Abstract
Human monocyte differentiation antigen CD14 is a pattern recognition receptor that enhances innate immune responses to infection by sensitizing host cells to bacterial lipopolysaccharide (LPS; endotoxin), lipoproteins, lipoteichoic acid and other acylated microbial products. CD14 physically delivers these lipidated microbial products to various Toll-like receptor signaling complexes that subsequently induce intracellular proinflammatory signaling cascades upon ligand binding. The ensuing cellular responses are usually protective to the host, but can also result in host fatality through sepsis. In this work, we have determined the X-ray crystal structure of human CD14. The structure reveals a bent solenoid typical of leucine rich repeat proteins with an amino terminal pocket that presumably binds acylated ligands including LPS. Comparison of human and mouse CD14 structures show great similarity in overall protein fold. However, compared to mouse CD14, human CD14 contains an expanded pocket and alternative rim residues that are likely to be important for LPS binding and cell activation. The X-ray crystal structure of human CD14 presented herein may foster additional ligand bound structural studies, virtual docking studies, and drug design efforts to mitigate LPS induced sepsis and other inflammatory diseases.
Keywords: CD14, Toll-like receptor 4 (TLR4), MD-2, lipopolysaccharide (LPS), sepsis
1. Introduction
CD14 was first characterized as a membrane cell surface differentiation marker of myeloid lineage cells (1). Membrane CD14 is a GPI anchored 55 kDa glycoprotein (2, 3) that is highly expressed on monocytes, macrophages and neutrophils with lower surface expression observed on a variety of other hematopoietic and stromal cells. Soluble forms of CD14 exist in serum, cerebrospinal, and other body fluids. Soluble CD14 is generated by at least three different mechanisms which include bypassing of GPI addition, cleavage of the GPI anchor by phospholipase D, or direct proteolytic cleavage from the cell surface (reviewed in (4)).
CD14 is best known as a pattern recognition receptor of the innate immune system that plays a prominent role in sensitizing cells to the presence of Gram-negative bacterial lipopolysaccharide (LPS; endotoxin) (4). CD14 is thought to sensitize cells to LPS by delivering this agonist to the Toll-like receptor 4 (TLR4) receptor signaling complex. Membrane CD14 shuttles LPS to TLR4 complexes in a two dimensional search (4); whereas, soluble CD14 enables cells that lack endogenous CD14, including most epithelial and endothelial cells, to respond to LPS (reviewed in (5)). LPS-induced TLR4 activation initiates host inflammation by driving cellular production of pro-inflammatory cytokines, chemokines and cell adhesion molecules. At the local site of infection, pro-inflammatory signaling resulting from CD14 shuttling is protective, leading to local clearance of invading bacteria. However, widespread infection and activation of this pro-inflammatory signaling system can cause fatality through sepsis (reviewed in (6, 7)). In response to LPS, CD14 deficient mice have blunted pro-inflammatory cytokine production and lack the lethargy, respiratory, and ruffled fur symptoms associated with murine septic shock. CD14 deficient mice also survive challenge with otherwise lethal doses of LPS (8).
Lipopolysaccharide is the major lipid present in the outer membrane of Gram-negative bacteria (9). LPS is an amphipathic molecule and in enteric bacteria is comprised of a hexaacylated lipid A region with a di-glucosamine backbone flanked by 1 and 4′ phosphates. The length, number, and saturation of the fatty acid tails in LPS can vary between different species of bacteria (10) as can the length and sugar content of the core and O-antigen regions (11). The binding of LPS by CD14 is slow unless this binding reaction is catalyzed by LPS binding protein (LBP) (12, 13). LBP is a 60 kDa human serum glycoprotein produced by liver hepatocytes (14, 15) that disaggregates and catalytically transfers LPS to CD14 (16). In concert with LBP, CD14 binds monomeric LPS (21) and shuttles it to the TLR4 complex. This effectively concentrates low levels of LPS and increases the sensitivity of the system (4, 12, 13, 17, 18). The major LPS acceptor of the TLR4 receptor complex is a 25kDa co-receptor called MD-2, which is physically associated with TLR4. The binding of LPS to the TLR4-MD2 complex facilitates dimerization resulting in a homodimeric receptor signaling complex composed of two TLR4 monomers, two MD-2 proteins and two LPS molecules (19). Cell signaling is facilitated by the intracellular signaling domain of TLR4 which upon dimerization creates a platform for intracellular signaling adaptors (20). Small gel filtration mixing studies have demonstrated the catalytic role of CD14 in delivering LPS to the TLR4-MD2 complex (21-24).
As a pattern recognition receptor, CD14 is capable of binding a wide variety of natural and synthetic acylated ligands in addition to LPS. For example, CD14 and LBP can work together to opsonize whole bacteria and apoptotic cells, clearing infection and reducing inflammation (25-29). CD14 is also able to bind and shuttle certain host phospholipids (30-32). Further, CD14 has been shown to enhance cellular inflammatory responses to a variety of acylated bacterial agonists of TLR2 including lipoteichoic acid, peptidoglycan, mycobacterial lipoarabinomannan, atypical lipopolysaccharides, and lipoproteins (33-40). Given this central role in TLR agonist delivery, CD14 is an obvious drug target for the treatment of sepsis (reviewed in (6, 7)).
The molecular interactions involved in the binding and delivery of structurally diverse ligands presumably requires multiple protein-ligand and protein-protein interaction sites on CD14 that are currently undefined. Additionally, the mechanisms involved in delivery of these ligands to multiple Toll-like receptor complexes are also unresolved. To address this gap in knowledge, many groups have purified various recombinant forms of soluble CD14 using bacteria, yeast, insect, and human cellular expression systems (12, 41-51), often with the goal of structure determination (49, 50). Currently, only the unliganded crystal structure of mouse CD14, purified from SF9 insect cells, is known (52). Mouse CD14 possesses an N-terminal hydrophobic cavity that provides a putative binding site for LPS and other acylated ligands. Despite previous efforts, no structure of human CD14 has been determined and the structural similarity between mouse and human CD14 is unresolved. As a first step towards this goal, we report the crystal structure of human CD14 (4GLP.pdb). Since septic shock is the most common cause of death in intensive care units (53), information garnered from the structure of human CD14 may foster ligand binding structural studies or drug development efforts.
2. Materials and methods
2.1. Expression
Human CD14 (aa 1-336) was amplified from genomic DNA by PCR using a sense primer, 5′-TTGGAATTCGCCGCCACCATGGAGCGCGCGTCCTGCTTGTTGC- 3′containing an EcoRI site, Kozak sequence, and a portion of the N-terminal secretion signal of human CD14, and an antisense primer, 5′- TTGTCTAGAACTACCGCGGGGGACGAGGGCAGTTCCAG GGACCAGGAAGG- 3′ containing a XbaI cleavage site followed by a thrombin digestion site. The PCR products were digested and cloned into a modified pDisplay vector via EcoRI and NheI sites (a kind gift from Dr. David Kranz, University of Illinois, Urbana-Champaign). This vector contains the coding sequence of the Fc domain of human immunoglobulin G1 downstream of a NheI restriction enabling generation of an Fc fusion protein. The antisense PCR primer was designed to include a thrombin cleavage site (LVPRGS) allowing for removal of the Fc fusion during purification. Site directed mutagenesis was completed through primer extension to yield the C306S mutation. The final construct sequence was confirmed by automated sequencing (UIUC Sequencing Center). After DNA amplification in E. coli DH5α cells, HEK 293F cells (Invitrogen) were transfected, cultured, and stably selected as previously described (54).
2.2. Purification
Human soluble CD14 (aa 1-336, C306S) was purified in four chromatographic steps. First, protein G affinity purification was used to harvest the human CD14 Fc fusion (CD14-Fc) from HEK 293F culture supernatant as previously described with the following exceptions (54). Two liters of HEK 293F supernatant from stably transfected CD14-Fc expressing cells was harvested seven days following seeding to 0.3 × 106 cells mL−1 in serum-free Freestyle 293F media (Invitrogen Life Technologies) under G418 (0.25 mg mL−1) selection. Recombinant protein G sepharose beads (GE Healthcare; 2 mL 50% slurry) were added to the filtered supernatant with stirring overnight at 4°C. Protein G beads bound to CD14-Fc were harvested by centrifugation at 2,500 × g, 15 min, 4°C and packed into a disposable PD-10 column (GE Healthcare). The column was washed with 0.02 M sodium phosphate, pH 7.0 and eluted in ten column volumes of 0.1 M glycine-Cl, pH 2.3 with 1 mL neutralizing 1 M Tris-HCl, pH 9 buffer. Thrombin (Novagen) was used to remove the Fc tag by overnight incubation at 22°C.
