Abstract
Our understanding of the mechanisms by which aging is produced is still very limited. Here, we have determined the sera metabolite profile of 117 wild-type mice of different genetic backgrounds ranging from 8-129 weeks of age. This has allowed us to define a robust metabolomic signature and a derived metabolomic score that reliably/accurately predicts the age of wild-type mice. In the case of telomerase-deficient mice, which have a shortened lifespan, their metabolomic score predicts older ages than expected. Conversely, in the case of mice that over-express telomerase, their metabolic score corresponded to younger ages than expected. Importantly, telomerase reactivation late in life by using a TERT based gene therapy recently described by us, significantly reverted the metabolic profile of old mice to that of younger mice, further confirming an anti-aging role for telomerase. Thus, the metabolomic signature associated to natural mouse aging accurately predicts aging produced by telomere shortening, suggesting that natural mouse aging is in part produced by presence of short telomeres. These results indicate that the metabolomic signature is associated to the biological age rather than to the chronological age. This constitutes one of the first aging-associated metabolomic signatures in a mammalian organism.
High throughput metabolite profiling technologies can be used to determine the metabolic status of whole organisms under conditions of interest, as well as to systematically define phenotypic patterns associated to mutant genotypes (Raamsdonk et al. 2001; Allen et al. 2003; Nicholson & Wilson 2003; Lewis et al. 2010). Here, we set to determine the metabolic profile of wild-type mice at different ages with the goal of obtaining a metabolomic signature of natural mouse aging, which could serve as a predictive model to estimate the biological age, as well as that could allow to identify new biomarkers of aging. Furthermore, we reasoned that analysis of this age-related metabolomic signature in short-lived telomerase-deficient (Terc−/−) and long-lived TERT transgenics mice could serve to assess the contribution of telomere shortening and telomerase status to the metabolomic changes associated to natural mouse aging (Blasco et al. 1997; Lee et al. 1998; Herrera et al. 1999; Garcia-Cao et al. 2006; Tomas-Loba et al. 2008). A liquid chromatography-mass spectrometry (UPLC™-MS) platform-based metabolomics approach was used to explore the relative levels of serum metabolites during mouse aging. To circumvent the potential influence of genetic background and gender in the metabolomic profile associated to mouse aging, 117 serum samples were analyzed from wild-type mice of different genetic backgrounds (see materials and methods), and these samples included sera from both male (51) and female (66) mice with ages ranging from 8 to 129 weeks old. Furthermore, we also determined the metabolomic profiles of middle aged (1 year) and old (2 years) control wild-type mice compared to that of 2 yrs old mice treated with a TERT based gene therapy recently shown by us to reactivate telomerase and delay aging (see materials and methods) (Bernardes de Jesus et al. 2012). The only phenotypic distinction we have analyzed in the current manuscript is age. This could be viewed as one limitation of the current study since co-founding effects (such as disease or obesity) were not taken into account and could all render data interpretation difficult.
The global metabolite profile was processed to create a multivariate model correlating the spectral data with the age of the wild-type samples. A predictive PLS model (Projection to Latent Structure; see materials and methods) was built and validated (Figure S1), obtaining a robust metabolomic model of aging for wild-type mice, which fits well a linear model (r2 = 0.944) (closed black diamonds in Figure 1a). Variable Influence on Projection (VIP) parameters and Spearman’s rank correlation coefficients (ρ) were examined for each metabolite, identifying the most influential biomarkers in the PLS model and ageing - correlative. Table 1 highlights a selection of 48 biomarkers for which there is a significant correlation of wild-type samples with age (Spearmańs rank correlation test p-values < 0.05). These biomarkers include phospholipids, fatty acids, and organic acids, in agreement with the fact that the extraction methodology provides a maximum coverage over those families of compounds.
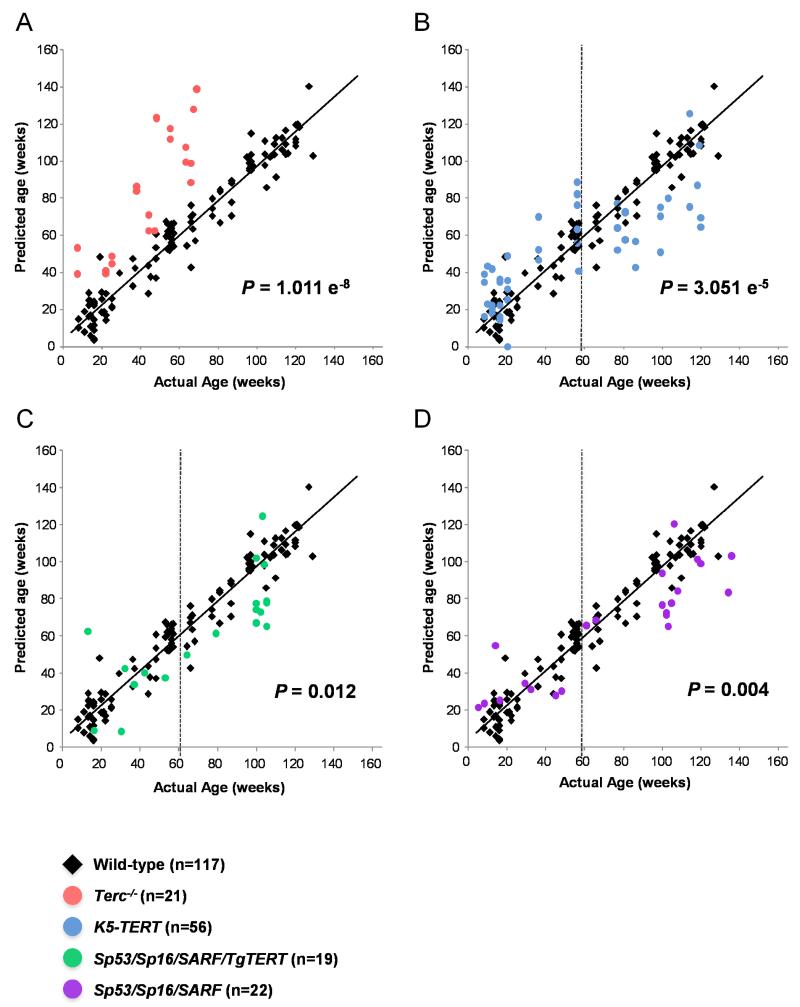
Figure 1. Age predictions with respect to the wild-type mice metabolomic signature.
The wild-type mice metabolomic ageing changes (black diamonds) fits a linear model: Predicted Age =0.934·Actual Age + 3.971 (r2=0.944). A) G1 Terc−/− samples (red circles); B) K5-Tert samples (blue circles); C) Sp53/Sp16/Sp19Arf/K5-Tert samples (green circles); D) Sp53/Sp16/Sp19Arf samples (purple circles). p-values for Fig. 1 b-d correspond only to mice over 60 months of age.
Table 1.
