Abstract
Pathogens use numerous methods to subvert host immune responses, including the modulation of host IL-10 production by diverse cell types. However, the B cell sources of IL-10 and their overall influence on innate and cellular immune responses have not been well characterized during infections. Using Listeria as a model pathogen, infection drove the acute expansion of a small subset of regulatory B cells (B10 cells) that potently suppress inflammation and autoimmunity through the production of IL-10. Unexpectedly, spleen bacteria loads were 92–97% lower in B10 cell-deficient CD19−/− mice, in mice depleted of mature B cells, and in mice treated with CD22 mAb to preferentially deplete B10 cells before infection. By contrast, the adoptive transfer of wild type B10 cells reduced bacterial clearance by 38-fold in CD19−/− mice through IL-10-dependent pathways. B10 cell depletion using CD22 mAb significantly enhanced macrophage phagocytosis of Listeria and their production of IFN-γ, TNF-α, and nitric oxide ex vivo. Accelerated bacteria clearance following B10 cell depletion significantly reduced Ag-specific CD4+ T cell proliferation and cytokine production, but did not alter CD8+ T cell responses. B10 cell regulatory function during innate immune responses was nonetheless dependent on cognate interactions with CD4+ T cells since B10 cells deficient in IL-10, MHC-II or IL-21 receptor expression did not influence Listeria clearance. Thus, Listeria manipulates immune responses through a strategy of immune evasion that involves the preferential expansion of endogenous B10 cells that regulate the magnitude and duration of both innate and cellular immune responses.
Keywords: B cells, Listeria monocytogenes, innate immunity, regulatory B cells, B10 cells
INTRODUCTION
Infectious organisms use numerous methods to subvert host immune responses, including the modulation of host cytokine production (1, 2). Listeria monocytogenes is a frequently used model for both infection and immune response evasion in mice (3). This facultative Gram-positive bacterium also causes listeriosis in humans. Listeria typically replicates within the cytosol of macrophages and epithelial cells, which protects the bacteria from deletion during humoral immune responses. Listeria and several other pathogens also induce serum IL-10 (4), a potent cytokine that can facilitate pathogen survival by negatively regulating both innate and acquired host immune responses (5–9). Consequently, IL-10−/− mice are resistant to infection with Listeria and other pathogens (7, 10, 11). Multiple cell types are capable of producing IL-10, including activated macrophages, T and B lymphocytes, mast cells, dendritic cells, and keratinocytes (5, 12, 13). How these distinct IL-10 producing cell subsets individually dictate the quality, quantity, and direction of host immune responses is generally unknown.
In addition to the classical immune responses that are induced during pathogen infections, host immunoregulatory pathways may also become activated to limit the magnitude and duration of immune responses and to prevent excessive immunopathology. B cell IL-10 production has been implicated in the negative regulation of immune responses against bacteria, helminths and parasitic protozoa. B cell-derived IL-10 can suppress Salmonella immunity by inhibiting neutrophils, natural killer cells, and inflammatory T cells (14). IL-10 produced by adoptively transferred peritoneal cavity B-1 cells also inhibits Borrelia hermsii clearance in B cell-deficient mice (15). B cells from Brudia pahangi infected mice produce IL-10, which down regulates B cell expression of the B7-1 and B7-2 costimulatory molecules (16). Schistosoma mansoni infection also induces spleen IL-10-secreting B cells in mice, which protects against allergic hypersensitivity (17, 18). B cells also produce IL-10 following Leishmania major infection, which can inhibit dendritic cell IL-12 production in vitro (8). Despite these individual findings, the B cell sources of IL-10 have not been well characterized in most cases and their overall influence on innate and cellular immune responses during infections has not been examined.