Next, the products of the thrombin cleavage reaction were separated by passing the sample through an ÄKTAprime™ plus FPLC fitted with two tandem 1mL Hi-Trap Protein A high performance columns (GE Healthcare). The columns were run at a 1ml min−1 flow rate in 0.02 M Tris HCl, pH 8.5. The flow through fractions containing free human CD14 were collected and injected on the two tandem Protein A columns three consecutive times to remove Fc. Flow through fractions containing soluble CD14 were pooled and concentrated to 1mL using an Amicon Ultra-15 unit (Millipore).
Since glycosylation contributes to protein heterogeneity, we removed N-linked glycans from CD14 by a one hour incubation at 37°C in 1xG7 buffer (0.05 M sodium phosphate, pH 7.5) containing 1,000 U peptide N-glycosidase F (PNGaseF) (New England Biolabs). This deglycosylated CD14 was further purified by anion exchange chromatography using two tandem 5 mL Hi-Trap Q anion exchange columns (GE Healthcare) and a linear NaCl gradient (0.02 M - 1 M) in 0.02 M Tris-HCl, pH 8.5 at 4 °C using a 1 mL min−1 flow rate. CD14 containing fractions were concentrated using an Amicon Ultra-15 unit (Millipore) to 0.5 mL. Finally, size-exclusion chromatography was performed using a Superdex 200 column (GE Healthcare) equilibrated with 0.02 M Tris-HCl, pH 8.5 and 0.1 M NaCl at a flow rate of 0.4 mL min−1. The fractions containing CD14 were pooled and concentrated to 10 mg ml−1 using an Amicon Ultra-4 unit (Millipore) as measured by Pierce BCA assay (Rockford, IL). Buffer conditions were determined using a solubility screen (55). This four column purification process yielded ~2 mg L−1 soluble, human CD14 with >90% purity by SDS-PAGE gel (Sup. Figs. 1A and 1B).
2.3. Crystallization and Optimization
Human CD14 (aa 1-336; C306S) was crystallized using the hanging-drop vapor-diffusion method. Crystallization conditions were initially screened using commercially available sparse matrix kit conditions including Hampton Research Crystal Screens I and II, Emerald Biosystems Wizard I and II, Hampton Research Natrix, and Hampton Research Salt Reaction sparse matrix screening kits, and a handmade crystal screen targeting the previously published mouse CD14 crystallization condition (1 μl protein solution and 1 μl of crystallization buffer containing 100 mM sodium HEPES (pH 7.5), 1.9 M Li2SO4, and 5 mM NiCl2) (52). Each condition was tested on a 12 mm x 0.22 mm siliconized glass cover slide (Hampton Research) over a VDX48 plate with sealant (Hampton Research) by mixing 1 μl of each screening solution with 1 μl protein solution (10 mg ml−1 protein in 0.02 M Tris-HCl, pH 8.5 and 0.1 M NaCl) equilibrated against 300 μl screening solution in the reservoir. Sparse matrix screening in hanging drop vaporization trays at 22°C, 18°C, and 4°C lead to three conditions with single crystals: Emerald Biosystems Wizard I #28 (20% PEG 3,000, 0.1 M HEPES, pH 7.5, and 0.2 M NaCl) after 2 - 4 days at 22°C; Hampton Research Crystal Screen I #18 (20% PEG 8,000, 0.1 M Na cacodylate*3H20, pH 6.5, and 0.2 M Mg(OAc)*4H20) after 6 days at 18°C; and Emerald Biosystems Wizard II #28 (20% PEG 8,000, 0.1 M MES, pH 6.0, and 0.2 M Ca(OAc)2) after 3 weeks at 22 °C (56-58). The best initial diffraction resolution (6.2 Å) was obtained using Emerald Biosystems Wizard II #28.
Extensive work was conducted to improve diffraction resolution. During crystal optimization, various additives, detergents, proteases, and cryoprotectants were screened, utilizing multiple crystal harvesting time points (59-61). Further post-crystallization optimization work included in situ proteolysis, dehydration via serial transfer on cover slips or over the reservoir for different durations, macromolecular crystal annealing, flash annealing, and reductive methylation of surface lysine residues using dimethylamine-borane complex (62, 63). Treatment with each of these additional techniques failed to improve the diffraction resolution (data not shown).
Single crystals of purified deglycosylated CD14 were grown at 22°C to a maximum dimension over ~40 days on a 22 mm x 0.22 mm siliconized glass cover slide (Hampton Research) over a VDX24 plate with sealant (Hampton Research) by mixing 1.5 μl 30% PEG 6,000, 0.1 M MES, pH 6.0, and 0.2 M Ca(OAc)2 crystallization solution with 1.5 μl protein solution (10 mg ml−1 protein in 0.02 M Tris-HCl, pH 8.5 and 0.1 M NaCl) equilibrated against 1 mL screening solution in the reservoir. Crystals were harvested from the drop and plunged directly into liquid N2 without additional cryoprotection.
2.4. Data collection and structure determination
A native diffraction data set was collected at 4.0 Å resolution from a single vitrified crystal in the optimized 20% PEG 6,000, 0.1 M MES, pH 6.0, and 0.2 M Ca(OAc)2 crystallization solution on the LS-CAT beamline, Sector 21 ID-F (Advanced Photon Source, Argonne, IL). Diffraction data were indexed and integrated using XDS followed by scaling and merging with XSCALE. The structure solution was determined via molecular replacement with Phaser in the CCP4 suite, using the molecular coordinates of mouse CD14 (PDB ID 1WWL), modified with CHAINSAW. Iterative cycles of model fitting were completed in COOT, followed by refinement in REFMAC. The final structure solution of human CD14 was deposited (PDB ID 4GLP; http://www.pdb.org/pdb/search/structidSearch.do?structureId=4GLP) and verified with Molprobity in PHENIX. Data collection and refinement statistics are summarized in Table 1.
Table I.
Data collection and refinement statistics
| Human CD14 | |
|---|---|
| Data collection | |
| Space group | P 32 2 1 |
| No. of mol. in asym. unit | 1 |
| Cell dimensions | |
| a, b, c (Å) | a = 147.52, b = 147.52, c = 44.07 |
| β (°) | β = 90 |
| Resolution (Å) | 4.0 |
| Rsyma,b (%) | 8.6 (84.2) |
| I/σa | 17.20 (3.19) |
| Completenessa (%) | 99.6 (100.0) |
| Refinement | |
| Resolution range (Å)a | 27.9-4.0 (4.1-4.0) |
| Total reflectionsa | 53002 (3887) |
| Unique reflectionsa | 4780 (341) |
| R factor (%)c | 28.5 |
| Rfree (%)c | 32.1 |
| No. atoms in protein | 2346 |
| Average B factor | 10.9 |
| Wilson B factor | 39.2 |
| r.m.s. deviation | |
| Bonds (Å) | 0.010 |
| Angles (°) | 1.640 |
Numbers in parentheses correspond to the highest resolution shell. r.m.s.d., Root-mean-square deviations from ideal geometry.
Rmerge = ∑h∑I ∣Iih-<Ih>∣/∑h∑I <Ih>, where <Ih> is the mean intensity of the observations Iih of reflection h.
R factor = ∑ (∥Fobs∣-Fcalc∥)/∑∣Fobs∣; Rfree is the R factor for a subset (5%) of reflections that was selected prior to refinement calculations and not included in the refinement.
2.5. LPS Binding Assay
The binding of LPS by PNGaseF deglycoslated human CD14 (34 kDa) was detected by a native PAGE gel shift method (64). 10 μg human CD14 was mixed with a 2 molar ratio of the Ra chemotype of E. coli LPS (3.79 kDa; Sigma-Aldrich), with or without a 1:100 molar ratio of CD14:human LBP (60 kDa; kindly provided by Dr. Jerrold Weiss, University of Iowa). The reactions were incubated for 1 h at 37°C in a total volume of 5 μL in PBS buffer containing 0.01M EDTA. 5 μL bromophenol blue with glycine dye was added and each sample was loaded in a 4-20% Mini-PROTEAN TGX precast gel, pH 6.8 (Bio-rad) run at 100V in the recommended running buffer without SDS at 4°C for 3 hours (Sup. Fig. 2).