Selection of ageing serum metabolic biomarkers.
| Rt (min) |
m/z (Da/e) |
Ion | ID | Spearman’s rho |
P-value | VIP |
|---|---|---|---|---|---|---|
|
| ||||||
| 7.11 | 774.5646 | [M+H]+ | ND | −0.64987 | 5.23 × 10−11 | 3.335 |
| 6.90 | 719.5687 | [M+H]+ | ND | −0.54365 | 1.56 × 10−7 | 2.999 |
| 3.67 | 378.2400 | [M-H]− | Sphingosine 1 - phosphate | −0.50817 | 1.28 × 10−6 | 0.917 |
| 6.52 | 608.4578 | ND | ND | −0.49980 | 2.03 × 10−6 | 1.264 |
| 6.54 | 717.5526 | ND | ND | −0.47733 | 6.62 × 10−6 | 2.166 |
| 6.78 | 745.5492 | [M+FA]− | SM(d18:1/16:1) | −0.46557 | 1.19 × 10−5 | 1.492 |
| 7.40 | 776.5789 | [M+H]+ | ND | −0.39461 | 2.67 × 10−4 | 3.393 |
| 4.74 | 480.3082 | [M-H]− | LysoPE(18:0) | −0.38019 | 4.64 × 10−4 | 0.944 |
| 3.81 | 542.3239 | [M+H]+ | LysoPC(20:5) | 0.37572 | 5.47 × 10−4 | 1.460 |
| 0.62 | 160.1316 | [M+H]+ | 2-aminooctanoic acid | 0.36586 | 7.83 × 10−4 | 1.896 |
| 0.82 | 204.1236 | [M+H]+ | L-Acetylcarnitine | −0.32805 | 2.79 × 10−3 | 1.161 |
| 0.60 | 147.0769 | [M+H]+ | Glutamine | −0.31736 | 3.89 × 10−3 | 1.445 |
| 7.50 | 826.5595 | [M+FA]− | PC(18:2/18:2) | −0.31388 | 4.32 × 10−3 | 0.929 |
| 2.98 | 481.2043 | ND | ND | −0.31254 | 4.50 × 10−3 | 1.044 |
| 4.25 | 502.2935 | [M+FA-MFA]− | LysoPE (20:3) | −0.29817 | 6.86 × 10−3 | 1.738 |
| 6.02 | 305.2474 | [M-H]− | Eicosatrienoic acid | 0.29803 | 6.89 × 10−3 | 0.916 |
| 7.88 | 722.5129 | [M-H]− | PE (P-16:0/20:4) | −0.29592 | 7.31 × 10−3 | 1.221 |
| 7.31 | 774.5367 | [M+FA]− | PE (14:0/18:2) | −0.29171 | 8.23 × 10−3 | 1.528 |
| 0.84 | 150.0588 | [M+H]+ | L-Methionine | −0.28323 | 0.0104 | 0.857 |
| 5.38 | 269.2268 | ND | ND | 0.28115 | 0.0110 | 1.054 |
| 7.17 | 747.5637 | [M+FA]− | SM(d18:1/16:0) | −0.27966 | 0.0115 | 1.060 |
| 4.17 | 452.2764 | [M-H]− | LysoPE(16:0) | −0.27951 | 0.0115 | 1.134 |
| 6.76 | 721.5845 | [M+H]+ | ND | −0.27871 | 0.0118 | 0.877 |
| 7.70 | 768.5907 | [M+H]+ | ND | −0.27472 | 0.0131 | 0.970 |
| 6.61 | 447.3831 | ND | ND | −0.26165 | 0.0183 | 1.231 |
| 7.29 | 773.5794 | [M+FA]− | SM(36:2) | −0.25655 | 0.0208 | 1.373 |
| 0.60 | 132.0771 | [M+H]+ | Creatine | −0.25449 | 0.0219 | 1.383 |
| 7.91 | 778.5623 | [M+FA]− | PC (16:0/16:0) | −0.24690 | 0.0263 | 1.108 |
| 7.74 | 772.5875 | [M+H]+ | PC(17:0/18:2) | −0.24509 | 0.0274 | 1.162 |
| 6.97 | 778.5341 | ND | ND | 0.24449 | 0.0278 | 1.136 |
| 0.61 | 116.0710 | [M+H]+ | Proline | −0.24321 | 0.0287 | 1.263 |
| 4.63 | 536.3651 | ND | ND | 0.24095 | 0.0302 | 1.096 |
| 7.38 | 744.5543 | [M+H]+ | PC(15:0/18:2) | −0.24050 | 0.0306 | 1.347 |
| 7.60 | 716.5233 | [M+H]+ | PE(16:0/18:2) | −0.23397 | 0.0355 | 1.164 |
| 7.60 | 748.5272 | [M+H]+ | PE(P-16:0/22:6) | −0.22895 | 0.0398 | 1.203 |
| 0.55 | 167.0198 | [M-H]− | Uric acid | 0.22832 | 0.0404 | 1.605 |
| 7.46 | 764.5225 | [M+H]+ | PE(16:0/22:6) | −0.22750 | 0.0411 | 1.109 |
| 5.84 | 325.3203 | ND | ND | 0.22342 | 0.0450 | 1.125 |
| 7.42 | 800.5449 | [M+FA]− | PC(34:3) | −0.22217 | 0.0462 | 0.964 |
| 5.39 | 296.2590 | [M+ACN+H]+ | Palmitoleic acid | 0.21926 | 0.0492 | 0.944 |
| 0.57 | 124.0079 | [M+H]+ | Taurine | 0.25912 | 0.0195 | 0.718 |
| 4.22 | 279.2307 | [M+H]+ | Linolenic acid | 0.27863 | 0.0118 | 0.717 |
| 5.99 | 283.2627 | [M+H]+ | Oleic acid | 0.24274 | 0.0290 | 0.739 |
| 6.61 | 652.4550 | ND | ND | −0.33682 | 2.11 × 10−3 | 0.797 |
| 7.47 | 850.5606 | [M+H]+ | PC(16:0/22:6) | −0.24003 | 0.0309 | 1.090 |
| 7.50 | 832.5889 | [M+H]+ | PC(40:7) | 0.27525 | 0.0129 | 0.604 |
| 7.99 | 808.5871 | [M+H]+ | PC(38:5) | 0.27729 | 0.0122 | 0.860 |
| 8.11 | 766.5396 | [M+H]+ | PE(38:4) | −0.38345 | 4.10 × 10−4 | 1.135 |
Peaks are listed for which there is significant correlation of wild-type samples with age [Spearman’s rank correlation test p-values < 0.05]. Rt (min), chromatographic retention time; m/z (Da/e) [m is the neutral molecular mass value, e is the elemental charge], mass-to-charge ratio of ion detected; Ion, most intense species observed; ID, metabolite name (lipid names follow the LIPID MAPS classification system for lipids www.lipidmaps.org); Spearman’s rho, Spearman’s rank correlation coefficient; p-value, p-value derived from the Spearman’s rank correlation coefficient; VIP, Variable Influence on Projection parameter. ND, not determined.