A subset of immunoregulatory B cells has been functionally identified in mice and humans by their ability to express IL-10 within 5 h of ex vivo stimulation (13, 19, 20). These rare IL-10-competent B cells have been functionally labeled as “B10 cells” to distinguish them from other regulatory B cell subsets that are known to exist (21, 22). Regulatory B10 cells limit inflammation and disease in mouse models of contact hypersensitivity, experimental autoimmune encephalomyelitis, lupus, allergy, and collagen-induced arthritis (20, 23–26). Regulatory B10 cells are found in the spleens of naïve mice at low frequencies (1–5%), where they predominantly represent a subset of the CD1dhiCD5+CD19hi B cell subpopulation (13, 20, 27) that shares overlapping cell surface markers with multiple phenotypically-defined B cell subsets (23, 28, 29). Agonistic CD40 signals or LPS can also “mature” additional CD1dhiCD5+ B10 progenitor (B10pro) cells to acquire IL-10 competence (21, 30, 31). Cognate interactions between T cells and B10 cells induce Ag-specific regulatory B10 effector cells that secrete Il-10 and regulate autoimmunity in vivo (32). IL-10-competent regulatory B cells that parallel mouse B10pro and B10 cells have also been identified in healthy and autoimmune humans (33). The capacity of human and mouse B10pro and B10 cells to express IL-10 is central to their negative regulation of inflammation, autoimmunity, and adaptive and innate immune responses (13, 20, 26, 30, 32, 34–36). B cell expression of cytoplasmic IL-10 protein parallels both their expression of IL-10 transcripts and secretion of IL-10 as measured by ELISA (13, 20, 26, 30, 32, 34–36). The current studies demonstrate that Listeria infection induces acute B10 cell expansion and IL-10 production in mice, which inhibits macrophage activation. As a consequence, bacteria loads are increased, thereby promoting T cell expansion and the development of cellular immunity. Conversely, B10 cell-deficiency or depletion uniquely reshapes the course and magnitude of both innate and cellular immune responses during infections.
MATERIALS AND METHODS
Mice and immunotherapy
C57BL/6, Il-10−/− (B6.129P2-Il10tmlCgn/J), and Tiger (B6.129S6-Il10tm1Flv/J) mice (37) were from The Jackson Laboratory (Bar Harbor, ME). A gene dose-dependent decrease in IL-10 production was not observed in homozygous Tiger mice, which occurs with T cells (37). MHC-II−/− (B6.129-H2-Ab1tm1GruB2mtmJaeN17) mice (38) were from Taconic Farms, Inc. (Hudson, NY). CD19−/− mice (39) were backcrossed with C57BL/6 mice for 14 generations. CD22−/−CD154Tg mice (31) were homozygous at both genetic loci. OT-II transgenic (Thy1.2+) mice (40) were crossed to Thy1.1+ mice to generate OT-II/Thy1.1-expressing T cells. The OVA-specific TCR transgenic line (OT-I) was as described (41). CD20−/−and IL-10−/−CD20−/− mice were as described (20). IL-21−/− mice were as described (42). All mice were bred in a specific pathogen-free barrier facility and used at 8–12 wk of age.
Total B cells were depleted in vivo using sterile, endotoxin-free CD20 mAb (MB20-11, IgG2c) as described (43). B10 cells were depleted in vivo using sterile, endotoxin-free CD22 (MB22-10, IgG2c) mAb as described (26, 27). CD20, CD22 or isotype-matched control mAb (250 μg in 200 μl PBS) were injected into the peritoneal cavity. All animal studies and procedures were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and were approved by the Duke University Institutional Animal Care and Use Committee.
Cell preparation and immunofluorescence analysis
Single-cell leukocyte suspensions from spleens and peripheral lymph nodes (pooled bronchial, axillary, and inguinal) were generated by gentle dissection. For macrophage assays, splenocytes were adhered to 6-well plates for 3 h. The plates were then washed twice with sterile PBS. Adherent macrophages (>90% F4/80+CD11b+) were removed by scraping and stimulated with LPS (1 μg/ml) for 4 or 24 h.
