizuka, 2008; Rockwell et al., 2012b). Structurally, phytochromes contain a C-terminal transmitter region and an N-terminal sensory module consisting of three domains: PAS (Per/ARNT/Sim), GAF (cGMP phosphodiesterase/adenylate cyclase/FhlA), and PHY (phytochrome-specific). Phytochromes covalently bind a linear tetrapyrrole (bilin) ligand as a chromophore in the N-terminal sensory region. Plant and cyanobacterial phytochromes (Phys and Cphs) bind the chromophores phytochromobilin and phycocyanobilin (PCB), respectively, while bacterial and fungal phytochromes (Bphs and Fphs) bind the chromophore biliverdin (Bhoo et al., 2001; Lamparter et al., 2004; Rockwell et al., 2006). In plants, phytochromes are responsible for regulation of seedling de-etiolation, flowering, and shade avoidance (Franklin and Quail, 2010; Chen and Chory, 2011).
Phytochromes convert between two relatively stable states: a red-light absorbing Pr state that predominantly exists in the dark or far-red light, and a far-red absorbing Pfr state that predominates in red light (Rockwell et al., 2006). As shown in Figure 1, conversion between these two states involves a cis-trans isomerization around a carbon double bond of the bilin chromophore, resulting in a conformational change in the associated phytochrome protein. This conformational change in phytochrome results in light-dependent binding or dissociation with downstream interacting proteins.
Cryptochromes
Cryptochrome photoreceptors are blue/UV-A light-responsive proteins (Lin and Shalitin, 2003). These proteins were first characterized in plants (Ahmad and Cashmore, 1993), and have subsequently been found in all kingdoms of life. Structurally, cryptochromes contain an N-terminal DNA photolyase homology region (PHR) that binds flavin adenine dinucleotide (FAD), and a C-terminal (CCT) domain essential for signal transduction in plants (Yang et al., 2000; Sancar, 2004). Despite substantial sequence homology with DNA photolyases within the PHR domain, cryptochromes do not retain DNA repair activity (Sancar, 2008). Functionally, cryptochromes have diverse roles in plants and animals. In plants, cryptochromes are essential in light-dependent regulation of growth and development, including de-etoliation and flowering, and are also involved in regulating the circadian clock; in animals, cryptochromes are central components of the circadian clock (Sancar, 2003; Lin and Todo, 2005; Chaves et al., 2011; Liu et al., 2011).
The precise photosensory mechanism by which cryptochromes convert between signaling states remains unclear (Liu et al., 2010; Losi and Gärtner, 2011). In vitro studies in large part with Arabidopsis cryptochrome 1 (AtCRY1) have led to the proposal of a trp-triad-dependent photoreduction that switches the signaling state of this protein. In this hypothesis, cryptochrome is proposed to bind oxidized FAD in the ground state, and light stimulation results in formation of the neutral radical semiquinone (FADH•), thought to be the signaling state, via intra-protein electron transfer between aromatic residues and the FAD cofactor (Giovani et al., 2003; Kottke et al., 2006; Banerjee et al., 2007; Bouly et al., 2007). Further studies have shown that conversion from the cofactor ground state (oxidized FAD) to the photoexcited state (FADH•) causes a conformational change in full length AtCRY1 (Chaves et al., 2011; Kondoh et al., 2011). Alternate hypotheses also have been proposed as mechanisms for photoactivation (Liu et al., 2010).
LOV (Light, Oxygen, or Voltage) domains
Similar to cryptochromes, proteins of the light-oxygen-voltage (LOV) domain family, which falls in the larger class of PAS (PER-ARNT-SIM) sensory domains, also bind a flavin chromophore. The LOV domain can be found as a single domain or associated with a wide range of effector domains, resulting in diverse roles for LOV-containing proteins (Losi and Gärtner, 2008). In plants, LOV domain proteins have functional roles in phototropism, stomatal translocation, and chloroplast movements (Liscum and Briggs, 1995; Christie et al., 2002; Cho et al., 2007). In bacteria, they are involved in regulation of stress responses, cell attachment, development, and virulence (Gaidenko et al., 2006; Purcell et al., 2007; Swartz et al., 2007). Consistent with such a variety of domain structures and roles, the changes that occur in LOV domain proteins during signal transduction are also quite diverse, such as unwinding of a C-terminal α–helix (Harper et al., 2003) or light dependent dimerization (Möglich and Moffat, 2007).
Despite this functional diversity, studies of different LOV domains suggest a conserved photoactivation mechanism. The photoactivation mechanism of the LOV2 domain of Avena sativa (oat) phototropin 1 (AsLOV2) has been one of the most extensively studied. Upon photoexcitation, a covalent thioether bond is formed between the FMN isoalloxazine ring and a highly conserved cysteine residue of the LOV domain (Salomon et al., 2000; Christie et al., 2012b). Adduct formation triggers undocking and unwinding of a C-terminal α-helix (Jα) that is docked to the monomeric protein in the dark state (Harper et al., 2003; Halavaty and Moffat, 2007).
LOV domains from diverse photoreceptors appear to undergo similar changes in the LOV core as AsLOV2, with light stimulation triggering formation of a covalent bond between the flavin cofactor and a conserved cysteine of LOV, resulting in altered interactions between the LOV core and other domains (Zoltowski and Gardner, 2011). Photoactivation of Neurospora crassa VVD, which leads to protein dimerization, alters interactions between the LOV core and an N-terminal extension (Zoltowski et al., 2007; Vaidya et al., 2011), while photoactivation of EL222, a LOV-containing DNA binding protein from Erythrobacter litoralis HTCC2594, results in dissociation from the LOV core of a C-terminal helix-turn-helix domain that mediates DNA binding (Nash et al., 2011). Photoactivation of the LOV domain of Bacillus subtilis YtvA, which exists as a dimer, causes small changes throughout the LOV domain and Jα helix, resulting in a subtle shift in the orientation of two LOV domain subunits relative to each other (Moglich and Moffat, 2007). In structural studies of the Vaucheria frigida Aureochrome 1 LOV domain, both N and C-terminal extensions were found associated with the LOV core, raising the possiblity that effector domains could be fused to either end of LOV for photoregulation (Mitra et al., 2012). Many of these studies examined photosensory domains alone or with small N- or C-terminal extensions, rather than in the context of the full-length proteins. Structural studies of full-length LOV proteins with diverse effector domains, as has been carried out with EL222 (Nash et al., 2011), can provide valuable mechanistic insights into ways to engineer LOV-containing proteins for optogenetic applications.
Engineered systems for controlling cell function
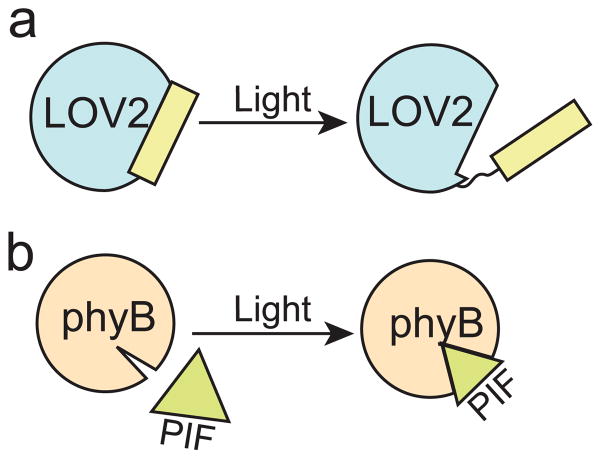
Genetically-encoded light-responsive tools have been utilized in a number of innovative ways to control biological function, allowing regulation of transcription, enzymatic activity, and protein subcellular localization in a light-dependent manner. Generally, there are two strategies for controlling biological processes with light using photoreceptor proteins: allosteric regulation (Fig. 2a), in which a photosensory domain is engineered to control enzyme activity or access to a binding site, or dimerization (Fig. 2b), in which modular protein interactions are used to control interactions of tethered target proteins. These different strategies can also be combined, as several recently developed technologies have used allosteric regulation to engineer proteins that can be used as modular dimerizers.