We then used this wild-type mouse metabolomic signature of aging to predict the ages of first generation (G1) mice deficient in telomerase activity due to deletion of the telomerase RNA component or Terc (G1 Terc−/− mice). These mice show premature aging and decreased median and maximum longevity already at the first generation (Blasco et al. 1997; Lee et al. 1998; Herrera et al. 1999; Garcia-Cao et al. 2006). Interestingly, G1 Terc−/− mice showed a significantly older (p-value = 1.011 e−8) metabolic age when compared to wild-type mice of the same chronological age (red circles in Figure 1a). Thus, the metabolomic signature associated to physiological wild-type mouse aging, accurately predicts aging produced by short telomeres owe to telomerase deficiency. These results suggest that physiological mouse aging is largely associated to the presence of short telomeres or to cellular conditions favoring telomere protection, such an increased load of tumor suppressors (Tomas-Loba et al. 2008). Indeed, the increase in the percentage of short telomeres has shown to be correlated to the realized lifespan in mice (Vera et al. 2012). Next, we used our wild-type mouse-aging metabolomic signature to predict the ages of transgenic mice over-expressing the telomerase reverse transcriptase subunit or TERT in various stratified epithelia (K5-TERT mice). We have previously shown that K5-TERT mice have increased tissue fitness and a delayed aging (Gonzalez-Suarez et al. 2001; Gonzalez-Suarez et al. 2005). Serum metabolomic profiles were not significantly different when comparing wild-type and TERT over-expressing mice of all age groups (5-140 weeks; p-value = 0.534) (blue circles, Figure 1b). However, when we compared metabolic profiles of only adult/old mice (ages ranging from 60-140 weeks old), mice over-expressing TERT showed a significantly younger (p-value = 3.051 e−5) metabolic age than wild-type animals (Figure 1b). To circumvent the cancer promoting activity of telomerase, we also determined the metabolomic profile of our recently generated TERT over-expressing mice with enhanced cancer resistance owe to increased expression of the tumor suppressors p53, p16 and p19ARF (Sp53/Sp16/SARF/TgTERT). Sp53/Sp16/SARF/TgTERT mice show a delayed aging and a 40% extension of the median lifespan (Tomas-Loba et al. 2008). Serum metabolomic profiles of Sp53/Sp16/SARF/TgTERT mice identified significant differences in metabolic age compared wild-type mice of the same chronological age, although the differences were not very robust (p-value = 0.031) when the complete range of ages (8-140 weeks old) was evaluated (green circles in Figure 1c). Again, when we compared the metabolic profile of adult/old Sp53/Sp16/SARF/TgTERT mice ranging in age from 60-140 weeks old, Sp53/Sp15/SARF/TgTERT mice showed a significantly younger (p-value = 0.012) metabolic age compared to wild-type mice of the same chronological age (green circles in Figure 1c). Delayed organismal aging and extended lifespan have also been demonstrated through improved damage protection by increasing the activity of the p53/ARF pathway in Sp53/Sp16/SARF mice (Matheu et al . 2007). Comparison of serum metabolomic profiles from Sp53/Sp16/SARF and wild-type mice identified modest but significant differences (p-value = 0.045) when the complete range of ages (8-140 weeks old) was evaluated (purple circles, Figure 1d). These differences became more significant (p-value = 0.004) when only adult/old mice (ages ranging between 60-140 weeks old) were compared (purple circles, Figure 1d), indicating that old Sp53/Sp16/SARF mice have significantly younger metabolic ages than age-matched wild-type mice. Together, these results support an impact of telomerase activity and telomere length, as well as of the p53/p16/p19ARF tumor suppressors, on the metabolomic profile associated to natural mouse aging.
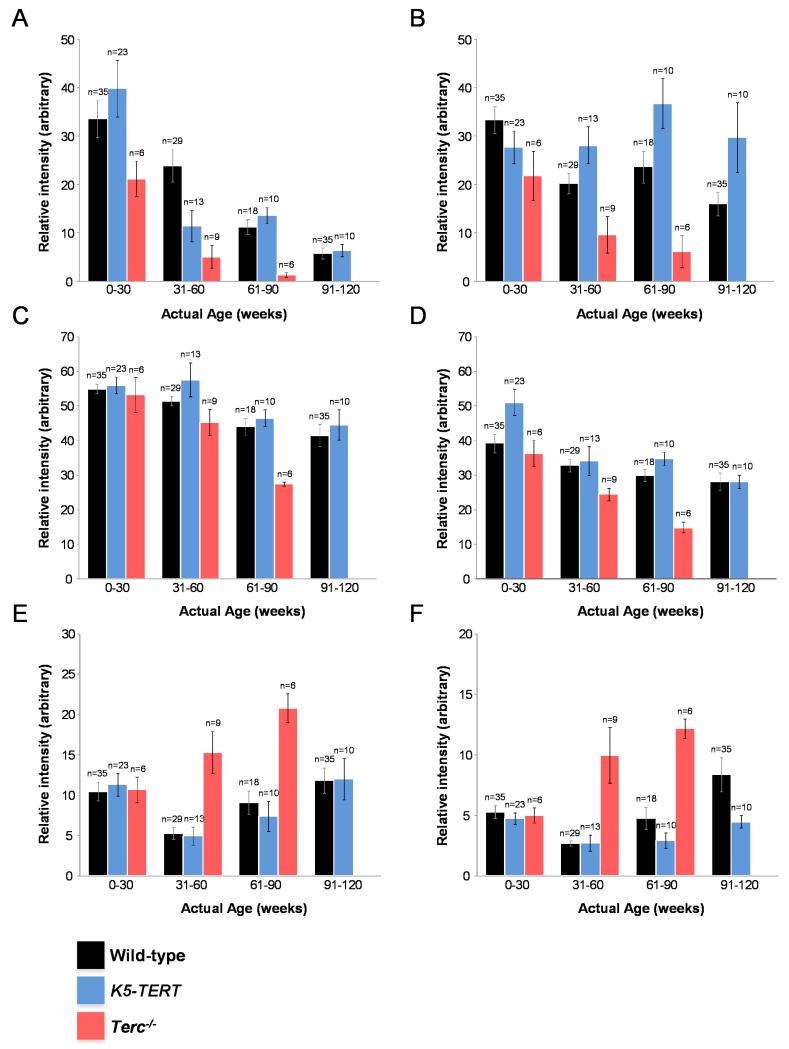
Next, we followed the behavior of different metabolites associated to mouse aging. Figure 2 shows representative examples of selected biomarkers in sera from wild-type, G1 Terc−/− and TERT transgenic mice at various ages. Figures 2a,c,d, corresponding to a not determined metabolite (Rt = 7.4 min; m/z = 776.5798), sphingomyelin SM(d18:1/16:1), and glycerophosphoethanolamine PE(P-16:0/22:6) respectively, exemplify the metabolic discrimination of G1 Terc−/− mice from the other sample groups. In particular, the relative decrease in abundance of these biomarkers occurs on a much faster timescale for the G1 Terc−/− mutants than for wild-type mice or TERT transgenics. Figure 2b illustrates a biomarker, lysoglicerophosphocholine (LysoPC(18:3), showing distinct temporal evolutions in the 3 sample groups. In particular, its relative intensity remains more or less constant over time for the TERT transgenics, while a marked decrease with age is observed for wild-type and G1 Terc−/− mice, the relative change once more occurring much quicker in the latter group. Opposite trends (increased rather than decreased relative intensities) are shown in Figures 2e,f, corresponding to glycerophosphoethanolamines PE(16:0/18:2) and PE(16:0/22:6), respectively. Both of these biomarkers increase rapidly with age in the Terc−/− cohorts, while a less pronounced change is observed in WT mice. The intensities of these biomarkers do not show significant changes over time in the TERT transgenic group. Interestingly, some of the metabolites identified here (ie, sphingomyelin, glycerophosphoethanolamines) have been previously linked to neurodegenerative disorders and various aspects of aging and disease (Klunk et al. 1997; Cutler & Mattson 2001).
Figure 2. Metabolite profile plots (mean ± 1 standard error of the mean).
A) Rt = 7.4 min, m/z = 776.5789 (not determined); B) LysoPE(20:3); C) SM(d18:1/16:1); D) PE(P-16:0/22:6); E) PE(16:0/18:2); F) PE(16:0/22:6). WT samples (black); K5-Tert samples (blue); Terc−/− samples (red).