FITC-, PE-, PE Cy5-, PE Cy7-, or allophycocyanin (APC)-conjugated CD1d (1B1), CD4 (H129.19), CD8 (53-6.7), CD5 (53-7.3), CD11b (M1/70), CD11c (N418), CD19 (6D5), F4/80 (BM8), CD49b (DX-5), CD80 (16-10A1), CD86 (GL1), Ly6C (AL21), Ly6G (1A8), and Thy1.1 (HIS5.1) mAbs were from BD Biosciences (San Diego, CA). Intracellular cytokine staining was performed using cytofix/cytoperm (BD Biosciences) with mAbs reactive with IL-10 (JES5-16E3), IFN-γ (XMG1.2), and TNF-α (MP6-XT22) (from eBioscience, San Diego, CA). Macrophages were identified by cell surface F4/80 and CD11b expression. For two- to six-color immunofluorescence analysis, single-cell suspensions (106 cells) were stained at 4°C using predetermined optimal concentrations of mAb for 20 min as described (44). Background staining was assessed using nonreactive, isotype-matched control mAbs (Caltag Laboratories, San Francisco, CA). Cells with the forward and side light scatter properties of lymphocytes or macrophages were analyzed using a BD FACSCanto II (BD Biosciences). Dead cells were detected using a LIVE/DEAD Fixable Violet Dead Cell Stain Kit (Life Technologies) before cell surface staining. All flow cytometry experiments were gated on viable, single lymphocytes or macrophages. For macrophage intracellular cytokine staining, splenocytes were harvested 1 d following infection and incubated with brefeldin A (1 μg/ml; eBioscience) for 5 h in the absence of stimulation. For Ag-specific T cell intracellular cytokine staining, lymphocytes were stimulated in vitro with OVA329-339 or OVA257-264 (10 μg/ml; American Peptide, Sunnyvale, CA) in the presence of brefeldin A (1 μg/ml) for 5 h before staining.
B10 cell analysis
Intracellular IL-10 expression was visualized by immunofluorescence staining and analyzed by flow cytometry as described (45). Briefly, isolated leukocytes or purified cells were resuspended (2 × 106 cells/ml) in complete medium (RPMI 1640 media containing 10% FCS, 200 μg/ml penicillin, 200 U/ml streptomycin, 4 mM L-glutamine, and 5 × 10−5 M 2-ME, all from Life Technologies, Grand Island, NY) with LPS (10 μg/ml, Escherichia coli serotype 0111:B4; Sigma-Aldrich), phorbol 12-myristate 13-acetate (50 ng/ml; Sigma-Aldrich), ionomycin (500 ng/ml; Sigma-Aldrich, St. Louis, MO), and monensin (2 μM; eBioscience) for 5 h in 48-well flat-bottom plates. In some experiments, the cells were incubated for 48 h with an agonistic anti-mouse CD40 mAb (1 μg/ml; HM40-3; BD Biosciences) as described (45). For IL-10 detection, Fc gamma receptors II and III were blocked with mouse Fc receptor mAb (2.4G2; BD Biosciences), and dead cells were identified using a LIVE/DEAD Fixable Violet Dead Cell Stain Kit (Life Technologies) before cell surface molecule immunofluorescence staining. Cells stained for cell surface molecules were fixed and permeabilized using a Cytofix/Cytoperm kit (BD Biosciences), according to the manufacturer’s instructions, and stained with APC-conjugated anti-mouse IL-10 mAb.
Infection, recovery and labeling of bacteria
Mice were infected i.v. with Listeria strain 10403S engineered to express chicken ovalbumin (OVA; kindly provided by Dr. Hao Shen, University of Pennsylvania, Philadelphia, PA) (40, 46). For immunizations, bacteria were inoculated into brain heart infusion media supplemented with erythromycin (5 μg/ml) overnight at 37°C. An aliquot (1 ml) of the overnight bacteria culture was washed twice with sterile PBS and diluted in PBS for infections as described (47). Mice received diluted bacteria (8 × 105 cfu, 0.1 LD50 in 200 μl) through the lateral tail vein. The inoculum was subsequently serially diluted on brain heart infusion plates and counted to verify the infection dose. At this infection dose, there are no symptoms of weight loss or core body temperature changes.