Figure 2. Strategies for engineered optogenetic regulation.
a) Allosteric regulation. Photoreceptors or domains such as AsLOV2-Jα that undergo a large structural change upon light binding can be attached to or inserted within other proteins to control enzyme activity or binding interactions. b) Optical dimerizers. Modular light-interacting domains are used to control interactions and localization of fused target proteins (e.g. phyB and PIF3).
Allosteric regulation
Light sensing domains have been used to allosterically control protein function. AsLOV2 has been used most prominently for this purpose, as this domain is small, modular, well characterized, and undergoes a large structural change upon photoexcitation. Generally, there have been two strategies for photoregulation using AsLOV2: (1) AsLOV2-Jα is attached to a target protein such that conformational changes in LOV2 induce conformational changes in the target, and (2) steric occlusion—a domain is attached to AsLOV2-Jα in such a way that binding to an effector is blocked in the dark but permitted in the light upon dissociation of Jα from the LOV core. Taking the former approach, the AsLOV2-Jα domain was inserted at different sites within the enzyme dihydrofolate reductase, resulting in light-regulated dihydrofolate reductase activity (Lee et al., 2008). AsLOV2-Jα has also been used to control activity of Lipase A (Krauss et al., 2010). In another study, AsLOV2-Jα was attached to an N-terminal helix of the bacterial trp repressor, TrpR (Strickland et al., 2008). The fusion protein, ‘LovTAP’, was found to bind DNA preferentially in light, demonstrating that changes arising in the Jα-helix upon photoexcitation were transferred to induce a conformational change in TrpR.
Prior studies of the interaction of AsLOV2 and the C-terminal Jα-helix have demonstrated that the Jα-helix is not simply bound to the LOV core in the dark and released in light, but populates both states in light and dark, though at different ratios (Yao et al., 2008). Mutations that destabilize docking to the LOV core were identified and found to lead to light-independent constitutive activity (Harper et al., 2004). In a followup to the LovTAP study (Strickland et al., 2008), the authors identified mutations in LOV-Jα that stabilized LOV docking, increasing the dynamic range of the DNA-binding photoswitch from a 5-fold to 70-fold difference in DNA binding affinity between lit and dark states (Strickland et al., 2010).
While these initial studies demonstrated light-regulated enzyme activity and binding, they did not demonstrate fine spatial control of biological activity. In a breakthrough study, AsLOV2 was used in mammalian cells to control the activity of Rac1, a GTPase that regulates actin cytoskeletal dynamics (Wu et al., 2009). Rac1 was fused at the C-terminus of LOV-Jα, sterically blocking its binding to effector proteins in the dark. Blue light irradiation resulted in unfolding of the Jα-helix, relieving the steric inhibition and facilitating the interaction of Rac1 with effectors. Using focused laser excitation directed to subcellular regions, the authors demonstrated conditional formation of lamellipodia only at illuminated sites. This work provided an exciting demonstration that cellular behavior can be controlled with light with high spatial and temporal resolution.
Although AsLOV2-Jα has been most extensively used, other domains have also been explored for optogenetic applications. Rotational movement upon photoactivation of a dimeric B. subtilis YtvA LOV domain was used to engineer a photocontrollable histidine kinase, in which a PAS domain from the oxygen sensor FixL of Bradyrhizobium japonicum was replaced by YtvA-LOV (Moglich et al., 2009). This fusion protein was further exploited for control of gene expression in bacteria, with the engineering of plasmids allowing induction (pDawn) or repression (pDusk) of gene expression with light (Ohlendorf et al., 2012). Other light-controlled transcriptional systems have also been engineered. A fusion between the cyanobacterial phytochrome, Cph1, and the E. coli ENVZ histidine kinase resulted in a red-light-regulated system to control transcription in E. coli (Levskaya et al., 2005). This work was followed by development of a green/red switchable system, using the cyanobacteriochrome CcaS and its response regulator CcaR (Tabor et al., 2011). The light-dimerizing Neurospora crassa VVD protein was fused to a DNA binding domain and activation domain and used to control transcription in mammalian cells (Wang et al., 2012). A recent paper also extends light-inducible transcriptional tools for use in Neurospora, using the endogenous light-regulated vvd promoter (Hurley et al., 2012).
Optical Dimerization Systems
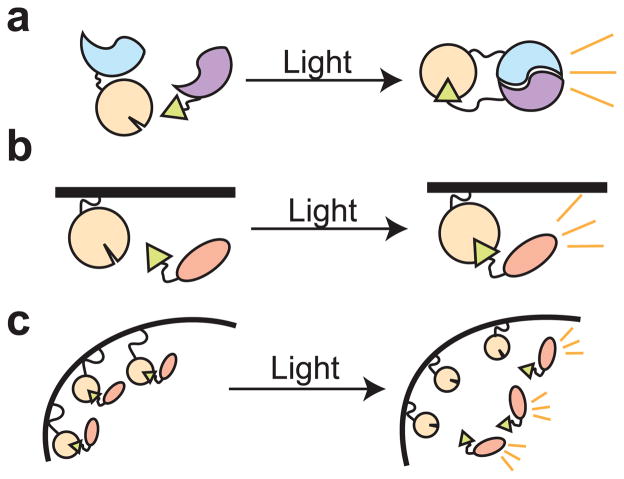
Another strategy for controlling cell function is through the use of modular domains that mediate protein dimerization, allowing inducible recruitment or sequestering of proteins to or from their sites of action (Spencer et al., 1993). Optical dimerizers utilize natural or engineered light-dependent interacting domains and can be controlled by different wavelengths of light. In general, these types of tools require less engineering, as the interacting domains are more modular and thus can be more easily appended to different target proteins for specific applications.
As shown in Figure 3, a variety of strategies exist for using optical dimerizers. These strategies rely on use of the modular domains to bring targets together to elicit a functional activity (recruitment), or alternatively, the domains may be used to keep a molecule in a nonfunctional state until release (sequestering). For example, dimerizers can be employed to unite a non-functional split protein into a functional whole (Fig. 3a), a strategy that has been utilized for light-regulated control of gene transcription or DNA recombination (Shimizu-Sato et al., 2002; Yazawa et al., 2009; Kennedy et al., 2010; Hughes et al., 2012a; Wang et al., 2012). Similarly, dimerizers can be recruited to a specific subcellular location such as the plasma membrane, initiating enzymatic activity or a signaling cascade (Fig 3b). Such strategies are used extensively in nature, where proteins are often kept in subcellular locales where they are unable to function, before recruitment to their sites of action (e.g. nucleus, plasma membrane). Alternatively, proteins may be sequestered from their usual sites of action, resulting in loss of protein activity (Fig. 3c). Optimally, a dimerization pair that interacts in the dark, such as CRY1 and phyB (Hughes et al., 2012b), could be used to sequester proteins in an inactive state until light stimulation. In the following sections we describe several of the existing optical dimerization systems.