2.6. Bioactivity Assay
Various concentrations of the Ra chemotype of E. coli LPS (Sigma) were pre-incubated for 1 h at 37 °C at a 2:1 molar ratio LPS:CD14 in RPMI media containing 1% human serum albumin. For some reactions, LBP was added as a catalyst at a 1:100 molar ratio of LBP:CD14. The bioactivity of CD14 in these reactions was assessed using SW620 human epithelial cells which express TLR4/MD-2 and lack detectable levels of membrane CD14 (42). SW620 cells were cultured in RPMI complete media containing 1% bovine serum albumin and seeded in a 96-well plate at a density of 1×106 cells/well and allowed to adhere for 1 h at 37°C, 5% CO2. To remove serum, SW620 cells were carefully washed 4 times with PBS, 2 times with Gibco FreeStyle 293 Expression Media (Invitrogen), and 2 times with RPMI containing 1% human serum albumin. Preincubated LPS samples were added to the washed SW620 epithelial cells. Supernatant was harvested 6 hours later and IL-8 production was measured using a human IL-8 Cytoset sandwich ELISA kit (Invitrogen).
3. Results
3.1 Generation of Bioactive Human CD14 Suitable for Crystallization
A number of important parameters were considered in generating a soluble form of human CD14 suitable for crystallization. Since soluble forms of CD14 are naturally heterogeneous at the C-terminus, we generated an expression construct which lacks the C-terminal protein sequence required for GPI anchoring. This was done with the knowledge that C-terminal truncation of CD14 does not affect LPS associated bioactivity, which resides entirely within the amino-terminal half of the protein (65, 66). Additionally, since our CD14 construct is truncated between C306 and C352, which are predicted to form the final disulfide bond, we mutated the unpaired C306 residue to serine. CD14 was expressed as an Fc tagged fusion protein and purified over protein A. An engineered thrombin cleavage site enabled removal of the Fc tag, and further purification steps resulted in highly pure soluble CD14 protein suitable for crystallization (Sup. Fig. 1A and 1B). To further ensure improved protein homogeneity for crystallization, we removed potentially heterogeneous N-linked glycans using PNGase F. This enzymatic deglycosylation was preferred because mutation of the N-linked glycosylation sites in CD14 prevents cellular secretion (52). As observed by others, PNGase F treatment reduced the molecular weight of CD14 by approximately 10kD ((67, 68) (Sup Fig 1C).
To ensure that our soluble human CD14 is biologically active, we measured the ability of the purified protein to bind LPS and to mediate MD-2/TLR-4 dependent chemokine production. Binding was assessed by mixing a 2:1 molar excess of the Ra chemotype of E. coli LPS with CD14 in the absence and presence of a catalytic amount of LBP. CD14 formed a complex with LPS as evidenced by altered mobility on a gradient native PAGE gel relative to CD14 alone (Sup. Fig. 2). We also assessed the ability of our purified soluble CD14 to induce LPS-mediated activation of human epithelial SW620 cells. These cells naturally express the TLR4/MD-2 signaling complex, but lack endogenous CD14 such that LPS induced activation is greatly enhanced by addition of active forms of soluble CD14 (41, 42). To this end, purified soluble CD14 was pre-incubated with increasing concentrations of LPS in the presence of LBP to facilitate CD14-LPS complex formation. Preincubated LPS, LBP and CD14 potently stimulated IL-8 production of SW620 cells and enabled detection of 100 pg/mL LPS, with maximal cell stimulation observed at a 10 ng/mL LPS (Sup. Fig. 3). These LPS-induced IL-8 production levels were comparable to that mediated by human serum which naturally contains LBP and CD14 catalytic activity. In contrast, either LPS alone or LPS preincubated with LBP poorly activated the cells. Taken together, these results show that the soluble human CD14 (aa 1-336, C306S) used herein binds LPS and is biologically active.
3.2 Crystal Structure of Human CD14
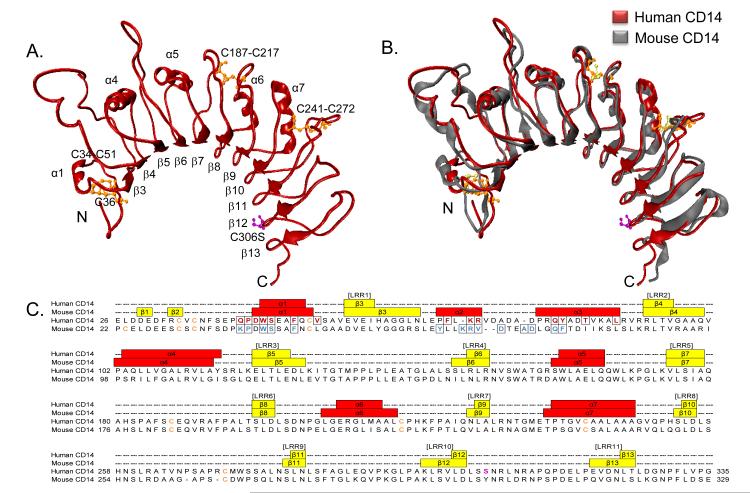
Human CD14 crystallized in the trigonal space group P3221 (Table I). The crystal structure begins with aa 26, indicating complete removal of the leader sequence by HEK 293F cells during secretion along with low electron density for the first six amino acids, aa 20-25. The overall X-ray crystal structure of human CD14 reveals a bent solenoid that is formed by 10 leucine rich repeats, each creating a single turn (Fig. 1A). This is consistent with the leucine rich repeat sequence motifs of human CD14 (69), and the expected bent solenoid fold generally characteristic of leucine rich repeat containing proteins (70). The structure also reveals five alpha helices on the convex side, as well as eleven beta strands that are coordinated in a parallel beta sheet on the concave side of the bent solenoid. The pairwise proximity between cysteine residues in the crystal structure at positions 34 and 51, 187 and 217, as well as 241 and 272 are consistent with the expected disulfide bonds of human CD14, which have been shown to be important for proper protein folding (49). Interestingly, recombinant purified human CD14 lacking the C306S mutation, a mutation originally designed to avoid unnatural disulfide binding, did not crystallize and formed aggregates in the crystallization drop (data not shown).
Fig. 1. Human CD14 X-ray crystal structure.
A, The X-ray crystal structure of human CD14 aa 26-335 is shown in ribbon (red). Alpha helices (α1, α4, α5, α6, α7), beta strands (β3-β13), and the positions of cysteine residues (orange) are indicated. The site directed mutagenesis of C306S is shown in magenta. B, Secondary structure alignment of human CD14 (red) and mouse CD14 (gray) was created using the default parameters of MultiSeq in VMD. Disulfide bonds in human CD14 (orange) and mouse CD14 (yellow) are indicated. The site directed mutagenesis of C306S is shown in human CD14 (magenta). C, A structure based sequence alignment of human and mouse CD14 was created using the default parameters of MultiSeq in VMD. MultiSeq output was used to assign human CD14 secondary structure elements upon visualization in VMD, and mouse CD14 secondary structure positions were derived from the header notation of 1WWL.pdb. Numbered leucine rich repeat regions, which are conserved in location between both species, are shown in brackets ([LRR]). Alpha helices (red rectangles), beta strands (yellow rectangles), turns and loops (black dashed lines) are indicated. Cysteine residues (orange) and residues located in the rim of the N-terminal pocket are boxed for human CD14 (maroon) and mouse CD14 (blue). Since amino acid numbering is not standardized in previous publications, we have chosen to compare mouse and human CD14 crystal structures by starting amino acid numbering at the first methionine residue.
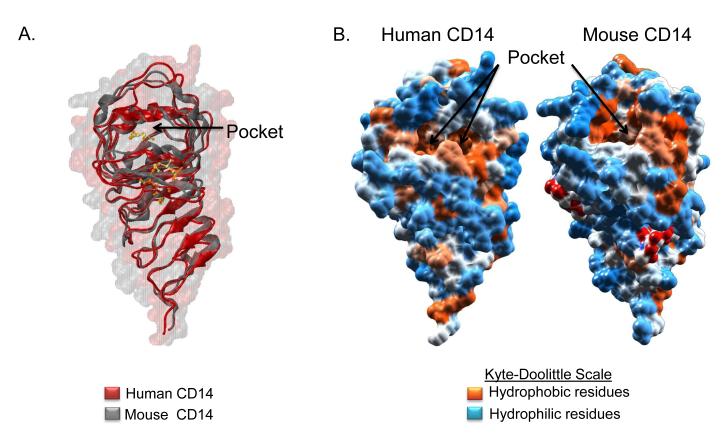
Another key structural feature present in the crystal structure of human CD14 is an N-terminal pocket or cavity (Fig. 2A). Although we cannot know with certainty the position of each side chain at this resolution, the overall cavity of human CD14 is comprised largely of hydrophobic residues present in α1 through α5 and β1 through β6 along with their connecting loops. It is reasonable to assume that this hydrophobic pocket serves as the binding site for the lipid chains of LPS and other acylated CD14 ligands (Fig. 2B), especially as the regions making up the pocket are entirely contained within the N-terminal region (aa 20-171) of CD14 that has been shown to be sufficient for bioactivity (65, 66).