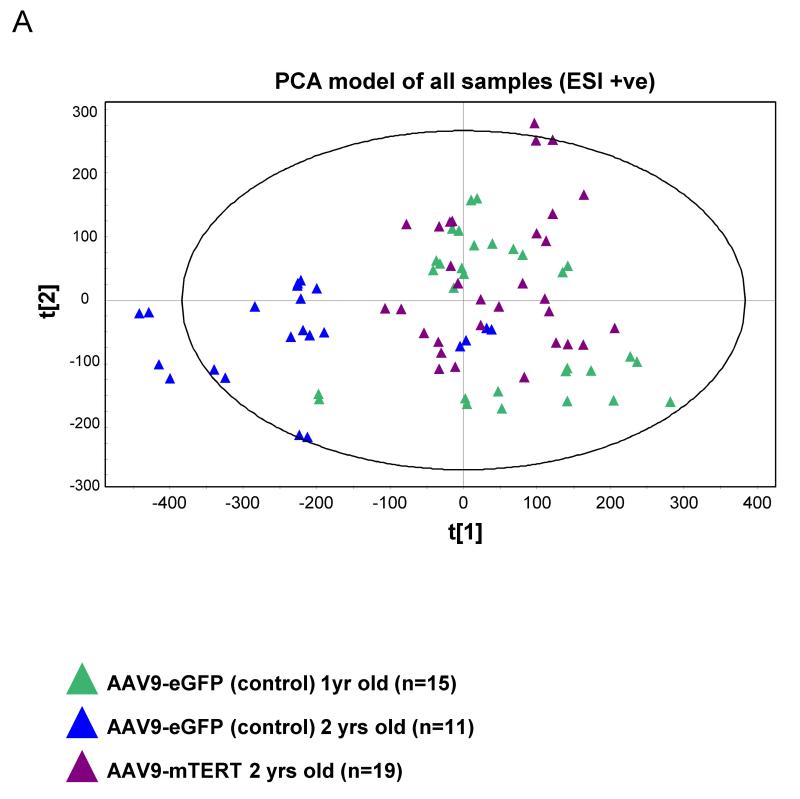
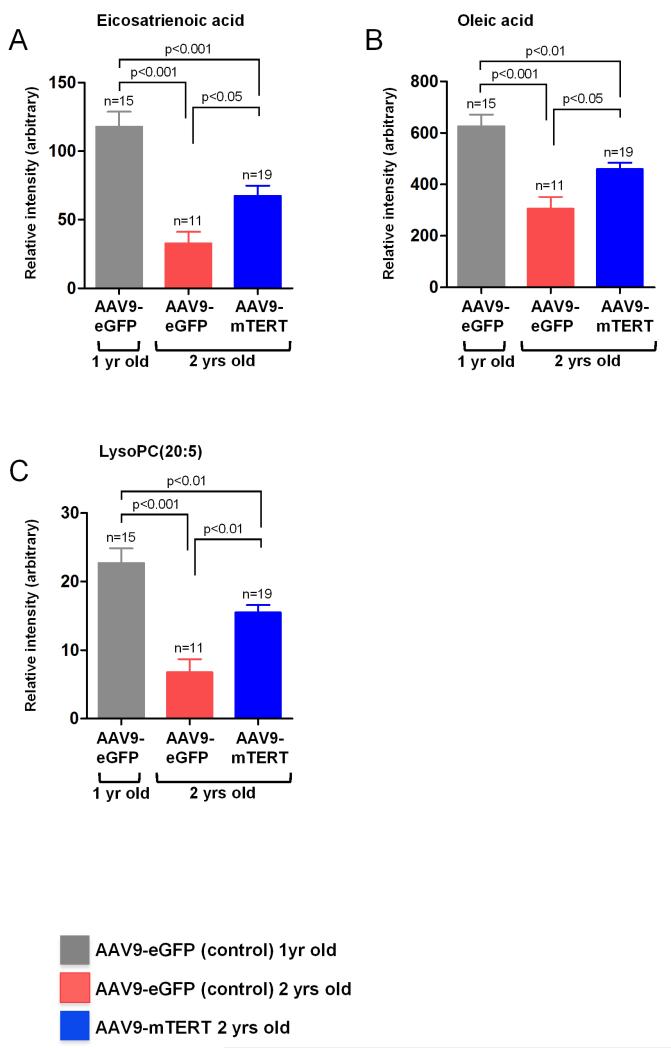
Recently, we reported that telomerase expression late in life by using a TERT based gene therapy strategy was sufficient to delay age-associated decay and to significantly extend mouse longevity (Bernardes de Jesus et al. 2012). Among other beneficial effects of TERT treatment, we observed that mice treated with AAV9-TERT showed improved metabolic parameters including a better performance in glucose and insulin tolerance tests. To dissect the effects of TERT treatment on metabolism, here we studied the metabolomics profiles of 1 and 2 year old mice treated with TERT (AAV9-mTERT) or with a control vector (AAV9-eGFP). To this end, we first compared the metabolomics profile of 1 yrs old and 2 yrs old control mice treated with AAV9-eGFP, which allowed to establish a set of metabolites that changed associated with aging (Table 2). Interestingly, we found that 2 months after TERT treatment, old mice (2 yrs old) showed a reversal of the metabolic signature associated to aging (Fig. 3). In particular, 2-year old mice treated with AAV9-mTERT showed a metabolic signature, which resembled the signature presented by the 1 yr old control mice. Therefore, expression of telomerase for 2 months in old mice partially rescues the age-associated metabolic decline (Figures 3 and 4). These results suggest metabolic changes related to the ageing process that can be reverted after treatment with AAV9-mTERT. Interestingly, some of the metabolites identified in the gene therapy TERT experiment were similar to those identified in the metabolic signature of aging. When we specifically studied some of these metabolites in the different experimental groups (Figure 4) we confirmed that they decreased associated with aging and that this decline was significantly reversed by TERT treatment.
Table 2.
Selection of biomarkers distinguishing one and two year old AAV9-GFP and comparison of two year old AAV9-GFP and AAV9-TERT treated mice.