Figure 3. Basic schemes for control of cell function by optical dimerizers.
a) Split protein reconstitution. Two non-functional parts of a protein (blue and purple half-circles) are brought together by optical dimerizers to restore protein activity in a light-dependent manner. b) Subcellular recruitment. A protein is recruited via dimerizers to a region of the cell where it is active. In the absence of recruitment the protein is nonfunctional. c) Sequestration. A protein is sequestered in an inactive state by a dimerizer, then released to sites of action with light.
PhyB/PIF
An interaction between phyB and a transcription factor, PIF3, was the first system described for optical control of protein-protein interactions in cells (Shimizu-Sato et al., 2002). As depicted in Figure 1, PIF3 interacts with phyB in the Pfr state, but not in the Pr state. This natural light-dependent interaction was used to regulate gene expression in yeast by linking phyB (residues 1-621) and PIF3 to separable activation and binding domains of the transcription factor Gal4. In the absence of red light illumination, phyB and PIF3 did not interact and thus the transcription factor was split in two separate non-functional parts. However, red light illumination promoted the binding of phyB to PIF3, reconstituting a functional Gal4 transcription factor and promoting transcription, which could be reversibly disassociated with far-red light illumination.
Additional studies used the same phyB/PIF3 interaction to control activity of other proteins. In one study, the interaction was used to induce actin assembly in vitro (Leung et al., 2008). Light-induced dimerization of phyB and PIF3 promoted interaction of GDP-bound Cdc42 and WASP, resulting in red light inducible actin assembly, through Arp2/3 activation, that could be reversed with application of far-red light. In another study, phyB and PIF3 were used to reconstitute a split S. cerevisiae VMA intein with light, allowing light-mediated control of protein splicing (Tyszkiewicz and Muir, 2008).
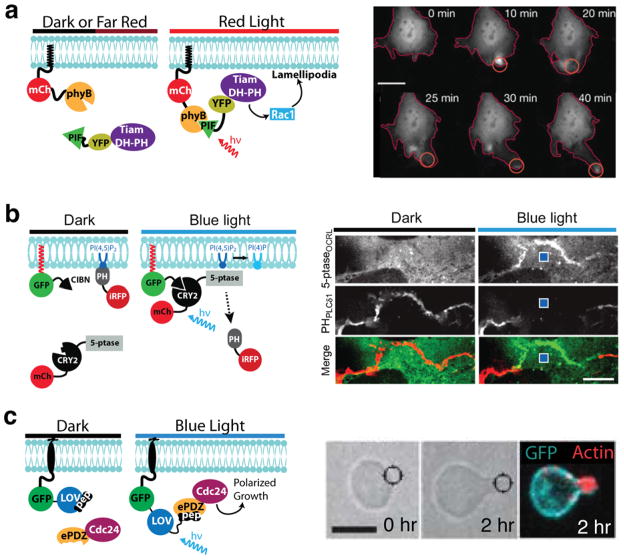
A key advantage of optogenetics is the ability to control processes with subcellular spatial precision. A prime demonstration of this was provided using a different Arabidopsis phyB protein interaction, phyB/PIF6, where authors demonstrated rapid and reversible translocation of proteins to the plasma membrane of mammalian cells using light (Levskaya et al., 2009). Using the N-terminal region of PIF6 (residues 1-100) and phyB (residues 1-908), the authors were able to rapidly recruit target proteins to the plasma membrane (τ = 1.3 s). Most importantly, as phyB can be reversibly photoswitched with two different light wavelengths (Fig. 1), application of farred light resulted in rapid reversal of binding (τ = 4 s). Using an approach pioneered with chemical dimerizers to activate small GTPases (Inoue et al., 2005), the study also demonstrated light dependent recruitment of catalytic domains (DH-PH domains) of guanine nucleotide exchange factors (GEFs) to the plasma membrane. Upon membrane recruitment of the catalytic domain of the Rac GEF Tiam (Tiam-DH-PH), the authors demonstrated activation of Rac and formation of cellular protrusions extending in the direction of the light (Fig. 4a). These breakthrough experiments demonstrated the power of optical dimerizers to alter cell function, not only with high temporal resolution, but with subcellular spatial precision as well.
Figure 4. Use of optogenetic systems for fine spatial control of cell function.
a) Focal activation of Rac-mediated cell protrusion with the phyB/PIF6 system. The catalytic domain of the Rac GEF Tiam (Tiam-DH-PH) is recruited to the plasma membrane via the phyB/PIF6 interaction, where it acts through Rac1 to form lamellipodia. Localized lamellipodia formation in NIH3T3 cells was induced by globally irradiating the cell with infrared (750 nm) light and spot illumination with a red (650 nm) laser. Scale bar 20 μm. Figure adapted with permission from Macmillan Publishers Ltd.: Nature (Levskaya et al., 2009), copyright 2009. b) Local recruitment of phosphatidylinositol 5′-phosphatase (5-ptase) to the plasma membrane using the CRY2/CIBN system. Upon focal light stimulation within COS-7 cells, cytosolic mCh-CRY2-5-ptase is recruited to a nearby region of the plasma membrane via interaction with CIBN. At the plasma memrane, the phosphatase depletes PI(4,5)P2 locally, visualized using the PI(4,5)P2 biosensor iRFP-PHPLCδ1. Images at right were taken of COS-7 cells expressing the constructs before and 10s after delivery of a 100-ms blue-light pulse (location marked by blue square). Scale bar: 5 μm. Figure reprinted with permission from The National Academy of Sciences, Proceedings of the National Academy of Sciences, USA (Idevall-Hagren et al., 2012), copyright 2012. c) Light-directed polarized yeast growth. Yeast cells expressing Cdc24-ePDZb1 and membrane localized Mid2-GFP-LOVpep were exposed to mating pheromone to induce cell cycle arrest. After 30 min, Cdc24-ePDZb1 was spot recruited using a blue laser and cells were imaged after two hours. Figure adapted with permission from Macmillan Publishers Ltd., Nature Methods (Strickland et al., 2012), copyright 2012.
It should be noted that in the above studies, use of phyB in yeast and mammalian cells required the addition of an exogenously supplied bilin cofactor. In plants, assembly of the light-sensitive holo form of phyB requires binding to phytochromobilin, a compound that is made in plants but is not present in other higher eukaryotes or yeast. A similar bilin compound, phycocyanobilin (PCB), can be extracted from Spirulina and supplied exogenously in the yeast media (Li and Lagarias, 1994; Shimazu-Sato et al., 2002), allowing formation of holo phyB. Mammals also do not make the bilin chromophore required for assembly of holo-phyB, and the Levskaya et al. (2009) work established that PCB can also be externally applied to mammalian cells. Alternatively, researchers have co-expressed biosynthetic enzymes that allow generation of the bilin cofactor within cells, though this approach has so far only extended to bacteria (Gambetta and Lagarias, 2001; Levskaya et al., 2005). The ability to produce the bilin cofactor in mammalian cells will certainly extend use of this system to more complex organism studies, rather than just cell culture.