Fig. 2. Comparison of mouse and human CD14 N-terminal ligand binding pocket.
A, Mouse (gray) and human (red) CD14 ribbon structures are overlaid over mouse (translucent gray) and human (translucent red) CD14 space filling structures drawn in VMD. CD14 structures are rotated 90° relative to Fig. 1B. The N-terminal hydrophobic binding pocket is indicated (black arrow). B, Space filling hydrophobicity structures were drawn in Chimera for human CD14 (left) and mouse CD14 (right). The Kyte-Doolittle scale was used to compare hydrophobicity by showing hydrophobic (orange), neutral (white), and hydrophilic (blue) residues as well as glycosylation sites (red).
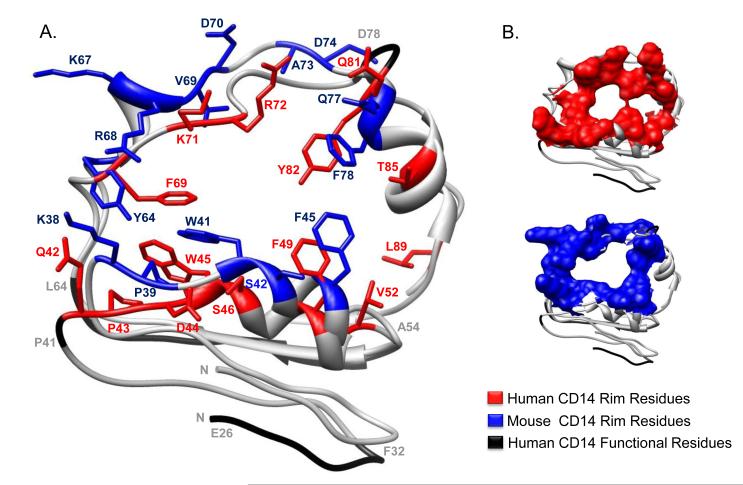
It is instructive to examine the rim residues of the CD14 hydrophobic pocket as these residues likely engage portions of LPS, or other acylated CD14 ligands, that are not accommodated by the pocket itself. Positively charged residues are located in the rim at K71 and R72 and just outside the rim at R80, K87, and R92 (Fig. 3A). Hydrophobic residues at W45, F49, V52, F69, Y82, and L89 encircle the rim and their side chains overlay the entrance to the pocket. When measured using the program Chimera, the pocket diameter of human CD14 from W45 to L89 is 15.7 Å wide. Interestingly, the hydrophobic pocket entrance is subdivided by an interaction between the α1 residue F49 and the α3 residue Y82 (Fig. 3B).
Fig. 3. Comparison of mouse and human CD14 pocket rim residues.
A, The structure of the rim of the predicted N-terminal ligand binding pocket of both mouse and human CD14 are overlaid. Rim residues of human CD14 are denoted in red and those of mouse CD14 are denoted in blue. Regions important for LPS binding, as determined from previous mutagenesis, blocking antibody, and epitope mapping studies are indicated in black with gray text. B, A space filling representation of the residues located on the rim of the predicted N-terminal ligand binding pocket are human and mouse CD14 are displayed in red and blue, respectively. Regions important for LPS binding are displayed in black ribbon.
3.3 Comparison of Human and Mouse CD14 Structure
Human soluble CD14 purifies and crystallizes as a monomer (Sup. Figs. 1A and 1B). In contrast, the murine protein crystalized as a dimer with the dimer interface comprised of C-terminal beta strand interactions (52). The mouse and human CD14 structures are comparable at the C-terminus because both were truncated at similar positions to make them more amenable to crystallization (52). Different crystallization conditions, constructs, and cellular expression systems may account for the differences observed in crystal packing between the two proteins. Nevertheless, it is unlikely that the C-terminal dimerization of truncated mouse CD14 is biologically relevant (52), as numerous biophysical studies support the idea that bioactive soluble CD14 is a monomeric protein (4, 12, 13, 21-24).
Human and mouse CD14 crystal structures contain a bent solenoid fold and asparagine ladder, which are structural features typical of leucine rich repeat proteins (70). In fact, both proteins are highly superimposable with a r.m.s.d. of 1.089 Å (Fig. 1B). The secondary structure of human CD14 includes alpha helices and beta strands that are equivalent to α1, α4, α5, α6, α7, and β3-β13 of the published mouse CD14 structure (Figs. 1B and 1C) (52). Although helical coils exist in human CD14 at locations that are synonymous to helices α2 and α3 of mouse CD14, the structure resolution falls just outside the range needed to determine helical packing with certainty (Figs. 1A and 1C; (71, 72). Human and mouse CD14 both contain a large N-terminal hydrophobic pocket consistent with their similar roles in the binding and delivery of various lipidated molecules including LPS (Figs. 2A and 2B). Similar to human CD14, the main pocket and sub-pockets present in mouse CD14 are comprised of hydrophobic residues within α1-5, β1-6, and their connecting loops (52). Using a structure based sequence alignment, created using MultiSeq in VMD, we compared the general amino acid characteristics present in the residues at the rim of the proposed binding pocket in both structures (73). The authors of the mouse CD14 crystal structure identified hydrophilic and hydrophobic amino acids in rim residue positions indicated in blue ((52); Fig. 3A). Many of these residues, including P39, W41, F45, and Q77 are conserved with rim residues at the pocket entrance of human CD14. Thus, the N-terminal hydrophobic pocket found in our crystal structure revealed a conservation of pocket size and hydrophobicity with that of mouse CD14.
Despite the wide ranging structural similarities between mouse and human CD14, differences do exist. For example, although the amino acids found at equivalent positions of mouse and human CD14 generally have similar physical properties, there is variation in the charge of certain rim residues. Namely, the positively charged K38 residue present in the mouse protein is not retained in the human protein. Likewise, the negatively charged D44 residue in human CD14 is not found in mouse CD14. Finally, the diameter and subdivision of the entrance to the pocket differs between species. The mouse crystal structure reveals an 8.0 Å wide entrance to the pocket that is bounded on one side by a hydrophobic interaction between F45 and F78 ((52); right side of Fig. 3A). Residues F45, L49, and I81 in the mouse CD14 structure create additional hydrophobic interactions which shield this side of the pocket. These hydrophobic interactions of the mouse CD14 pocket are not present in the human CD14 structure whose pocket extends to include the rim residues T85 and L89 (Fig. 3A). The biological significance of these differences between the pocket and associated rim of mouse and human CD14 is difficult to define without structural information from ligand bound proteins.
4. Discussion
CD14 sensitizes cells to LPS by delivering this bioactive lipid to MD-2; an essential component of the TLR4 signaling complex. All members of the MD-2 related lipid-recognition protein superfamily are characterized by two β sheets organized in an αβ cup fold to create a centralized hydrophobic LPS binding cavity (74). In the crystal structures of human and mouse MD-2 bound to lipid IVa, all four acyl chains of the ligand are buried inside and occupy the majority of the hydrophobic pocket volume (75, 76). Crystal structures of MD-2 in the context of the entire mouse and human TLR4/MD-2/LPS homodimeric signaling complex reveals a narrow and deep (1720 Å3 volume) hydrophobic binding pocket that completely sequesters five of the six fatty acid chains (77, 78). The remaining unbound acyl chain lies along the surface of MD-2 and, together with the F126 loop of MD-2, creates a new hydrophobic patch that promotes homodimer formation by association with TLR4 from an adjacent TLR4/MD-2/LPS complex (19, 77).
Compared to MD-2, CD14 has been reported to afford less protection against enzymatic removal of secondary fatty acids of bound LPS (79). This probably reflects that fact that at about 820 Å3, the hydrophobic pockets of both human and mouse CD14 are considerably smaller than that of MD-2 (52) (Figs. 2A and 2B). Similar to MD-2, previous circular dichroism, tryptophan fluorescence, and NMR studies suggest that LPS binding does not induce large structural changes in human CD14 (50, 80). Given these volume constraints and pocket rigidity, it is unlikely that CD14 can accommodate all the acyl chains of LPS. This may contribute to the reduced affinity of CD14 for LPS compared to MD-2 (22, 23).