| Rt (min) |
m/z (Da/e) |
Ion | ID | AAV9-eGFP 2yr old/ 1yr old |
2yr old AAV9-mTERT/ 2yr old AAV9-eGFP |
||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Fold- change |
p-value | Fold- change |
p-value | ||||
|
| |||||||
| 6.15 | 717.5180 | [M+FA]− | SM32:2 | 0.36 | 1.12 × 10−6 | 2.63 | 1.30 × 10−6 |
| 3.88 | 564.3295 | [M+FA]− | LysoPC(18:2) | 0.60 | 1.62 × 10−6 | 1.52 | 1.34 × 10−4 |
| 6.42 | 283.2631 | [M-H]− | (18/0) Stearic acid | 0.48 | 1.86 × 10−6 | 1.84 | 4.96 × 10−6 |
| 4.41 | 528.3083 | ND | ND | 0.26 | 2.01 × 10−6 | 3.26 | 2.50 × 10−6 |
| 3.62 | 512.2981 | [M+FA]− | LysoPC(14:0) | 0.50 | 3.01 × 10−6 | 1.90 | 3.71 × 10−6 |
| 5.81 | 255.2321 | [M-H] | (16/0) Palmitic acid | 0.44 | 3.62 × 10−6 | 1.75 | 1.21 × 10−5 |
| 4.10 | 540.3247 | [M+FA]− | LysoPC(16:0) | 0.77 | 3.75 × 10−6 | 1.32 | 1.98 × 10−5 |
| 5.20 | 277.2150 | [M-H]− | (18/3n-6) Gamma-Linolenic acid | 0.41 | 4.15 × 10−6 | 1.85 | 7.34 × 10−5 |
| 4.37 | 297.2425 | ND | ND | 0.39 | 4.35 × 10−6 | 1.91 | 7.04 × 10−4 |
| 5.80 | 305.2474 | [M-H]− | (20/3n-9) 5,8,11-Eicosatrienoic acid | 0.28 | 6.07 × 10−6 | 2.07 | 5.61 × 10−3 |
| 5.20 | 301.2155 | [M-H]− | (20/5n-3) 5,8,11,14,17-Eicosapentaenoic acid | 0.32 | 7.38 × 10−6 | 2.23 | 2.33 × 10−4 |
| 4.00 | 500.2761 | [M-H]− | LysoPE(20:4) | 0.51 | 1.08 × 10−5 | 1.79 | 1.42 × 10−4 |
| 3.02 | 423.1231 | ND | ND | 0.21 | 1.42 × 10−5 | 4.53 | 4.72 × 10−5 |
| 2.59 | 300.2017 | ND | ND | 0.47 | 1.50 × 10−5 | 2.11 | 2.30 × 10−4 |
| 3.68 | 586.3120 | [M+FA]− | LysoPC(20:5) | 0.30 | 1.77 × 10−5 | 2.28 | 1.98 × 10−4 |
| 5.37 | 321.2028 | ND | ND | 0.25 | 1.81 × 10−5 | 2.65 | 7.38 × 10−4 |
| 3.94 | 552.3198 | [M+FA-MFA]− | LysoPC(22:6) | 0.53 | 1.86 × 10−5 | 1.80 | 1.34 × 10−5 |
| 5.03 | 277.2156 | [M-H]− | (18/3n-3) Alpha-Linolenic acid | 0.32 | 3.78 × 10−5 | 2.27 | 6.49 × 10−5 |
| 7.57 | 944.5649 | ND | ND | 1.61 | 3.82 × 10−5 | 0.57 | 1.79 × 10−4 |
| 2.66 | 326.7101 | ND | ND | 0.41 | 4.14 × 10−5 | 2.70 | 1.51 × 10−4 |
| 3.93 | 588.3280 | [M+FA]− | LysoPC(20:4) | 0.53 | 4.26 × 10−5 | 1.96 | 3.45 × 10−6 |
| 5.99 | 603.2928 | ND | ND | 0.35 | 4.38 × 10−5 | 1.57 | 2.31 × 10−5 |
| 4.30 | 566.3456 | [M+FA]− | LysoPC(18:1) | 0.61 | 4.68 × 10−5 | 2.95 | 3.04 × 10−5 |
| 2.58 | 195.1178 | ND | ND | 0.36 | 5.15 × 10−5 | 2.35 | 1.51 × 10−4 |
| 2.66 | 652.4113 | ND | ND | 0.47 | 5.71 × 10−5 | 2.02 | 8.87 × 10−6 |
| 4.00 | 524.2758 | [M-H]− | LysoPE(22:6) | 0.53 | 5.76 × 10−5 | 1.98 | 1.11 × 10−4 |
| 2.60 | 388.2538 | ND | ND | 0.53 | 5.95 × 10−5 | 2.46 | 7.18 × 10−5 |
| 5.67 | 329.2474 | [M-H]− | (22/5n-3) 7,10,13,16,19-Docosapentaenoic acid | 0.33 | 6.06 × 10−5 | 1.50 | 3.44 × 10−3 |
| 5.99 | 281.2479 | [M-H]− | (18/1 n-9) Oleic acid | 0.49 | 6.09 × 10−5 | 1.82 | 2.49 × 10−6 |
| 4.02 | 612.3282 | [M+FA]− | LysoPC(22:6) | 0.58 | 6.12 × 10−5 | 1.31 | 4.63 × 10−4 |
| 3.97 | 564.3266 | [M+FA]− | LysoPC(18:2) | 0.71 | 9.93 × 10−5 | 1.74 | 5.97 × 10−5 |
| 6.01 | 331.2632 | [M-H]− | (22/4n-6) 7,10,13,16-Docosatetraenoic acid | 0.47 | 1.03 × 10−4 | 1.68 | 8.65 × 10−6 |
| 4.02 | 588.3294 | [M+FA]− | LysoPC(20:4) | 0.60 | 1.08 × 10−4 | 1.35 | 4.59 × 10−2 |
| 3.93 | 476.2758 | [M-H]− | LysoPE(18:2) | 0.59 | 1.22 × 10−4 | 2.03 | 2.96 × 10−4 |
| 6.11 | 281.2470 | [M-H]− | (18/1n-x) | 0.40 | 2.88 × 10−4 | 2.33 | 9.40 × 10−5 |
| 5.99 | 621.3031 | ND | ND | 0.36 | 3.01 × 10−4 | 1.83 | 3.35 × 10−5 |
| 6.66 | 638.4756 | [M+FA]− | PC(0-24:0/0:0) | 0.52 | 4.04 × 10−4 | 2.31 | 9.07 × 10−5 |
| 6.80 | 733.5489 | [M+FA]− | SM(d18:1/15:0) | 0.57 | 4.23 × 10−4 | 2.02 | 1.01 × 10−7 |
| 5.83 | 329.2474 | [M-H]− | (22/5n-6) 4,7,10,13,16-Docosapentaenoic acid | 0.42 | 4.35 × 10−5 | 1.92 | 5.42 × 10−5 |
| 5.49 | 327.2314 | [M-H]− | (22/6n-3) 4,7,10,13,16,19-Docosahexaenoic acid | 0.48 | 6.43 × 10−4 | 1.88 | 6.99 × 10−6 |
| 4.39 | 494.3241 | [M+FA-MFA]− | LysoPC(17:0) | 0.62 | 7.43 × 10−4 | 2.63 | 1.30 × 10−6 |
| 5.55 | 303.2318 | [M-H]− | (20/4n-6) Arachidonic acid | 0.50 | 1.04 × 10−3 | 1.52 | 1.34 × 10−4 |
Peaks are listed for which there is significant correlation of wild-type samples with age [test p-values < 0.05]. Rt (min), chromatographic retention time; m/z (Da/e) [m is the neutral molecular mass value, e is the elemental charge], mass-to-charge ratio of ion detected; Ion, most intense species observed; ID, metabolite name (lipid names follow the LIPID MAPS classification system for lipids www.lipidmaps.org); ND, not determined.
Figure 3. PCA scores plot discriminating two years old AAV9-GFP from one year old AAV9-GFP and two years old AAV9-mTERT treated mice.
Duplicate sample injection data are shown (duplicate sample extracts were injected in each ionization mode as quality controls).
Figure 4. Profile plot of biomarkers.
Raw intensity data, including average group intensities, number of samples and p-values, corresponding to the highlighted biomarkers.
The existence of a metabolomic signature suggests that aging is accompanied by predictable metabolomic changes. This is akin to other aging-associated features that reflect on the appearance of tissues or in the performance of physiological systems. It is conceivable that physiological decline in important metabolic organs, such as liver, muscle, pancreas or adipose tissue, will reflect on the serum metabolite pattern. The identification of this signature is a first step to understand the metabolic alterations associated to aging and guide anti-aging interventions targeting metabolism. Recently, a study in humans demonstrated that metabolic profiles are age dependent and might reflect different aging processes, such as incomplete mitochondrial fatty acid oxidation (Yu et al. 2012), demonstrating the importance of age-related metabolomic profiles.