GIGANTEA/FKF1
A blue light-dependent interaction between the LOV-domain-containing protein FKF1 and GIGANTEA, proteins that control flowering in Arabidopsis, was used to induce protein dimerization (Yazawa et al., 2009). The group used the interaction for light-induced recruitment of a constitutively active Rac1 protein, resulting in lamellipodia formation, and to bring together a split transcription factor in mammalian cells, resulting in blue light induction of transcription. A recent paper further explored use of the FKF1/GIGANTEA interaction to induce transcription in mammalian cells, bringing together an engineered zinc finger protein and a VP-16 transcriptional activation domain (Polstein and Gersbach, 2012). The FKF1/GIGANTEA interaction is long lived, making this system potentially useful for applications where reversibility is not desired.
CRY2/CIB1
A cryptochrome-based dimerization system was demonstrated using Arabidopsis cryptochrome 2 (CRY2) and its interaction partner CIB1 (Kennedy et al., 2010). The CRY2/CIB interaction was used to recruit target proteins to the plasma membrane in mammalian cells, where they responded to light within seconds of blue light illumination and the interaction naturally reversed within several minutes (t1/2 ~ 5.5 min). In two demonstrations, the authors used the system to bring together split protein fragments to reconstitute functional activity. In the first case, the CRY2/CIBN dimerizers were used to bring together split fragments of Cre DNA recombinase. While no recombinase activity was observed in the dark, blue light stimulation resulted in dose-dependent induction of DNA recombination. In the second demonstration, the authors used CRY/CIB to bring together a split Gal4 transcriptional activator, using a similar approach as that of Quail and colleagues with phyB/PIF3 (Shimizu-Sato et al., 2002). In follow up studies examining light-induced control of DNA transcription in yeast, the authors used the CRY/CIB dimerizers to bring together a split LexA-VP16 transcription factor, which showed robust activation in light (~50-fold activation of a reporter over dark levels after several hours) with low background (Hughes et al., 2012a). These experiments demonstrated the advantages of optogenetic tools for user-controlled, dose-dependent regulation of enzyme activity.
While the above studies demonstrated regulation of localization by global illumination of cells, as well as temporal and dose dependent control of enzyme activity using light, they did not demonstrate use of the CRY/CIB dimerizers for subcellular control of protein localization. However, a recent publication demonstrates use of CRY2/CIB for local recruitment of phosphatidylinositol 5′-phosphatase to the plasma membrane in COS-7 cells (Idevall-Hagren et al., 2012). Once recruited, the enzyme dephosphorylated phosphoinositide (PI(4,5)P2) in a spatially restricted manner only in the localized region where it had been recruited (Fig. 4b). In these studies, the phosphatase was fused to CRY2 and expressed in the cytosol, where it was recruited to the membrane by binding to a prenylated CIBN. In this configuration, with the photoreceptor module expressed in the cytosol, it might have been expected that stimulated CRY2 would diffuse rapidly throughout the cell. Surprisingly, local recruitment could be achieved: when illuminated, CRY2 was preferentially recruited only to plasma membrane-bound CIB in the nearby vicinity. With the phosphatidylinositol 5′-phosphatase recruitment studies, Idevall-Hagren and colleagues also demonstrated similar reversibility of the system as seen previously (Kennedy et al., 2010), with the phosphatase returning to the cytosol within minutes (t1/2 = 6.8 ± 1 min), followed by recovery of inositol phosphate (PI(4,5)P2) after a short lag due to resynthesis. Comparison of the optical dimerizers with chemical dimerizers demonstrated that the optical system performed at over an order of a magnitude faster (t1/2 = 3.1 ± 0.2 s) than a previously used rapamycin-based system (Idevall-Hagren et al., 2012).
Engineered optical dimerizers using AsLOV2
Recently, two groups used AsLOV2 to control binding of a peptide epitope, generating engineered LOV-domain-based optical dimerizers (Lungu et al., 2012; Strickland et al., 2012). In both cases, a peptide epitope was fused to the C-terminus of AsLOV2-Jα. In one system, designated TULIPs (Tunable, Light-controlled Interacting Proteins), the caged peptide (LOVpep) interacts with an engineered PDZ domain (Strickland et al., 2012). In the dark, the peptide is sterically hindered from PDZ domain binding, whereas light stimulation results in release of the steric interference and permits interaction. Mutants that alter photocycle duration and Jα helix docking to the LOV core were examined to determine their effects on TULIP caging and kinetics. As measured by plasma membrane recruitment experiments, mutant variants showed a range of dimerization affinities, dark-state recoveries, and altered dissociation kinetics, demonstrating the ability to ‘tune’ this system for specific applications using mutagenesis. The group established functionality in mammalian cells and in yeast, where the system was used to inducibly control the yeast mating pathway. In this demonstration, light-dependent mating pathway activation was achieved by triggering recruitment of Ste5, a scaffold protein involved in the mating MAP kinase cascade, to the plasma membrane. In a separate experiment, polarity of yeast mating projections could be controlled at precise subcellular locations via membrane recruitment of Cdc24 (Fig. 4c).
Employing a similar strategy as with TULIPs, Kuhlman and colleagues designed an optical dimerizer that cages a vinculin binding peptide, ipaA (Lungu et al., 2012). Initial constructs showed significant (49-fold) differences in binding affinity in lit versus dark states when tested in vitro, but the caged ipaA retained substantial binding to vinculin in the dark. When these constructs were tested in yeast for control of transcription (via reconstitution of a split Gal4 transcription factor controlling cell growth), the yeast showed no phenotypic differences in light vs dark, likely due to the high dark binding. Addition of a mutation in vinculin that reduces affinity to ipaA substantially reduced dark state binding affinity while maintaining lit state affinity, resulting in a phenotypic difference in transcriptional activation when tested in yeast. These results suggest that engineered optical dimerizers may require significant tuning of binding affinities for optimal function within cells.
The Road Ahead: Discovery, Optimization, and Challenges
Adapting the new set of optogenetic tools for cellular control presents a number of challenges. Here we describe the road ahead for optogenetics research, discussing strategies for optimizing existing tools, the need to identify new photoreceptor modules for the optogenetic toolkit, and future engineering challenges.
Optimization of photoreceptors for optogenetic applications
While existing optogenetic tools can be successfully adopted as is for new applications (e.g. adoption of the CRY2/CIB system for phosphoinositide studies in mammalian cells (Idevall-Hagren et al., 2012)), further engineering and optimization of these proteins will greatly improve their broad adoption and general utility. Identification of mutant photoreceptor variants with altered dark reversion rates and spectral sensitivities, for example, will improve versatility and allow tuning for specific applications. For example, to control some processes, such as activation of DNA recombination or transcription, a dark reversion rate of minutes or hours may be more useful than a dark reversion rate of seconds. Alternatively, in the absence of systems that can be inducibly shut off such as Phy/PIF, localized activation will be enhanced by a fast dark reversion rate, such that proteins that diffuse away from a site of illumination will rapidly revert to the dark state. Several prior studies have demonstrated that photocycle kinetics of photoreceptors can be tuned by mutagenesis (Christie et al., 2007; Zoltowski et al., 2009; Raffelberg et al., 2011). For example, single amino acid changes were found to yield an 85-fold increase in YtvA dark recovery rate (Raffelberg et al., 2011), or a 10-fold increase in AsLOV2 dark recovery rate (Christie et al., 2007). While mutant variants can be identified by random selection or using structural guidance, natural photoreceptor variants also possess diverse photokinetics and spectral tuning (Man et al., 2003; Jentzsch et al., 2009; Pathak et al., 2009; Narikawa et al., 2011; Rockwell et al., 2012a; Rockwell et al., 2012b) and may provide starting points for protein engineering.