Given the physical limitations of the hydrophobic pocket, we hypothesize that CD14 stabilizes additional fatty acid chains on a hydrophobic cluster located near α1 just outside of the pocket entrance. A similar hydrophobic cluster is present in a comparable location in the crystal structure of mouse CD14 ((52); Fig. 2B). In human CD14, the hydrophobic cluster is comprised of the amino acids F32, F49, V52, S53, A54, V55, and L89 (Fig. 2B). This cluster and associated acyl chains of LPS could perhaps create a new protein-protein interaction surface that may help facilitate the transfer of LPS to another molecule such as MD-2.
Our crystal structure confirms that the hydrophobic pocket encompasses the N-terminal half (aa 20-171) of CD14 that has been shown to be the bioactive portion of the protein (28). Extensive deletion mutagenesis, alanine scanning mutagenesis and epitope mapping of inhibitory anti-CD14 antibodies have revealed four separate regions in the N-terminal half of CD14 that are critical for LPS binding and cell activation (Fig. 3B). These four functionally important regions are delineated by amino acids 26-32, 41-44, 56-64 and 78-83 which are physically located near β1, within the loop preceding α1, across β3 and within α3, respectively (28, 81-83),(84). Additionally, LPS binding has been shown to protect aa 57-64 from endoprotease AspN digestion (80). The residues in the first three regions near α1, β1 and β3 appear to fulfill critical roles in forming the pocket entrance, capping the leucine rich repeat beta sheet, and/or stabilizing proper folding of the solenoid.
Residues in the fourth region encompass a Y82 loop located on the opposite side of the pocket entrance. Although Y82 appears to weakly block the entrance of the pocket through interaction with F49, we believe that upon binding LPS, these hydrophobic residues may change position to accommodate the acyl chains within the pocket. Although the corresponding interaction is represented by F45 and F78 in mouse CD14, additional hydrophobic residues present in mouse CD14, but absent in human CD14, close off the pocket entrance (Figs. 3A and 3B). Overall, our human CD14 crystal structure allows for the resolution of an N-terminal hydrophobic pocket predicted by multiple studies to be the LPS binding site. Compared to mouse CD14, our work provides an alternative definition of rim residues, reveals an expanded pocket entrance, and identifies a unique hydrophobic bridge feature that incorporates residues shown to be important in LPS binding and cell activation (Figs. 3A and 3B).
Hydrophilic residues at the opening of MD-2 have been shown to be important for LPS binding by properly aligning the negatively charged 1 and 4′ phosphates of LPS (42, 77). Similar to MD-2, charged rim residues in CD14 may be important for orienting or binding LPS. For example, previous work has shown that mutation of E47, which falls near the front of the rim in α1, to either lysine or arginine can block binding of P. gingivalis LPS (85). Solution NMR spectroscopy analyzing soluble CD14 aa 20-171 bound to the Re chemotype of LPS did not have structural assignments available upon publication, but does show high average temperature factor evidence for hydrophilic residues at the rim suggesting local flexibility to accommodate LPS binding (50). Further work to develop a ligand bound CD14 structure would help to clarify the roles of both the hydrophobic binding pocket and the hydrophilic rim residues required for ligand binding.
A number of natural and synthetic acylated agonists and antagonists are known to be shuttled by CD14. For example, CD14 delivers tri- and di-acylated agonists to complexes formed by members of the TLR2 subfamily (33-40, 86). Natural antagonists of LPS, including R. sphaeroides pentacylated lipid A, tetracylated lipid IVa and monophosphoryl lipid A, an approved vaccine adjuvant in Europe, are shuttled by CD14 to MD-2/TLR4 (78, 87-90). Eritoran, a synthetic LPS antagonist, that is structurally similar to lipid IVa, is also delivered to MD-2 by CD14 (91, 92). More recently, synthetic LPS inhibitors derived from either diacylated sugars or tetraacylated sulfate containing compounds have been identified and shown to competitively block LPS binding to CD14 (93-95). It is highly likely that the hydrophobic binding pocket in human CD14 accommodates these natural and synthetic agonists and antagonists especially as they contain fewer acyl chains than hexa-acylated enteric bacterial LPS (89, 96). Since TLR4 inhibitors are largely LPS mimics that are either delivered by CD14 or directly compete with LPS for interaction with CD14, the structural information presented here may further the design of drugs for the treatment of sepsis and other inflammatory diseases.
Supplementary Material
Acknowledgements
We would like to thank Danielle Gray and Yi Gui Gao at the George L. Clark X-Ray Facility at the University of Illinois Urbana-Champaign, Raven Huang, Chio Mui Chan, and Diana Rañoa at the University of Illinois Urbana-Champaign, and Amy Sarjeant at the Northwestern Molecular Structure Education and Research Center (IMSERC) for technical advice; Theresa Gioannini and Jerrold Weiss at the University of Iowa for purified LBP; David Kranz at the University of Illinois Urbana-Champaign for the modified pDisplay vector; and Keith Brister, Joseph Brunzelle, David Smith, and Zdzislaw Wawrzak at the Life Sciences Collaborative Access Team (21 ID–F/G at Argonne National Laboratory, Advanced Photon Source) for technical assistance during crystallographic data collection.
Footnotes
NIH R01 AI052344 (R.I.T.); NIH T32 GM007283 (S.L.K.)
7. References
- 1.Bazil V, Horejsi V, Hilgert I, McMichael MJ, editors. Leucocyte Typing III. White Cell Differentiation Antigens. The Workshop: Myeloid Panel Antibodies Recognizing the 53-kDa Molecular Weight Monocyte Antigen (CD14); Oxford. Oxford University Press; 1982. pp. 611–613. [Google Scholar]
- 2.Haziot A, Chen S, Ferrero E, Low MG, Silber R, Goyert SM. The monocyte differentiation antigen, CD14, is anchored to the cell membrane by a phosphatidylinositol linkage. Journal of immunology. 1988;141:547–552. [PubMed] [Google Scholar]
- 3.Simmons DL, Tan S, Tenen DG, Nicholson-Weller A, Seed B. Monocyte antigen CD14 is a phospholipid anchored membrane protein. Blood. 1989;73:284–289. [PubMed] [Google Scholar]
- 4.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science (New York, N.Y.) 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 5.Tapping RI, Tobias PS. Soluble CD14-mediated cellular responses to lipopolysaccharide. Chemical immunology. 2000;74:108–121. doi: 10.1159/000058751. [DOI] [PubMed] [Google Scholar]
- 6.Salomao R, Brunialti MK, Rapozo MM, Baggio-Zappia GL, Galanos C, Freudenberg M. Bacterial sensing, cell signaling, and modulation of the immune response during sepsis. Shock (Augusta, Ga.) 2012;38:227–242. doi: 10.1097/SHK.0b013e318262c4b0. [DOI] [PubMed] [Google Scholar]
- 7.Opal SM, Cohen J. Clinical gram-positive sepsis: does it fundamentally differ from gram-negative bacterial sepsis? Critical care medicine. 1999;27:1608–1616. doi: 10.1097/00003246-199908000-00039. [DOI] [PubMed] [Google Scholar]
- 8.Haziot A, Ferrero E, Kontgen F, Hijiya N, Yamamoto S, Silver J, Stewart CL, Goyert SM. Resistance to endotoxin shock and reduced dissemination of gram-negative bacteria in CD14-deficient mice. Immunity. 1996;4:407–414. doi: 10.1016/s1074-7613(00)80254-x. [DOI] [PubMed] [Google Scholar]