The age-biomarkers identified here include lipids and other small molecules such as creatine, methionine and uric acid. Whether these biomarkers play a role in aging or are due to secondary phenomena will require further investigation. Aberrant lipid homeostasis has been linked to a number of age-related diseases, including cardiovascular disease, inflammation, metabolic syndrome and type II diabetes (Maxfield & Tabas 2005). Creatine deficiency is associated with muscle loss (sarcopenia), a common condition in the elderly (Pearlman & Fielding 2006; Brosnan & Brosnan 2007). Moreover, creatine synthesis makes major demands of methionine (Brosnan & Brosnan 2007), another biomarker identified in this study whose levels decrease with age. Uric acid, which was found to increase with age, correlates with several risk factors, including renal dysfunction, hypertension, insulin resistance, and cardiovascular disease (Feig et al. 2008). A recent study using three different long-lived mouse models (caloric restriction, insulin receptor substrate 1 knockout, and Ames dwarf) has identified several plasma biomarkers of aging using 1H NMR spectroscopy (Wijeyesekera et al. 2012). In this study a panel of metabolic differences was generated for each model relative to their controls, and then the three models were compared to one another. Although the age-biomarkers identified by this approach differed between the three models, they also included lipids, such as phosphatidylcholine, and small molecules (methionine, choline, creatine) that are coincident with our present findings. Taken together, these results suggest that changes in lipid homeostasis and one-carbon metabolism may be associated to the aging process.
Finally, the fact that metabolomic changes associated to natural mouse aging can also predict aging provoked by accelerated telomere shortening owe to telomerase deficiency provides strong evidence that telomere loss significantly contributes to natural mouse aging. This is further substantiated by our findings that TERT treatment is able to reverse some of the metabolic changes associated with aging. In a similar manner, the metabolomic signature described here can be used to enquire the contribution of other molecular pathways (ie, tumor suppression) to natural mouse aging, as well as to estimate the impact of genetic modifications or treatments on mouse aging.
MATERIALS AND METHODS
Mice and samples
Mice were generated and housed at the Spanish National Cancer Research Center barrier area, where pathogen-free procedures are employed in all mouse rooms (Specific Pathogen Free (SPF) health status is assured through a comprehensive health surveillance program). Mice were under controlled room temperature and fed ad libitum with an irradiated diet (n° 2018, Harlan). All genetic backgrounds were kept under the same conditions. Wild-type mice used to establish a metabolomic signature of aging were from the following mouse lines maintained at the CNIO: AZD (SuperSIRT+/+; 100% C57BL/6 background)(Pfluger et al. 2008), AIU (ATM+/+; mixed C57BL/6-129sv background)(Barlow et al. 1996), RFC (Terc+/+; 100% C57BL/6 background)(Blasco et al. 1997), RSZ (wild-type controls for Sp53/Sp16/SARF/TgTert and Sp53/Sp16/SARF transgenics both of a mixed 75% C57BL/6-25% DBA/2 background)(Tomas-Loba et al. 2008), RPR (wild-type controls for K5-TRF2/K5-TERT; mixed 75% C57BL/6-25% DBA/2 background) (Munoz et al. 2005), RPP (wild-type controls for K5-TRF2/G1Terc−/−; 100% C57BL/6 background) (Munoz et al. 2005), RPT (XPF+/+; mixed C57BL/6-129sv background) (Tian et al. 2004) and RXS (wild-type controls for K5-TERT both mixed 75% C57BL/6-25% DBA/2 and 100% C57BL/6 backgrounds)(Gonzalez-Suarez et al. 2001). Mutant mouse lines used for comparative purposes were Sp53/Sp16/SARF and Sp53/Sp16/SARF/TgTert (mixed 75% C57BL/6-25% DBA/2 background), K5-TERT (mixed 75% C57BL/6-25% DBA/2 background) and G1 Terc−/− (pure C57BL/6 background) (Gonzalez-Suarez et al. 2001; Matheu et al. 2007; Tomas-Loba et al. 2008).
Mice studied for TERT based gene therapy have been previously described (Bernardes de Jesus et al. 2012). Separate groups of mice were tail-vein injected with 2×1012 vg (viral genomes)/animal of either AAV9-GFP at 1 year old (AAV9-GFP, n=15 [47% males]); or AAV9-GFP and AAV9-mTERT at 2 years old (AAV9-GFP, n=11 [36% males]; AAV9-mTERT, n=19 [58% males]. Sera were collected 2 months post telomerase expression.
Samples were extracted from facial vein before 12am after overnight fasting and collected in tubes without any anticoagulant. The samples were collected in a Specific Pathogen Free (SPF) area of CNIO and kept at room temperature until leaving this area (15 to 20min). Samples were maintained in ice for 10 additional minutes and centrifuged at 4°C. The serum supernatant was then aliquoted and frozen at °80C, to minimize freeze-thaw degradation, until they were analyzed. For all the samples the procedure was the same. Procedures for sample extraction and storage were similar to those employed in other wide-coverage metabolomics approaches (Dunn et al. 2011). Housed mice followed the recommendations of the Federation of European Laboratory Animal Science Associations.
Metabolic Profiling
A global metabolite profiling UPLC®-MS methodology was employed where all endogenous metabolite related features, characterized by their mass-to-charge ratio m/z and retention time Rt, are included in a subsequent multivariate analysis procedure used to study metabolic correlation between spectral data and age in the WT samples. Using the model built, age predictions of transgenic samples were obtained. Where possible, Rt-m/z features corresponding to putative biomarkers were later identified. Sample preparation and LC/MS analysis was performed as described in detail previously (Barr et al. 2010). Briefly, liquid-liquid extraction of the metabolites was performed by protein precipitation of 50 μL defrosted serum with four volumes of methanol. After brief vortex mixing the samples were incubated overnight at °20 °C. Supernatants were collected after centrifugation at 13,000 rpm for 10 minutes, and transferred to vials for UPLC®-MS analysis. Chromatography was performed on a 1 mm i.d. × 100 mm ACQUITY 1.7 μm C8 BEH column (Waters Corp., Milford, USA) using an ACQUITY UPLC® system (Waters Corp., Milford, USA). The volume of sample injected onto the column was 2 μL. The eluent was introduced into the mass spectrometer (QTOF Premier™, Waters Corp., Milford, USA) by electrospray ionisation, with capillary and cone voltages set in the positive and negative ion modes to 3200 V and 30 V, and 2800 V and 50 V respectively. The nebulisation gas was set to 600 L/h at a temperature of 350 °C. The cone gas was set to 30 L/h and the source temperature set to 150 °C. All spectra were mass corrected in real time by reference to leucine enkephalin, infused at 50 μL/min through an independent reference electrospray, sampled every 10 s. An appropriate test mixture of standard compounds was analyzed before and after the entire set of randomized, duplicated sample injections in order to examine the retention time stability (generally < 6 s variation, injection-to-injection), mass accuracy (generally < 3 ppm for m/z 400-1000, and < 1.2 mDa for m/z 50-400) and sensitivity of the system throughout the course of the run which lasted a maximum of 26 h per batch of samples injected.
The analytical methodology was designed to provide maximum coverage over phospholipids, fatty acids, and organic acids, whilst offering relatively high-throughput with minimum injection-to-injection carryover effects.
Data analysis
All data were processed using the MarkerLynx application manager for MassLynx 4.1 software (Waters Corp., Milford, USA). The LC/MS data are peak-detected and noise-reduced in both the LC and MS domains such that only true analytical peaks are further processed by the software (e.g. noise spikes are rejected). A list of intensities (chromatographic peak areas) of the peaks detected is then generated for the first chromatogram, using the Rt-m/z data pairs as identifiers. This process is repeated for each LC-MS analysis and the data sorted such that the correct peak intensity data for each Rt-m/z pair are aligned in the final data table. The ion intensities for each peak detected are then normalized, within each sample, to the sum of the peak intensities in that sample. There were no significant differences (F < Fcrit) between the total intensities used for normalization and the sample groups being compared in the study. The resulting normalized peak intensities form a single matrix with Rt-m/z pairs for each file in the dataset. All processed data, positive and negative ion modes, were mean centered and unit variance scaled during multivariate data analysis.