To recover bacteria from infected mice, spleens were harvested and homogenized using a Tissuemizer (Fischer Scientific, Pittsburgh, PA) in 4 ml of sterile PBS. The spleen homogenates were serially diluted in sterile PBS 10-fold until a dilution of 105. A 100 μl aliquot of each dilution was spread on brain heart infusion plates supplemented with erythromycin (5 μg/ml) and incubated at 37°C for 24–48 h before the colonies were counted to determine bacteria numbers.
Bacteria were labeled as previously described (48, 49). Briefly, bacteria were incubated with 100 μM CFSE (Invitrogen) for 10 minutes at 37°C, washed in PBS and immediately used for the in vitro infection of macrophages. CFSE-labeling did not affect bacteria viability (data not shown) as described (50). The number of phagocytosed bacteria was determined using a similar protocol as that used for other Gram-positive bacteria. Briefly, macrophages were infected at a multiplicity of infection of 10 for 2 h before the macrophages were washed, harvested, and assessed for CFSE+ bacteria. Extracellular bacteria were eliminated from the analysis by adding trypan blue (0.2% final) prior to flow cytometry analysis as described (51).
Lymphocyte subset isolation and adoptive transfer experiments
Splenic B cells from naïve wild type, IL-10−/−, CD22−/−CD154Tg, CD20−/− or CD20−/−IL-10−/− mice were isolated using CD19 mAb-coated microbeads (Miltenyi Biotec, Auburn, CA) according to the manufacturer’s instructions. When necessary, B cells were enriched a second time using a fresh magnetic-activated cell sorting column to obtain >95% cell purities. CD1dhiCD5+ and CD1dloCD5− B cells were identified by immunofluorescence staining and purified (95–98%) using a FACSAria flow cytometer (BD Biosciences). B cell subset isolation did not induce detectable IL-10 production (data not shown). After purification, 106 cells were immediately transferred into appropriate recipient mice through a lateral tail vein. Without appropriate ex vivo stimulation, the donor B cells remained IL-10 negative following transfers into recipient mice. Splenic CD4+ or CD8+ T cells, and macrophages were purified using CD4+ T cell isolation kits, CD8+ T cell isolation kits, and CD11b mAb-coated microbeads (Miltenyi Biotec), respectively. When necessary, the cells were enriched a second time using a fresh magnetic-activated cell sorting column to obtain >95% cell purities. Isolated CD4+ or CD8+ T cells from naïve OT-II/Thy1.1+ or OT-I mice were labeled with CFSE (5 μM) for 10 min at 37°C, with 3×106 cells transferred i.v. into recipient mice.
ELISAs
For assessing IFN-γ production, purified macrophages were stimulated in vitro with media or LPS (1 μg/ml) for 4 or 24 h. Tissue culture supernatant fluid IFN-γ concentrations were measured by ELISA (BD Biosciences) according to the manufacturer’s instructions. For assessing IL-10 secretion, purified CD19+, CD1dhiCD5+, or CD1dloCD5− B cells (4 × 105/0.2 ml of complete medium) from naïve or Listeria infected mice were cultured in triplicate using 96-well flat-bottom tissue culture plates. Culture supernatant fluid IL-10 concentrations were quantified using IL-10 OptEIA ELISA kits (BD Pharmingen) following the manufacturer’s protocols.
Statistical analyses
Differences between means were assessed using the Student’s t-test (GraphPad Prism software, version 5.00, San Diego, CA). Significant differences between data sets that were not normally distributed or were of unequal variance were determined using the Mann–Whitney rank sum test. P values of <0.05 were considered significant.