- 9.Rietschel ET, Brade H, Brade L, Kaca W, Kawahara K, Lindner B, Luderitz T, Tomita T, Schade U, Seydel U, et al. Newer aspects of the chemical structure and biological activity of bacterial endotoxins. Progress in clinical and biological research. 1985;189:31–51. [PubMed] [Google Scholar]
- 10.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annual review of biochemistry. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erridge C, Bennett-Guerrero E, Poxton IR. Structure and function of lipopolysaccharides. Microbes and infection / Institut Pasteur. 2002;4:837–851. doi: 10.1016/s1286-4579(02)01604-0. [DOI] [PubMed] [Google Scholar]
- 12.Hailman E, Lichenstein HS, Wurfel MM, Miller DS, Johnson DA, Kelley M, Busse LA, Zukowski MM, Wright SD. Lipopolysaccharide (LPS)-binding protein accelerates the binding of LPS to CD14. The Journal of experimental medicine. 1994;179:269–277. doi: 10.1084/jem.179.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tobias PS, Soldau K, Gegner JA, Mintz D, Ulevitch RJ. Lipopolysaccharide binding protein-mediated complexation of lipopolysaccharide with soluble CD14. The Journal of biological chemistry. 1995;270:10482–10488. doi: 10.1074/jbc.270.18.10482. [DOI] [PubMed] [Google Scholar]
- 14.Tobias PS, Soldau K, Ulevitch RJ. Isolation of a lipopolysaccharide-binding acute phase reactant from rabbit serum. The Journal of experimental medicine. 1986;164:777–793. doi: 10.1084/jem.164.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schumann RR, Leong SR, Flaggs GW, Gray PW, Wright SD, Mathison JC, Tobias PS, Ulevitch RJ. Structure and function of lipopolysaccharide binding protein. Science (New York, N.Y.) 1990;249:1429–1431. doi: 10.1126/science.2402637. [DOI] [PubMed] [Google Scholar]
- 16.Yu B, Wright SD. Catalytic properties of lipopolysaccharide (LPS) binding protein. Transfer of LPS to soluble CD14. The Journal of biological chemistry. 1996;271:4100–4105. doi: 10.1074/jbc.271.8.4100. [DOI] [PubMed] [Google Scholar]
- 17.Goyert SM, Ferrero E, Rettig WJ, Yenamandra AK, Obata F, Le Beau MM. The CD14 monocyte differentiation antigen maps to a region encoding growth factors and receptors. Science (New York, N.Y.) 1988;239:497–500. doi: 10.1126/science.2448876. [DOI] [PubMed] [Google Scholar]
- 18.Pugin J, Heumann ID, Tomasz A, Kravchenko VV, Akamatsu Y, Nishijima M, Glauser MP, Tobias PS, Ulevitch RJ. CD14 is a pattern recognition receptor. Immunity. 1994;1:509–516. doi: 10.1016/1074-7613(94)90093-0. [DOI] [PubMed] [Google Scholar]
- 19.Shimazu R, Akashi S, Ogata H, Nagai Y, Fukudome K, Miyake K, Kimoto M. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. The Journal of experimental medicine. 1999;189:1777–1782. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nunez Miguel R, Wong J, Westoll JF, Brooks HJ, O’Neill LA, Gay NJ, Bryant CE, Monie TP. A dimer of the Toll-like receptor 4 cytoplasmic domain provides a specific scaffold for the recruitment of signalling adaptor proteins. PloS one. 2007;2:e788. doi: 10.1371/journal.pone.0000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gioannini TL, Teghanemt A, Zhang D, Coussens NP, Dockstader W, Ramaswamy S, Weiss JP. Isolation of an endotoxin-MD-2 complex that produces Toll-like receptor 4-dependent cell activation at picomolar concentrations. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4186–4191. doi: 10.1073/pnas.0306906101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prohinar P, Re F, Widstrom R, Zhang D, Teghanemt A, Weiss JP, Gioannini TL. Specific high affinity interactions of monomeric endotoxin.protein complexes with Toll-like receptor 4 ectodomain. The Journal of biological chemistry. 2007;282:1010–1017. doi: 10.1074/jbc.M609400200. [DOI] [PubMed] [Google Scholar]
- 23.Teghanemt A, Prohinar P, Gioannini TL, Weiss JP. Transfer of monomeric endotoxin from MD-2 to CD14: characterization and functional consequences. The Journal of biological chemistry. 2007;282:36250–36256. doi: 10.1074/jbc.M705995200. [DOI] [PubMed] [Google Scholar]
- 24.Teghanemt A, Widstrom RL, Gioannini TL, Weiss JP. Isolation of monomeric and dimeric secreted MD-2. Endotoxin.sCD14 and Toll-like receptor 4 ectodomain selectively react with the monomeric form of secreted MD-2. The Journal of biological chemistry. 2008;283:21881–21889. doi: 10.1074/jbc.M800672200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jack RS, Grunwald U, Stelter F, Workalemahu G, Schutt C. Both membrane-bound and soluble forms of CD14 bind to gram-negative bacteria. European journal of immunology. 1995;25:1436–1441. doi: 10.1002/eji.1830250545. [DOI] [PubMed] [Google Scholar]
- 26.Grunwald U, Fan X, Jack RS, Workalemahu G, Kallies A, Stelter F, Schutt C. Monocytes can phagocytose Gram-negative bacteria by a CD14-dependent mechanism. Journal of immunology. 1996;157:4119–4125. [PubMed] [Google Scholar]
- 27.Fan X, Stelter F, Menzel R, Jack R, Spreitzer I, Hartung T, Schutt C. Structures in Bacillus subtilis are recognized by CD14 in a lipopolysaccharide binding protein-dependent reaction. Infection and immunity. 1999;67:2964–2968. doi: 10.1128/iai.67.6.2964-2968.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stelter F, Bernheiden M, Menzel R, Jack RS, Witt S, Fan X, Pfister M, Schutt C. Mutation of amino acids 39-44 of human CD14 abrogates binding of lipopolysaccharide and Escherichia coli. European journal of biochemistry / FEBS. 1997;243:100–109. doi: 10.1111/j.1432-1033.1997.00100.x. [DOI] [PubMed] [Google Scholar]
- 29.Schiff DE, Kline L, Soldau K, Lee JD, Pugin J, Tobias PS, Ulevitch RJ. Phagocytosis of gram-negative bacteria by a unique CD14-dependent mechanism. Journal of leukocyte biology. 1997;62:786–794. doi: 10.1002/jlb.62.6.786. [DOI] [PubMed] [Google Scholar]
- 30.Wurfel MM, Wright SD. Lipopolysaccharide-binding protein and soluble CD14 transfer lipopolysaccharide to phospholipid bilayers: preferential interaction with particular classes of lipid. Journal of immunology. 1997;158:3925–3934. [PubMed] [Google Scholar]
- 31.Yu B, Hailman E, Wright SD. Lipopolysaccharide binding protein and soluble CD14 catalyze exchange of phospholipids. The Journal of clinical investigation. 1997;99:315–324. doi: 10.1172/JCI119160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang PY, Kitchens RL, Munford RS. Phosphatidylinositides bind to plasma membrane CD14 and can prevent monocyte activation by bacterial lipopolysaccharide. The Journal of biological chemistry. 1998;273:24309–24313. doi: 10.1074/jbc.273.38.24309. [DOI] [PubMed] [Google Scholar]
- 33.Nakata T, Yasuda M, Fujita M, Kataoka H, Kiura K, Sano H, Shibata K. CD14 directly binds to triacylated lipopeptides and facilitates recognition of the lipopeptides by the receptor complex of Toll-like receptors 2 and 1 without binding to the complex. Cellular microbiology. 2006;8:1899–1909. doi: 10.1111/j.1462-5822.2006.00756.x. [DOI] [PubMed] [Google Scholar]
- 34.Dziarski R, Tapping RI, Tobias PS. Binding of bacterial peptidoglycan to CD14. The Journal of biological chemistry. 1998;273:8680–8690. doi: 10.1074/jbc.273.15.8680. [DOI] [PubMed] [Google Scholar]
- 35.Gupta D, Kirkland TN, Viriyakosol S, Dziarski R. CD14 is a cell-activating receptor for bacterial peptidoglycan. The Journal of biological chemistry. 1996;271:23310–23316. doi: 10.1074/jbc.271.38.23310. [DOI] [PubMed] [Google Scholar]
- 36.Savedra R, Jr., Delude RL, Ingalls RR, Fenton MJ, Golenbock DT. Mycobacterial lipoarabinomannan recognition requires a receptor that shares components of the endotoxin signaling system. Journal of immunology. 1996;157:2549–2554. [PubMed] [Google Scholar]