Multivariate data analysis
The first objective in the data analysis process was to create a multivariate model able to correlate the spectral data and WT samples’ age. The predictive multivariate modelling was carried out using the SIMCA - P+ (version 12.0.1; Umetrics, Sweden) software. Projection to Latent Structure (PLS) is a very useful statistical technique when there are a lot of highly correlated prediction variables (multicollinearity). This supervised pattern recognition method relates the two data matrices, e.g. Rt-m/z pair data (X matrix) and age (Y matrix), extracting the limited number of latent factors (which are linear combinations of the original prediction variables) that explain as much as possible the covariance the matrices and allowing that the response variable Y could be predicted from X. The performance of the PLS model was evaluated using the R2Y and Q2 parameters. R2Y provides an indication of how much of the variation within the data set can be explained by the model. Q2 parameter describes the predictive ability of model to estimate the Y data (a Q2 score between 0.6 – 1.0 is indicative of a reliable classifier)
Validation of PLS models
The best validation of a model is the analysis of an independent and representative set of samples that does not form part of the model-building population. However, the reduced sample size of wild-type mice over 100 weeks old, necessary for the prediction of the long-lived TERT transgenic mice, limited the splitting of the samples into a test and a validation group. As recommended by (Wold et al. 2001), we followed two ways of PLS model validation: cross validation and model re-estimation. Component-wise cross-validation was carried out, calculating R2Y and Q2 parameters by iteratively leaving out 1/7th of the samples from the model and predicting their group classification. Paired sample injections were randomly distributed over the cross-validation groups (7-fold), minimizing the risk of calculating falsely predictive components. Additionally, 200 parallel PLS models permuting randomly the Y data respecting their related X data set were generated. The comparison of the estimated values of R2Y and Q2 of the parallels and original model gives an indication of the statistical significance of the predictive power of the PLS model. If the original model is valid, randomization of the Y data would be expected to considerably reduce Q2. As can be seen in Supplementary figure 1, most of the models obtained through this randomization were non predictive (negative Q2).
Predictions of transgenic samples
PLS model from WT samples was used to predict ages of mice lacking telomerase activity (G1 Terc−/−), over-expressing telomerase reverse transcriptase (K5-Tert), simultaneously over-expressing K5-Tert, p53 and p19ARF, and over-expressing p53 and p19ARF.
Differences between transgenic and WT predicted ages were evaluated by calculating the unpaired student’s t-test p-values after data normalization with respect the real age.
Metabolite biomarker selection according to the age correlation
Variable Influence on Projection (VIP) in PLS models statistic represents the value of each X variable in fitting the PLS model, measuring the importance of those variables with respect to its correlation to the Y data. In the present work, Rt-m/z pair data with a larger VIP-value are the most influential for the model and those with VIP-values lower than 0.7 are considered to have small contribution to the prediction.
Additionally, Spearman’s rank correlation coefficient (ρ) for each selected metabolite biomarker was calculated. The Spearman correlation coefficient is a nonparametric measure of correlation between two variables, indicating with the sign the direction of association between the independent variable (Rt-m/z pair) and the dependent variable (Age), e.g. if Age-variable tends to increase when Rt-m/z pair increases, the Spearman correlation coefficient is positive. Only P-values derived from the Spearman’s rank correlation coefficient lower than 0.05 were considered in the metabolite biomarker selection.
Multivariate analysis of AAV9-eGFP and AAV9-mTERT mice
Principal components analysis PCA analysis of the data set was performed to enable easy visualisation of any metabolic clustering of the different groups of samples (Figures 3). In PCA, the data matrix is reduced to a series of principal components (PCs), each a linear combination of the original Rt-m/z pair peak areas. Each successive PC explains the maximum amount of variance possible, not accounted for by the previous PCs.
The orthogonal partial least-squares to latent structures discriminant analysis (OPLS-DA) method were used for the identification of metabolites contributing to the clustering observed in the PCA plots. The performance of the OPLS-DA model was evaluated using Q2Y parameter, which describes the predictive ability of model to estimate the Y data, and calculated by iteratively leaving out samples from the model and predicting their group classification. Paired sample injections were randomly distributed over the cross-validation groups (7-fold), minimizing the risk of calculating falsely predictive components. Appropriate filtration of the loading profile associated with the OPLS-DA predictive components resulted in a set of candidate biomarkers that were further evaluated by calculating group percentage changes and unpaired Student’s t-test p-values.
One-way ANOVA with Tukey’s Multiple Comparison Test has been used for statistical comparisons of Figure 4.
Metabolite biomarker identification
Exact molecular mass data from redundant m/z peaks corresponding to the formation of different parent (e.g. cations in the positive ion mode, anions in the negative ion mode, adducts, multiple charges) and product (formed by spontaneous “in-source” CID) ions were first used to help confirm the metabolite molecular mass. This information was then submitted for database searching, either in-house or using the online ChemSpider database (www.chemspider.com) where the Kegg, Human Metabolome Database and Lipid Maps data source options were selected. MS/MS data analysis highlights neutral losses or product ions, which are characteristic of metabolite groups and can serve to discriminate between database hits.
Supplementary Material
Supplementary figure 1. Validation of WT ageing - PLS model by permuting randomly the Age data respecting to the X data set. These re-estimations of the model evaluate the probability to get a good fit with random response data. R2 parameter (green triangles) estimates how well the model fits the data and Q2 parameter (blue squares) provides an estimate of how well the model predicts the Age data. Permutation of Age data reduces considerably Q2, being negative or non-predictive for most of the re-estimated models.
ACKNOWLEDGEMENTS
We are indebted to Cristina Alonso and Jonathan Barr from OWL for generation and analysis of the metabolomic profiles. Work in the laboratory of M.A.B. was funded by Spanish Ministry of Science and Innovation Projects SAF2008-05384 and CSD2007-00017, European Union FP7 Projects 2007-A-201630 (GENICA) and 2007-A-200950 (TELOMARKER), European Research Council Advanced Grant GA#232854, Körber Foundation, Fundación Botín and Fundación Lilly. Work in the laboratory of J.M.M is funded by grants from the MICINN (CIBERehd), the European Union (HEPADIP), the NIH (AT-1576, AT-4896) and ETORTEK-2008.
Footnotes
AUTHOR CONTRIBUTIONS
A. T-L generated the mice presented in Fig 1 and 2 of the study and collected the samples. B.B.J. generated the mice presented in Fig 3 and 4 of the study and collected the samples. M.A.B. and J.M.M. designed the experimental plan, analysed and interpreted the data. M.A.B. directed the project and wrote the paper.
DISCLOSURES
MAB is a co-founder of Life Length, S.L. a biotechnology company that commercializes telomere length tests; and JMM is a co-founder of OWL a metabolomics-based biotechnology company.