In addition to identifying mutations that affect photocycle kinetics, expanding the range of wavelengths available for optogenetic control is another engineering goal. A critical challenge for integrating optogenetic actuators and light-based reporters involves the significant spectral overlap between different systems, which can blur the distinction between excitation response and output fluorescence. For example, the excitation wavelengths of common fluorescent proteins (eGFP, CFP, and YFP) overlap with the wavelengths used to stimulate flavin-based actuators (i.e. LOV, cryptochrome, BLUF), thus these fluorescent probes cannot be visualized without actuating the process under investigation. Identification of variants with shifted absorption spectra would permit better integration of multiple fluorescent reporters and/or multiple optogenetic modules in a single system (Fig. 5), enabling more complex optical control over biological phenomena. Prior studies have indicated that mutation of residues situated near the chromophore can modulate spectral tuning in a variety of proteins (Chan et al., 1992; Lin et al., 1998; Takahashi and Ebrey, 2003; Nash et al., 2008; Arents et al., 2011). In this effort, natural photoreceptor variants can provide guidance. For example, natural variants of proteorhodopsins were found to show depth stratified spectral tuning that was determined by a single amino acid residue change, converting the protein’s absorption maxima from about 525 to 490 nm (Man et al., 2003). The success of site-directed mutagenesis and directed evolution for spectral tuning and further optimization of fluorescent proteins (Heim et al., 1994; Heim and Tsien, 1996; Shaner et al., 2004; Auldridge et al., 2012) suggests these approaches hold promise for engineering optogenetic actuators as well.
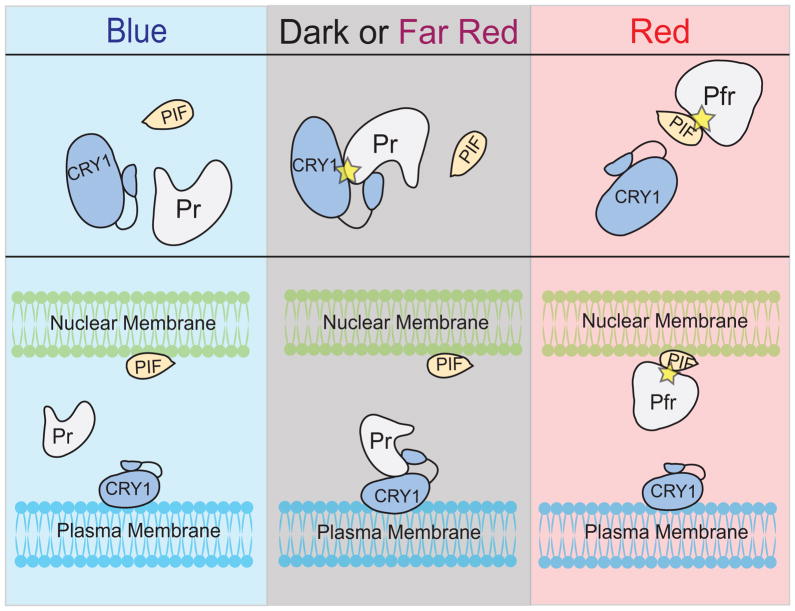
Figure 5. Integrating multiple optical tools for complex control.
In this hypothetical scheme, the phyB/PIF optical dimerizer system is combined with a CRY1/phyB system that is light-dissociated. Thus, in blue light, none of the proteins interact. In dark or far-red light, phyB interacts with CRY1, while in red light phyB interacts with PIF. Thus, a protein of interest tethered to phyB can be shuttled from one location to the other (for example, plasma membrane vs. nuclear membrane) via changes in light wavelength.
Discovery and exploration of new photoresponsive modules
While LOV, cryptochrome, and phytochrome domain-containing proteins from plants have dominated optogenetic tool development, numerous photosensory proteins exist throughout the biological kingdoms that are ripe for exploration. Identification of novel biological photoreceptors and characterization of their modes of action and biochemical properties will provide us with a vast choice of design modules for engineering. For example, 578 different LOV domain sequences, showing diverse domain architecture, were recently identified from metagenomic analysis of soil, marine, air, symbiotic and extreme environments (Pathak et al., 2012). As has been found with bacteriorhodopsin-based ion channels (Yizhar et al., 2011; Chow et al., 2012), natural variants often have quite different properties, such as different dark reversion rates or spectral sensitivities. LOV domains with novel features have been reported from organisms inhabiting extreme environments, such as thermophilic, acidic or saline environments (Pathak et al., 2009; Pathak et al., 2012). In nature, these sensory domains couple to effector or output domains in quite different ways (Losi and Gartner, 2008). Coupling novel sensory proteins to different or mutagenized effector domains will provide promising avenues for future engineering efforts. For example, a BLUF domain with a light-responsive adenylate cyclase effector domain was engineered to regulate synthesis of cyclic GMP in response to light (Ryu et al., 2010). As cyclic nucleotides are important secondary messengers in eukaryotic cells, optical control of these systems could allow regulation of a wide range of cellular activities.
Engineering challenges
A substantial challenge for optogenetics is the significant engineering required to develop robust, tightly-controlled systems. Allosteric designs, joining effector domains to photosensory domains for tight light control, require significant design, testing, and optimization. For example, the development of a photoactivatable Rac1 required sampling of numerous fusion junctions between the LOV2 domain and Rac1 to ensure adequate allosteric caging (Wu et al., 2009). While optical dimerizers are more modular than engineered allosteric systems, even these exhibit context-dependent behavior when fused to different proteins. For instance, while phyB/PIF3 dimerizers were found to work well in yeast (Shimizu-Sato et al., 2002; Tyszkiewicz and Muir, 2008; Hughes et al., 2012a), these domains did not work for a membrane recruitment assay in NIH3T3 mammalian cells (Levskaya et al., 2009). As another example, CRY2 only tolerated mCherry fused to its C-terminus, but not its N-terminus, in a CIB membrane recruitment assay in HEK293 cells, even though Gal4BD fused to the N-terminus of CRY2 showed light-dependent binding to CIB (Kennedy et al., 2010). In addition to engineering challenges with modular domains, each application also presents its own unique challenges. The creation of robust split-protein systems, for example, is difficult as it is not always clear how a protein should be divided for functional reconstitution or whether the protein can be successfully reconstituted to restore function at all. Thus, in particular when designing new allosteric control strategies, but also when using modular domains with new applications, testing of a variety of design strategies, including different domain orders or linker sizes, is critical.
Concluding Remarks
The light-regulated systems described in this review provide ways to non-invasively control cellular function with unprecedented spatiotemporal precision. Despite exciting new developments, the field of optogenetics is still in its infancy. The number of characterized tools is still somewhat limited, awaiting the discovery of new photosensory domains and photoexcitation mechanisms that will greatly expand the optogenetic toolkit. Further engineering and quantitative characterization of existing tools will provide users with the capacity to tune absorbance wavelengths, dark reversion rates, activation kinetics, and binding affinities for specific applications. The field will also be advanced by innovation in other engineering areas, such as the further development of hardware or software to enhance or regulate light delivery (Leifer et al., 2011; Milias-Argeitis et al., 2011; Toettcher et al., 2011; Dugue et al., 2012). With further development of optogenetic tools, optogenetics will likely extend well beyond neurobiology to become a transformative force in nearly any field requiring fast, conditional control of cellular biochemistry. The precise and non-invasive character of these tools will likely encourage a shift in experimental paradigm, emboldening researchers to explore novelty in their experimental designs to address previously intractable questions.