- 37.Yu W, Soprana E, Cosentino G, Volta M, Lichenstein HS, Viale G, Vercelli D. Soluble CD14(1-152) confers responsiveness to both lipoarabinomannan and lipopolysaccharide in a novel HL-60 cell bioassay. Journal of immunology. 1998;161:4244–4251. [PubMed] [Google Scholar]
- 38.Elass E, Coddeville B, Guerardel Y, Kremer L, Maes E, Mazurier J, Legrand D. Identification by surface plasmon resonance of the mycobacterial lipomannan and lipoarabinomannan domains involved in binding to CD14 and LPS-binding protein. FEBS letters. 2007;581:1383–1390. doi: 10.1016/j.febslet.2007.02.056. [DOI] [PubMed] [Google Scholar]
- 39.Lien E, Sellati TJ, Yoshimura A, Flo TH, Rawadi G, Finberg RW, Carroll JD, Espevik T, Ingalls RR, Radolf JD, Golenbock DT. Toll-like receptor 2 functions as a pattern recognition receptor for diverse bacterial products. The Journal of biological chemistry. 1999;274:33419–33425. doi: 10.1074/jbc.274.47.33419. [DOI] [PubMed] [Google Scholar]
- 40.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 41.Pugin J, Schurer-Maly CC, Leturcq D, Moriarty A, Ulevitch RJ, Tobias PS. Lipopolysaccharide activation of human endothelial and epithelial cells is mediated by lipopolysaccharide-binding protein and soluble CD14. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:2744–2748. doi: 10.1073/pnas.90.7.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tapping RI, Tobias PS. Cellular binding of soluble CD14 requires lipopolysaccharide (LPS) and LPS-binding protein. The Journal of biological chemistry. 1997;272:23157–23164. doi: 10.1074/jbc.272.37.23157. [DOI] [PubMed] [Google Scholar]
- 43.Haziot A, Rong GW, Bazil V, Silver J, Goyert SM. Recombinant soluble CD14 inhibits LPS-induced tumor necrosis factor-alpha production by cells in whole blood. Journal of immunology. 1994;152:5868–5876. [PubMed] [Google Scholar]
- 44.Stelter F, Pfister M, Bernheiden M, Jack RS, Bufler P, Engelmann H, Schutt C. The myeloid differentiation antigen CD14 is N- and O-glycosylated. Contribution of N-linked glycosylation to different soluble CD14 isoforms. European journal of biochemistry / FEBS. 1996;236:457–464. doi: 10.1111/j.1432-1033.1996.00457.x. [DOI] [PubMed] [Google Scholar]
- 45.Majerle A, Kidric J, Jerala R. Expression and refolding of functional fragments of the human lipopolysaccharide receptor CD14 in Escherichia coli and Pichia pastoris. Protein expression and purification. 1999;17:96–104. doi: 10.1006/prep.1999.1109. [DOI] [PubMed] [Google Scholar]
- 46.Bai J, Yin J, Wang ZD, Wang W, Wang ZZ, Song W. Clone and expression of human soluble CD14 and study of its function. Sheng wu gong cheng xue bao = Chinese journal of biotechnology. 2001;17:269–272. [PubMed] [Google Scholar]
- 47.Nomura S, Inamori K, Muta T, Yamazaki S, Sunakawa Y, Iwanaga S, Takeshige K. Purification and characterization of human soluble CD14 expressed in Pichia pastoris. Protein expression and purification. 2003;28:310–320. doi: 10.1016/s1046-5928(02)00705-2. [DOI] [PubMed] [Google Scholar]
- 48.Burkhardt M, LopezAcosta A, Reiter K, Lopez V, Lees A. Purification of soluble CD14 fusion proteins and use in an electrochemiluminescent assay for lipopolysaccharide binding. Protein expression and purification. 2007;51:96–101. doi: 10.1016/j.pep.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 49.Meng J, Parroche P, Golenbock DT, McKnight CJ. The differential impact of disulfide bonds and N-linked glycosylation on the stability and function of CD14. The Journal of biological chemistry. 2008;283:3376–3384. doi: 10.1074/jbc.M707640200. [DOI] [PubMed] [Google Scholar]
- 50.Albright S, Chen B, Holbrook K, Jain NU. Solution NMR studies provide structural basis for endotoxin pattern recognition by the innate immune receptor CD14. Biochemical and biophysical research communications. 2008;368:231–237. doi: 10.1016/j.bbrc.2008.01.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vida A, Bardoel B, Milder F, Majoros L, Sumegi A, Bacsi A, Vereb G, van Kessel KP, van Strijp JA, Antal-Szalmas P. Fusion of the Fc part of human IgG1 to CD14 enhances its binding to Gram-negative bacteria and mediates phagocytosis by Fc receptors of neutrophils. Immunology letters. 2012;146:31–39. doi: 10.1016/j.imlet.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 52.Kim JI, Lee CJ, Jin MS, Lee CH, Paik SG, Lee H, Lee JO. Crystal structure of CD14 and its implications for lipopolysaccharide signaling. The Journal of biological chemistry. 2005;280:11347–11351. doi: 10.1074/jbc.M414607200. [DOI] [PubMed] [Google Scholar]
- 53.Rangel-Frausto MS. Sepsis: still going strong. Archives of medical research. 2005;36:672–681. doi: 10.1016/j.arcmed.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 54.Guan Y, Ranoa DR, Jiang S, Mutha SK, Li X, Baudry J, Tapping RI. Human TLRs 10 and 1 share common mechanisms of innate immune sensing but not signaling. Journal of immunology. 2010;184:5094–5103. doi: 10.4049/jimmunol.0901888. [DOI] [PubMed] [Google Scholar]
- 55.Jancarik J, Pufan R, Hong C, Kim SH, Kim R. Optimum solubility (OS) screening: an efficient method to optimize buffer conditions for homogeneity and crystallization of proteins. Acta crystallographica. Section D, Biological crystallography. 2004;60:1670–1673. doi: 10.1107/S0907444904010972. [DOI] [PubMed] [Google Scholar]
- 56.McPherson A. Current approaches to macromolecular crystallization. European journal of biochemistry / FEBS. 1990;189:1–23. doi: 10.1111/j.1432-1033.1990.tb15454.x. [DOI] [PubMed] [Google Scholar]
- 57.Jancarik J, Kim S-H. Sparse matrix sampling: a screening method for crystallization of proteins. Journal of Applied Crystallography. 1991;24:409–411. [Google Scholar]
- 58.McPherson A. Use of polyethylene glycol in the crystallization of macromolecules. Methods in enzymology. 1985;114:120–125. doi: 10.1016/0076-6879(85)14008-5. [DOI] [PubMed] [Google Scholar]
- 59.Sauter C, Ng JD, Lorber B, Keith G, Brion P, Hosseini MW, Lehn J-M, Giege R. Effects of additives for the crystallization of proteins and nucleic acids. J. Crystal Growth. 1999;196:365–376. [Google Scholar]
- 60.McPherson A, Koszelak S, Axelrod H, Day J, Robinson L, McGrath M, Williams R, Cascio D. The effects of netural detergents on the crystallization of soluble proteins. J. Crystal Growth. 1986;76:547–553. [PubMed] [Google Scholar]
- 61.Garman E. ‘Cool’ crystals: macromolecular cryocrystallography and radiation damage. Current opinion in structural biology. 2003;13:545–551. doi: 10.1016/j.sbi.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 62.Heras B, Martin JL. Post-crystallization treatments for improving diffraction quality of protein crystals. Acta crystallographica. Section D, Biological crystallography. 2005;61:1173–1180. doi: 10.1107/S0907444905019451. [DOI] [PubMed] [Google Scholar]
- 63.Walter TS, Meier C, Assenberg R, Au KF, Ren J, Verma A, Nettleship JE, Owens RJ, Stuart DI, Grimes JM. Lysine methylation as a routine rescue strategy for protein crystallization. Structure (London, England : 1993) 2006;14:1617–1622. doi: 10.1016/j.str.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neumeister B, Faigle M, Sommer M, Zahringer U, Stelter F, Menzel R, Schutt C, Northoff H. Low endotoxic potential of Legionella pneumophila lipopolysaccharide due to failure of interaction with the monocyte lipopolysaccharide receptor CD14. Infection and immunity. 1998;66:4151–4157. doi: 10.1128/iai.66.9.4151-4157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Juan TS, Kelley MJ, Johnson DA, Busse LA, Hailman E, Wright SD, Lichenstein HS. Soluble CD14 truncated at amino acid 152 binds lipopolysaccharide (LPS) and enables cellular response to LPS. The Journal of biological chemistry. 1995;270:1382–1387. doi: 10.1074/jbc.270.3.1382. [DOI] [PubMed] [Google Scholar]