REFERENCES
- Allen J, Davey HM, Broadhurst D, Heald JK, Rowland JJ, Oliver SG, Kell DB. High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat Biotechnol. 2003;21:692–696. doi: 10.1038/nbt823. [DOI] [PubMed] [Google Scholar]
- Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shiloh Y, Crawley JN, Ried T, Tagle D, Wynshaw-Boris A. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- Barr J, Vazquez-Chantada M, Alonso C, Perez-Cormenzana M, Mayo R, Galan A, Caballeria J, Martin-Duce A, Tran A, Wagner C, Luka Z, Lu SC, Castro A, Le Marchand-Brustel Y, Martinez-Chantar ML, Veyrie N, Clement K, Tordjman J, Gual P, Mato JM. Liquid chromatography-mass spectrometry-based parallel metabolic profiling of human and mouse model serum reveals putative biomarkers associated with the progression of nonalcoholic fatty liver disease. J Proteome Res. 2010;9:4501–4512. doi: 10.1021/pr1002593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardes de Jesus B, Vera E, Schneeberger K, Tejera AM, Ayuso E, Bosch F, Blasco MA. Telomerase gene therapy in adult and old mice delays aging and increases longevity without increasing cancer. EMBO Mol Med. 2012;4:1–14. doi: 10.1002/emmm.201200245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Brosnan JT, Brosnan ME. Creatine: endogenous metabolite, dietary, and therapeutic supplement. Annu Rev Nutr. 2007;27:241–261. doi: 10.1146/annurev.nutr.27.061406.093621. [DOI] [PubMed] [Google Scholar]
- Cutler RG, Mattson MP. Sphingomyelin and ceramide as regulators of development and lifespan. Mech Ageing Dev. 2001;122:895–908. doi: 10.1016/s0047-6374(01)00246-9. [DOI] [PubMed] [Google Scholar]
- Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, Brown M, Knowles JD, Halsall A, Haselden JN, Nicholls AW, Wilson ID, Kell DB, Goodacre R. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 2011;6:1060–1083. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- Feig DI, Kang DH, Johnson RJ. Uric acid and cardiovascular risk. N Engl J Med. 2008;359:1811–1821. doi: 10.1056/NEJMra0800885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cao I, Garcia-Cao M, Tomas-Loba A, Martin-Caballero J, Flores JM, Klatt P, Blasco MA, Serrano M. Increased p53 activity does not accelerate telomere-driven ageing. EMBO Rep. 2006;7:546–552. doi: 10.1038/sj.embor.7400667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Suarez E, Geserick C, Flores JM, Blasco MA. Antagonistic effects of telomerase on cancer and aging in K5-mTert transgenic mice. Oncogene. 2005;24:2256–2270. doi: 10.1038/sj.onc.1208413. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Suarez E, Samper E, Ramirez A, Flores JM, Martin-Caballero J, Jorcano JL, Blasco MA. Increased epidermal tumors and increased skin wound healing in transgenic mice overexpressing the catalytic subunit of telomerase, mTERT, in basal keratinocytes. EMBO J. 2001;20:2619–2630. doi: 10.1093/emboj/20.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera E, Samper E, Martin-Caballero J, Flores JM, Lee HW, Blasco MA. Disease states associated with telomerase deficiency appear earlier in mice with short telomeres. EMBO J. 1999;18:2950–2960. doi: 10.1093/emboj/18.11.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klunk WE, Xu CJ, McClure RJ, Panchalingam K, Stanley JA, Pettegrew JW. Aggregation of beta-amyloid peptide is promoted by membrane phospholipid metabolites elevated in Alzheimer’s disease brain. J Neurochem. 1997;69:266–272. doi: 10.1046/j.1471-4159.1997.69010266.x. [DOI] [PubMed] [Google Scholar]
- Lee HW, Blasco MA, Gottlieb GJ, Horner JW, 2nd, Greider CW, DePinho RA. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- Lewis GD, Farrell L, Wood MJ, Martinovic M, Arany Z, Rowe GC, Souza A, Cheng S, McCabe EL, Yang E, Shi X, Deo R, Roth FP, Asnani A, Rhee EP, Systrom DM, Semigran MJ, Vasan RS, Carr SA, Wang TJ, Sabatine MS, Clish CB, Gerszten RE. Metabolic signatures of exercise in human plasma. Sci Transl Med. 2010;2:33ra37. doi: 10.1126/scitranslmed.3001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheu A, Maraver A, Klatt P, Flores I, Garcia-Cao I, Borras C, Flores JM, Vina J, Blasco MA, Serrano M. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–379. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- Maxfield FR, Tabas I. Role of cholesterol and lipid organization in disease. Nature. 2005;438:612–621. doi: 10.1038/nature04399. [DOI] [PubMed] [Google Scholar]
- Munoz P, Blanco R, Flores JM, Blasco MA. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat Genet. 2005;37:1063–1071. doi: 10.1038/ng1633. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Wilson ID. Opinion: understanding ‘global’ systems biology: metabonomics and the continuum of metabolism. Nat Rev Drug Discov. 2003;2:668–676. doi: 10.1038/nrd1157. [DOI] [PubMed] [Google Scholar]
- Pearlman JP, Fielding RA. Creatine monohydrate as a therapeutic aid in muscular dystrophy. Nutr Rev. 2006;64:80–88. doi: 10.1301/nr.2006.feb.80-88. [DOI] [PubMed] [Google Scholar]
- Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci U S A. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raamsdonk LM, Teusink B, Broadhurst D, Zhang N, Hayes A, Walsh MC, Berden JA, Brindle KM, Kell DB, Rowland JJ, Westerhoff HV, van Dam K, Oliver SG. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat Biotechnol. 2001;19:45–50. doi: 10.1038/83496. [DOI] [PubMed] [Google Scholar]
- Tian M, Shinkura R, Shinkura N, Alt FW. Growth retardation, early death, and DNA repair defects in mice deficient for the nucleotide excision repair enzyme XPF. Mol Cell Biol. 2004;24:1200–1205. doi: 10.1128/MCB.24.3.1200-1205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas-Loba A, Flores I, Fernandez-Marcos PJ, Cayuela ML, Maraver A, Tejera A, Borras C, Matheu A, Klatt P, Flores JM, Vina J, Serrano M, Blasco MA. Telomerase reverse transcriptase delays aging in cancer-resistant mice. Cell. 2008;135:609–622. doi: 10.1016/j.cell.2008.09.034. [DOI] [PubMed] [Google Scholar]
- Vera E, Bernardes de Jesus B, Foronda M, Flores JM, Blasco MA. The rate of increase of short telomeres predicts longevity in mammals. Cell Reports. 2012 doi: 10.1016/j.celrep.2012.08.023. In Press. [DOI] [PubMed] [Google Scholar]
- Wijeyesekera A, Selman C, Barton RH, Holmes E, Nicholson JK, Withers DJ. Metabotyping of long-lived mice using 1H NMR spectroscopy. J Proteome Res. 2012;11:2224–2235. doi: 10.1021/pr2010154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold S, Sjöström M, Eriksson L. PLS-regression: a basic tool of chemometrics. Chemometrics and Intelligent Laboratory Systems. 2001;58:109–130. [Google Scholar]
- Yu Z, Zhai G, Singmann P, He Y, Xu T, Prehn C, Romisch-Margl W, Lattka E, Gieger C, Soranzo N, Heinrich J, Standl M, Thiering E, Mittelstrass K, Wichmann HE, Peters A, Suhre K, Li Y, Adamski J, Spector TD, Illig T, Wang-Sattler R. Human serum metabolic profiles are age dependent. Aging Cell. 2012 doi: 10.1111/j.1474-9726.2012.00865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1. Validation of WT ageing - PLS model by permuting randomly the Age data respecting to the X data set. These re-estimations of the model evaluate the probability to get a good fit with random response data. R2 parameter (green triangles) estimates how well the model fits the data and Q2 parameter (blue squares) provides an estimate of how well the model predicts the Age data. Permutation of Age data reduces considerably Q2, being negative or non-predictive for most of the re-estimated models.