Acknowledgments
The authors thank Matthew Kennedy for helpful comments. This work was supported by a grant from the National Institutes of Health [R01 GM100225] to C.L.T.
References
- Ahmad M, Cashmore AR. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature. 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- Arents JC, Perez MA, Hendriks J, Hellingwerf KJ. On the midpoint potential of the FAD chromophore in a BLUF-domain containing photoreceptor protein. FEBS Lett. 2011;585:167–172. doi: 10.1016/j.febslet.2010.11.035. [DOI] [PubMed] [Google Scholar]
- Auldridge ME, Satyshur KA, Anstrom DM, Forest KT. Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein. J Biol Chem. 2012;287:7000–7009. doi: 10.1074/jbc.M111.295121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee R, Schleicher E, Meier S, Viana RM, Pokorny R, Ahmad M, Bittl R, Batschauer A. The signaling state of Arabidopsis cryptochrome 2 contains flavin semiquinone. J Biol Chem. 2007;282:14916–14922. doi: 10.1074/jbc.M700616200. [DOI] [PubMed] [Google Scholar]
- Bhoo SH, Davis SJ, Walker J, Karniol B, Vierstra RD. Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature. 2001;414:776–779. doi: 10.1038/414776a. [DOI] [PubMed] [Google Scholar]
- Bouly JP, Schleicher E, Dionisio-Sese M, Vandenbussche F, Van Der Straeten D, Bakrim N, Meier S, Batschauer A, Galland P, Bittl R, et al. Cryptochrome blue light photoreceptors are activated through interconversion of flavin redox states. J Biol Chem. 2007;282:9383–9391. doi: 10.1074/jbc.M609842200. [DOI] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Chan T, Lee M, Sakmar TP. Introduction of hydroxyl-bearing amino acids causes bathochromic spectral shifts in rhodopsin. Amino acid substitutions responsible for red-green color pigment spectral tuning. J Biol Chem. 1992;267:9478–9480. [PubMed] [Google Scholar]
- Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GT, Batschauer A, Ahmad M. The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol. 2011;62:335–364. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J. Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 2011;21:664–671. doi: 10.1016/j.tcb.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HY, Tseng TS, Kaiserli E, Sullivan S, Christie JM, Briggs WR. Physiological roles of the light, oxygen, or voltage domains of phototropin 1 and phototropin 2 in Arabidopsis. Plant Physiol. 2007;143:517–529. doi: 10.1104/pp.106.089839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow BY, Han X, Boyden ES. Genetically encoded molecular tools for light-driven silencing of targeted neurons. Prog Brain Res. 2012;196:49–61. doi: 10.1016/B978-0-444-59426-6.00003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM, Arvai AS, Baxter KJ, Heilmann M, Pratt AJ, O’Hara A, Kelly SM, Hothorn M, Smith BO, Hitomi K, et al. Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science. 2012a;335:1492–1496. doi: 10.1126/science.1218091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM, Corchnoy SB, Swartz TE, Hokenson M, Han IS, Briggs WR, Bogomolni RA. Steric interactions stabilize the signaling state of the LOV2 domain of phototropin 1. Biochemistry. 2007;46:9310–9319. doi: 10.1021/bi700852w. [DOI] [PubMed] [Google Scholar]
- Christie JM, Gawthorne J, Young G, Fraser NJ, Roe AJ. LOV to BLUF: flavoprotein contributions to the optogenetic toolkit. Mol Plant. 2012b;5:533–544. doi: 10.1093/mp/sss020. [DOI] [PubMed] [Google Scholar]
- Christie JM, Swartz TE, Bogomolni RA, Briggs WR. Phototropin LOV domains exhibit distinct roles in regulating photoreceptor function. Plant J. 2002;32:205–219. doi: 10.1046/j.1365-313x.2002.01415.x. [DOI] [PubMed] [Google Scholar]
- Davis SJ, Vener AV, Vierstra RD. Bacteriophytochromes: phytochrome-like photoreceptors from nonphotosynthetic eubacteria. Science. 1999;286:2517–2520. doi: 10.1126/science.286.5449.2517. [DOI] [PubMed] [Google Scholar]
- Deisseroth K. Optogenetics. Nat Methods. 2011;8:26–29. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugue GP, Akemann W, Knopfel T. A comprehensive concept of optogenetics. Prog Brain Res. 2012;196:1–28. doi: 10.1016/B978-0-444-59426-6.00001-X. [DOI] [PubMed] [Google Scholar]
- Franklin KA, Quail PH. Phytochrome functions in Arabidopsis development. J Exp Bot. 2010;61:11–24. doi: 10.1093/jxb/erp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidenko TA, Kim TJ, Weigel AL, Brody MS, Price CW. The blue-light receptor YtvA acts in the environmental stress signaling pathway of Bacillus subtilis. J Bacteriol. 2006;188:6387–6395. doi: 10.1128/JB.00691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovani B, Byrdin M, Ahmad M, Brettel K. Light-induced electron transfer in a cryptochrome blue-light photoreceptor. Nat Struct Biol. 2003;10:489–490. doi: 10.1038/nsb933. [DOI] [PubMed] [Google Scholar]
- Halavaty AS, Moffat K. N- and C-terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa. Biochemistry. 2007;46:14001–14009. doi: 10.1021/bi701543e. [DOI] [PubMed] [Google Scholar]
- Han X, Boyden ES. Multiple-color optical activation, silencing, and desynchronization of neural activity, with single-spike temporal resolution. PLoS One. 2007;2:e299. doi: 10.1371/journal.pone.0000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper SM, Christie JM, Gardner KH. Disruption of the LOV-Jalpha helix interaction activates phototropin kinase activity. Biochemistry. 2004;43:16184–16192. doi: 10.1021/bi048092i. [DOI] [PubMed] [Google Scholar]
- Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- Heim R, Prasher DC, Tsien RY. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci U S A. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- Hughes RM, Bolger S, Tapadia H, Tucker CL. Light-mediated control of DNA transcription in yeast. Methods. 2012a doi: 10.1016/ymeth.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes RM, Vrana JD, Song J, Tucker CL. Light-dependent, dark-promoted interaction between Arabidopsis cryptochrome 1 and phytochrome B proteins. J Biol Chem. 2012b;287:22165–22172. doi: 10.1074/jbc.M112.360545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM, Chen C, Loros JL, Dunlap JC. Light-inducible system for tunable protein expression in Neurospora crassa. G3. 2012 doi: 10.1534/g3.112.003939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idevall-Hagren O, Dickson EJ, Hille B, Toomre DK, De Camilli P. Optogenetic control of phosphoinositide metabolism. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1211305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M, Ishizuka T. Cyanobacteriochromes: a new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochem Photobiol Sci. 2008;7:1159–1167. doi: 10.1039/b802660m. [DOI] [PubMed] [Google Scholar]
- Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat Methods. 2005;2:415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentzsch K, Wirtz A, Circolone F, Drepper T, Losi A, Gartner W, Jaeger KE, Krauss U. Mutual exchange of kinetic properties by extended mutagenesis in two short LOV domain proteins from Pseudomonas putida. Biochemistry. 2009;48:10321–10333. doi: 10.1021/bi901115z. [DOI] [PubMed] [Google Scholar]
- Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh M, Shiraishi C, Muller P, Ahmad M, Hitomi K, Getzoff ED, Terazima M. Light-induced conformational changes in full-length Arabidopsis thaliana cryptochrome. J Mol Biol. 2011;413:128–137. doi: 10.1016/j.jmb.2011.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottke T, Batschauer A, Ahmad M, Heberle J. Blue-light-induced changes in Arabidopsis cryptochrome 1 probed by FTIR difference spectroscopy. Biochemistry. 2006;45:2472–2479. doi: 10.1021/bi051964b. [DOI] [PubMed] [Google Scholar]
- Krauss U, Lee J, Benkovic SJ, Jaeger KE. LOVely enzymes - towards engineering light-controllable biocatalysts. Microb Biotech. 2010;3:15–23. doi: 10.1111/j.1751-7915.2009.00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamparter T, Carrascal M, Michael N, Martinez E, Rottwinkel G, Abian J. The biliverdin chromophore binds covalently to a conserved cysteine residue in the N-terminus of Agrobacterium phytochrome Agp1. Biochemistry. 2004;43:3659–3669. doi: 10.1021/bi035693l. [DOI] [PubMed] [Google Scholar]
- Lee J, Natarajan M, Nashine VC, Socolich M, Vo T, Russ WP, Benkovic SJ, Ranganathan R. Surface sites for engineering allosteric control in proteins. Science. 2008;322:438–442. doi: 10.1126/science.1159052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leifer AM, Fang-Yen C, Gershow M, Alkema MJ, Samuel AD. Optogenetic manipulation of neural activity in freely moving Caenorhabditis elegans. Nat Methods. 2011;8:147–152. doi: 10.1038/nmeth.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DW, Otomo C, Chory J, Rosen MK. Genetically encoded photoswitching of actin assembly through the Cdc42-WASP-Arp2/3 complex pathway. Proc Natl Acad Sci U S A. 2008;105:12797–12802. doi: 10.1073/pnas.0801232105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levskaya A, Chevalier AA, Tabor JJ, Simpson ZB, Lavery LA, Levy M, Davidson EA, Scouras A, Ellington AD, Marcotte EM, Voigt CA. Engineering Escherichla coli to see light. Nature. 2005;438:441–442. doi: 10.1038/nature04405. [DOI] [PubMed] [Google Scholar]
- Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lagarias JC. Phytochrome assembly in living cells of the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1994;91:12535–12539. doi: 10.1073/pnas.91.26.12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gutierrez DV, Hanson MG, Han J, Mark MD, Chiel H, Hegemann P, Landmesser LT, Herlitze S. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc Natl Acad Sci U S A. 2005;102:17816–17821. doi: 10.1073/pnas.0509030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Shalitin D. Cryptochrome structure and signal transduction. Annu Rev Plant Biol. 2003;54:469–496. doi: 10.1146/annurev.arplant.54.110901.160901. [DOI] [PubMed] [Google Scholar]
- Lin C, Todo T. The cryptochromes. Genome Biol. 2005;6:220. doi: 10.1186/gb-2005-6-5-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SW, Kochendoerfer GG, Carroll KS, Wang D, Mathies RA, Sakmar TP. Mechanisms of spectral tuning in blue cone visual pigments. Visible and raman spectroscopy of blue-shifted rhodopsin mutants. J Biol Chem. 1998;273:24583–24591. doi: 10.1074/jbc.273.38.24583. [DOI] [PubMed] [Google Scholar]
- Liscum E, Briggs WR. Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell. 1995;7:473–485. doi: 10.1105/tpc.7.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Liu H, Zhong D, Lin C. Searching for a photocycle of the cryptochrome photoreceptors. Curr Opin Plant Biol. 2010;13:578–586. doi: 10.1016/j.pbi.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Liu B, Zhao C, Pepper M, Lin C. The action mechanisms of plant cryptochromes. Trends Plant Sci. 2011;16:684–691. doi: 10.1016/j.tplants.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losi A, Gartner W. Bacterial bilin- and flavin-binding photoreceptors. Photochem Photobiol Sci. 2008;7:1168–1178. doi: 10.1039/b802472c. [DOI] [PubMed] [Google Scholar]
- Losi A, Gartner W. Old chromophores, new photoactivation paradigms, trendy applications: flavins in blue light-sensing photoreceptors. Photochem Photobiol. 2011;87:491–510. doi: 10.1111/j.1751-1097.2011.00913.x. [DOI] [PubMed] [Google Scholar]
- Lungu OI, Hallett RA, Choi EJ, Aiken MJ, Hahn KM, Kuhlman B. Designing photoswitchable peptides using the AsLOV2 domain. Chem Biol. 2012;19:507–517. doi: 10.1016/j.chembiol.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man D, Wang W, Sabehi G, Aravind L, Post AF, Massana R, Spudich EN, Spudich JL, Beja O. Diversification and spectral tuning in marine proteorhodopsins. EMBO J. 2003;22:1725–1731. doi: 10.1093/emboj/cdg183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y, Zhang F. Molecular tools and approaches for optogenetics. Biol Psychiatry. 2012;71:1033–1038. doi: 10.1016/j.biopsych.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milias-Argeitis A, Summers S, Stewart-Ornstein J, Zuleta I, Pincus D, El-Samad H, Khammash M, Lygeros J. In silico feedback for in vivo regulation of a gene expression circuit. Nat Biotechnol. 29:1114–1116. doi: 10.1038/nbt.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra D, Yang X, Moffat K. Crystal structures of Aureochrome1 LOV suggest new design strategies for optogenetics. Structure. 2012;20:698–706. doi: 10.1016/j.str.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moglich A, Ayers RA, Moffat K. Design and signaling mechanism of light-regulated histidine kinases. J Mol Biol. 2009;385:1433–1444. doi: 10.1016/j.jmb.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moglich A, Moffat K. Structural basis for light-dependent signaling in the dimeric LOV domain of the photosensor YtvA. J Mol Biol. 2007;373:112–126. doi: 10.1016/j.jmb.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moglich A, Yang X, Ayers RA, Moffat K. Structure and function of plant photoreceptors. Annu Rev Plant Biol. 2010;61:21–47. doi: 10.1146/annurev-arplant-042809-112259. [DOI] [PubMed] [Google Scholar]
- Narikawa R, Suzuki F, Yoshihara S, Higashi S, Watanabe M, Ikeuchi M. Novel photosensory two-component system (PixA-NixB-NixC) involved in the regulation of positive and negative phototaxis of cyanobacterium Synechocystis sp PCC 6803. Plant Cell Physiol. 2011;52:2214–2224. doi: 10.1093/pcp/pcr155. [DOI] [PubMed] [Google Scholar]
- Nash AI, Ko WH, Harper SM, Gardner KH. A conserved glutamine plays a central role in LOV domain signal transmission and its duration. Biochemistry. 2008;47:13842–13849. doi: 10.1021/bi801430e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash AI, McNulty R, Shillito ME, Swartz TE, Bogomolni RA, Luecke H, Gardner KH. Structural basis of photosensivity in a bacterial light-oxygen-voltage/helix-turn-helix (LOV-HTH) DNA-binding protein. Proc Natl Acad Sci USA. 2011;108:9449–9454. doi: 10.1073/pnas.1100262108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlendorf R, Vidavski RR, Eldar A, Moffat K, Moglich A. From Dusk till Dawn: One-Plasmid Systems for Light-Regulated Gene Expression. J Mol Biol. 2012;416:534–542. doi: 10.1016/j.jmb.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Pathak GP, Ehrenreich A, Losi A, Streit WR, Gartner W. Novel blue light-sensitive proteins from a metagenomic approach. Env Microbiol. 2009;11:2388–2399. doi: 10.1111/j.1462-2920.2009.01967.x. [DOI] [PubMed] [Google Scholar]