- 66.Viriyakosol S, Kirkland TN. The N-terminal half of membrane CD14 is a functional cellular lipopolysaccharide receptor. Infection and immunity. 1996;64:653–656. doi: 10.1128/iai.64.2.653-656.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alexander S, Elder JH. Endoglycosidases from Flavobacterium meningosepticum application to biological problems. Methods in enzymology. 1989;179:505–518. doi: 10.1016/0076-6879(89)79151-5. [DOI] [PubMed] [Google Scholar]
- 68.Goyert SM, Ferrero EM, Seremetis SV, Winchester RJ, Silver J, Mattison AC. Biochemistry and expression of myelomonocytic antigens. Journal of immunology. 1986;137:3909–3914. [PubMed] [Google Scholar]
- 69.Ferrero E, Goyert SM. Nucleotide sequence of the gene encoding the monocyte differentiation antigen, CD14. Nucleic acids research. 1988;16:4173. doi: 10.1093/nar/16.9.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Enkhbayar P, Kamiya M, Osaki M, Matsumoto T, Matsushima N. Structural principles of leucine-rich repeat (LRR) proteins. Proteins. 2004;54:394–403. doi: 10.1002/prot.10605. [DOI] [PubMed] [Google Scholar]
- 71.Creighton TE. Protein structure. Stability of alpha-helices. Nature. 1987;326:547–548. doi: 10.1038/326547a0. [DOI] [PubMed] [Google Scholar]
- 72.Barlow DJ, Thornton JM. Helix geometry in proteins. Journal of molecular biology. 1988;201:601–619. doi: 10.1016/0022-2836(88)90641-9. [DOI] [PubMed] [Google Scholar]
- 73.Roberts E, Eargle J, Wright D, Luthey-Schulten Z. MultiSeq: unifying sequence and structure data for evolutionary analysis. BMC bioinformatics. 2006;7:382. doi: 10.1186/1471-2105-7-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Inohara N, Nunez G. ML -- a conserved domain involved in innate immunity and lipid metabolism. Trends in biochemical sciences. 2002;27:219–221. doi: 10.1016/s0968-0004(02)02084-4. [DOI] [PubMed] [Google Scholar]
- 75.Ohto U, Fukase K, Miyake K, Satow Y. Crystal structures of human MD-2 and its complex with antiendotoxic lipid IVa. Science (New York, N.Y.) 2007;316:1632–1634. doi: 10.1126/science.1139111. [DOI] [PubMed] [Google Scholar]
- 76.Ohto U, Fukase K, Miyake K, Shimizu T. Structural basis of species-specific endotoxin sensing by innate immune receptor TLR4/MD-2. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:7421–7426. doi: 10.1073/pnas.1201193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 78.Akashi S, Nagai Y, Ogata H, Oikawa M, Fukase K, Kusumoto S, Kawasaki K, Nishijima M, Hayashi S, Kimoto M, Miyake K. Human MD-2 confers on mouse Toll-like receptor 4 species-specific lipopolysaccharide recognition. International immunology. 2001;13:1595–1599. doi: 10.1093/intimm/13.12.1595. [DOI] [PubMed] [Google Scholar]
- 79.Gioannini TL, Teghanemt A, Zhang D, Prohinar P, Levis EN, Munford RS, Weiss JP. Endotoxin-binding proteins modulate the susceptibility of bacterial endotoxin to deacylation by acyloxyacyl hydrolase. The Journal of biological chemistry. 2007;282:7877–7884. doi: 10.1074/jbc.M605031200. [DOI] [PubMed] [Google Scholar]
- 80.McGinley MD, Narhi LO, Kelley MJ, Davy E, Robinson J, Rohde MF, Wright SD, Lichenstein HS. CD14: physical properties and identification of an exposed site that is protected by lipopolysaccharide. The Journal of biological chemistry. 1995;270:5213–5218. doi: 10.1074/jbc.270.10.5213. [DOI] [PubMed] [Google Scholar]
- 81.Juan TS, Hailman E, Kelley MJ, Wright SD, Lichenstein HS. Identification of a domain in soluble CD14 essential for lipopolysaccharide (LPS) signaling but not LPS binding. The Journal of biological chemistry. 1995;270:17237–17242. doi: 10.1074/jbc.270.29.17237. [DOI] [PubMed] [Google Scholar]
- 82.Viriyakosol S, Kirkland TN. A region of human CD14 required for lipopolysaccharide binding. The Journal of biological chemistry. 1995;270:361–368. doi: 10.1074/jbc.270.1.361. [DOI] [PubMed] [Google Scholar]
- 83.Viriyakosol S, Mathison JC, Tobias PS, Kirkland TN. Structure-function analysis of CD14 as a soluble receptor for lipopolysaccharide. The Journal of biological chemistry. 2000;275:3144–3149. doi: 10.1074/jbc.275.5.3144. [DOI] [PubMed] [Google Scholar]
- 84.Juan TS, Hailman E, Kelley MJ, Busse LA, Davy E, Empig CJ, Narhi LO, Wright SD, Lichenstein HS. Identification of a lipopolysaccharide binding domain in CD14 between amino acids 57 and 64. The Journal of biological chemistry. 1995;270:5219–5224. doi: 10.1074/jbc.270.10.5219. [DOI] [PubMed] [Google Scholar]
- 85.Shapiro RA, Cunningham MD, Ratcliffe K, Seachord C, Blake J, Bajorath J, Aruffo A, Darveau RP. Identification of CD14 residues involved in specific lipopolysaccharide recognition. Infection and immunity. 1997;65:293–297. doi: 10.1128/iai.65.1.293-297.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Takeuchi O, Kawai T, Muhlradt PF, Morr M, Radolf JD, Zychlinsky A, Takeda K, Akira S. Discrimination of bacterial lipoproteins by Toll-like receptor 6. International immunology. 2001;13:933–940. doi: 10.1093/intimm/13.7.933. [DOI] [PubMed] [Google Scholar]
- 87.Golenbock DT, Hampton RY, Qureshi N, Takayama K, Raetz CR. Lipid A-like molecules that antagonize the effects of endotoxins on human monocytes. The Journal of biological chemistry. 1991;266:19490–19498. [PubMed] [Google Scholar]
- 88.Muroi M, Tanamoto K. Structural regions of MD-2 that determine the agonist-antagonist activity of lipid IVa. The Journal of biological chemistry. 2006;281:5484–5491. doi: 10.1074/jbc.M509193200. [DOI] [PubMed] [Google Scholar]
- 89.Rietschel ET, Kirikae T, Schade FU, Mamat U, Schmidt G, Loppnow H, Ulmer AJ, Zahringer U, Seydel U, Di Padova F, et al. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1994;8:217–225. doi: 10.1096/fasebj.8.2.8119492. [DOI] [PubMed] [Google Scholar]
- 90.Mata-Haro V, Cekic C, Martin M, Chilton PM, Casella CR, Mitchell TC. The vaccine adjuvant monophosphoryl lipid A as a TRIF-biased agonist of TLR4. Science (New York, N.Y.) 2007;316:1628–1632. doi: 10.1126/science.1138963. [DOI] [PubMed] [Google Scholar]
- 91.Kim HM, Park BS, Kim JI, Kim SE, Lee J, Oh SC, Enkhbayar P, Matsushima N, Lee H, Yoo OJ, Lee JO. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 92.Hawkins LD, Christ WJ, Rossignol DP. Inhibition of endotoxin response by synthetic TLR4 antagonists. Current topics in medicinal chemistry. 2004;4:1147–1171. doi: 10.2174/1568026043388123. [DOI] [PubMed] [Google Scholar]
- 93.Piazza M, Rossini C, Della Fiorentina S, Pozzi C, Comelli F, Bettoni I, Fusi P, Costa B, Peri F. Glycolipids and benzylammonium lipids as novel antisepsis agents: synthesis and biological characterization. Journal of medicinal chemistry. 2009;52:1209–1213. doi: 10.1021/jm801333m. [DOI] [PubMed] [Google Scholar]
- 94.Piazza M, Yu L, Teghanemt A, Gioannini T, Weiss J, Peri F. Evidence of a specific interaction between new synthetic antisepsis agents and CD14. Biochemistry. 2009;48:12337–12344. doi: 10.1021/bi901601b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Piazza M, Calabrese V, Damore G, Cighetti R, Gioannini T, Weiss J, Peri F. A synthetic lipid A mimetic modulates human TLR4 activity. ChemMedChem. 2012;7:213–217. doi: 10.1002/cmdc.201100494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schromm AB, Brandenburg K, Loppnow H, Moran AP, Koch MH, Rietschel ET, Seydel U. Biological activities of lipopolysaccharides are determined by the shape of their lipid A portion. European journal of biochemistry / FEBS. 2000;267:2008–2013. doi: 10.1046/j.1432-1327.2000.01204.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.