- Pathak GP, Losi A, Gartner W. Metagenome-based screening reveals worldwide distribution of LOV-domain proteins. Photochem Photobiol. 2012;88:107–118. doi: 10.1111/j.1751-1097.2011.01024.x. [DOI] [PubMed] [Google Scholar]
- Polstein LR, Gersbach CA. Light-inducible spatiotemporal control of gene activation by customizable zinc finger transcription factors. J Am Chem Soc. 2012;134:16480–16483. doi: 10.1021/ja3065667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell EB, Crosson S. Photoregulation in prokaryotes. Curr Opin Microbiol. 2008;11:168–178. doi: 10.1016/j.mib.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Purcell EB, Siegal-Gaskins D, Rawling DC, Fiebig A, Crosson S. A photosensory two-component system regulates bacterial cell attachment. Proc Natl Acad Sci U S A. 2007;104:18241–18246. doi: 10.1073/pnas.0705887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffelberg S, Mansurova M, Gartner W, Losi A. Modulation of the photocycle of a LOV domain photoreceptor by the hydrogen-bonding network. J Am Chem Soc. 2011;133:5346–5356. doi: 10.1021/ja1097379. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Gulevich AG, Lagarias JC. Phycoviolobilin formation and spectral tuning in the DXCF cyanobacteriochrome subfamily. Biochemistry. 2012a;51:1449–1463. doi: 10.1021/bi201783j. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Lagarias JC. Mechanistic insight into the photosensory versatility of DXCF cyanobacteriochromes. Biochemistry. 2012b;51:3576–3585. doi: 10.1021/bi300171s. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu MH, Moskvin OV, Siltberg-Liberles J, Gomelsky M. Natural and engineered photoactivated nucleotidyl cyclases for optogenetic applications. J Biol Chem. 2010;285:41501–41508. doi: 10.1074/jbc.M110.177600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry. 2000;39:9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev. 2003;103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- Sancar A. Photolyase and cryptochrome blue-light photoreceptors. Adv Protein Chem. 2004;69:73–100. doi: 10.1016/S0065-3233(04)69003-6. [DOI] [PubMed] [Google Scholar]
- Sancar A. Structure and function of photolyase and in vivo enzymology: 50th anniversary. J Biol Chem. 2008;283:32153–32157. doi: 10.1074/jbc.R800052200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. A light-switchable gene promoter system. Nat Biotechnol. 2002;20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Controlling signal transduction with synthetic ligands. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- Strickland D, Lin Y, Wagner E, Hope CM, Zayner J, Antoniou C, Sosnick TR, Weiss EL, Glotzer M. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat Methods. 2012;9:379–384. doi: 10.1038/nmeth.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland D, Moffat K, Sosnick TR. Light-activated DNA binding in a designed allosteric protein. Proc Natl Acad Sci U S A. 2008;105:10709–10714. doi: 10.1073/pnas.0709610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland D, Yao X, Gawlak G, Rosen MK, Gardner KH, Sosnick TR. Rationally improving LOV domain-based photoswitches. Nat Methods. 2010;7:623–626. doi: 10.1038/nmeth.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz TE, Tseng TS, Frederickson MA, Paris G, Comerci DJ, Rajashekara G, Kim JG, Mudgett MB, Splitter GA, Ugalde RA, et al. Blue-light-activated histidine kinases: two-component sensors in bacteria. Science. 2007;317:1090–1093. doi: 10.1126/science.1144306. [DOI] [PubMed] [Google Scholar]
- Tabor JJ, Levskaya A, Voigt CA. Multichromatic control of gene expression in Escherichia coli. J Mol Biol. 2011;405:315–324. doi: 10.1016/j.jmb.2010.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Ebrey TG. Molecular basis of spectral tuning in the newt short wavelength sensitive visual pigment. Biochemistry. 2003;42:6025–6034. doi: 10.1021/bi020629+. [DOI] [PubMed] [Google Scholar]
- Toettcher JE, Gong D, Lim WA, Weiner OD. Light-based feedback for controlling intracellular signaling dynamics. Nat Methods. 2011;8:837–839. doi: 10.1038/nmeth.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyszkiewicz AB, Muir TW. Activation of protein splicing with light in yeast. Nat Methods. 2008;5:303–305. doi: 10.1038/nmeth.1189. [DOI] [PubMed] [Google Scholar]
- Vaidya AT, Chen CH, Dunlap JC, Loros JJ, Crane BR. Structure of a light-activated LOV protein dimer that regulates transcription. Sci Signal. 2011;4:ra50. doi: 10.1126/scisignal.2001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst MA, Hellingwerf KJ. Photoreceptor proteins, “star actors of modern times”: a review of the functional dynamics in the structure of representative members of six different photoreceptor families. Acc Chem Res. 2004;37:13–20. doi: 10.1021/ar020219d. [DOI] [PubMed] [Google Scholar]
- Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat Methods. 2012;9:266–269. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- Wu D, Hu Q, Yan Z, Chen W, Yan C, Huang X, Zhang J, Yang P, Deng H, Wang J, et al. Structural basis of ultraviolet-B perception by UVR8. Nature. 2012;484:214–219. doi: 10.1038/nature10931. [DOI] [PubMed] [Google Scholar]
- Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HQ, Wu YJ, Tang RH, Liu D, Liu Y, Cashmore AR. The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell. 2000;103:815–827. doi: 10.1016/s0092-8674(00)00184-7. [DOI] [PubMed] [Google Scholar]
- Yao X, Rosen MK, Gardner KH. Estimation of the available free energy in a LOV2-J alpha photoswitch. Nat Chem Biol. 2008;4:491–497. doi: 10.1038/nchembio.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE. Induction of protein-protein interactions in live cells using light. Nat Biotechnol. 2009;27:941–945. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Zhang F, Vierock J, Yizhar O, Fenno LE, Tsunoda S, Kianianmomeni A, Prigge M, Berndt A, Cushman J, Polle J, et al. The microbial opsin family of optogenetic tools. Cell. 2011;147:1446–1457. doi: 10.1016/j.cell.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang LP, Brauner M, Liewald JF, Kay K, Watzke N, Wood PG, Bamberg E, Nagel G, Gottschalk A, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- Zoltowski BD, Schwerdtfeger C, Widom J, Loros JJ, Bilwes AM, Dunlap JC, Crane BR. Conformational switching in the fungal light sensor Vivid. Science. 2007;316:1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoltowski BD, Vaccaro B, Crane BR. Mechanism-based tuning of a LOV domain photoreceptor. Nat Chem Biol. 2009;5:827–834. doi: 10.1038/nchembio.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoltowski BD, Gardner KH. Tripping the light fantastic: blue-light photoreceptors as examples of environmentally modulated protein-protein interactions. Biochemistry. 2011;50:4–16. doi: 10.1021/bi101665s. [DOI] [PMC free article] [PubMed] [Google Scholar]