RESULTS
B10 cells expand during L. monocytogenes infection
The effects of infection on spleen CD19+ B cells was assessed in wild type mice given L. monocytogenes engineered to express chicken OVA (40, 46). Following 3 d of infection, there was little change in spleen B cell or CD4+ T cell numbers relative to control mice given PBS (Fig. 1A). B cell numbers increased significantly by day 5 along with a 46% (p=0.03) reduction in CD4+ T cell frequencies, with normal cell numbers by 8 d post-infection, as described (52). Listeria infection alone did not induce measurable IL-10 expression in B cells cultured ex vivo with monensin alone (Fig. 1B). Therefore, changes in regulatory B10 cell frequencies and numbers during Listeria infection were assessed following ex vivo B cell stimulation with PMA, ionomycin, and monensin (PIM) for 5 h to induce cytoplasmic IL-10 accumulation that was visualized by immunofluorescence staining with flow cytometry analysis as described (45). LPS was added to the 5 h PIM cultures (LPIM) as it marginally enhances B10 cell enumeration (30). The advantage of using cytoplasmic IL-10 expression to enumerate B10 cells is that this technique demonstrates the frequency and numbers of B cells that are competent to express IL-10 and the level of IL-10 synthesis once induced.
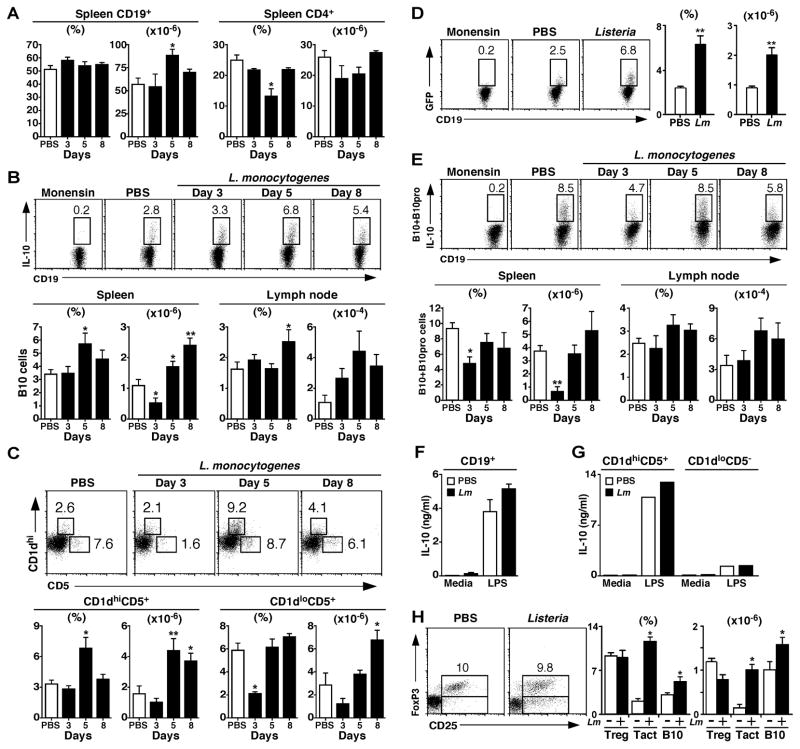
Figure 1. B10 cells expand following Listeria infection.
Wild type mice were given PBS or infected with 8×105 cfu of Listeria-OVA on d 0. (A) Spleen B and T cell numbers after Listeria infection. At the indicated times post-infection, CD19+ B cell and CD4+ T cell frequencies and numbers were determined by immunofluorescence staining with flow cytometry analysis. Data from PBS-treated control mice were unchanged across all time points (d 3, 5, and 8 post-infection) and were therefore pooled. Values represent mean (± SEM, n=3–6/group) cell frequencies or numbers. (B) B10 cells expand after Listeria infection. At the indicated times post-infection, spleen and lymph node lymphocytes were stimulated ex vivo with LPS, PMA, ionomycin, and monensin (LPIM) for 5 h and stained for cell surface CD19 and intracellular IL-10. Representative histograms show IL-10 expression by viable and single spleen CD19+ B cells. Cells from infected mice that were cultured with monensin alone served as negative staining controls. Numbers indicate the frequencies of cells within the indicated gates. Data from PBS-treated mice were unchanged across all time points (d 3, 5, and 8 post-infection) and were therefore pooled. Values in the bar graphs represent mean (± SEM, n=10) frequencies or numbers of IL-10+ B cells. (C) The spleen CD1dhiCD5+CD19+ B cell subset expands after Listeria infection. At the indicated times post-infection, CD1dloCD5+CD19+ and CD1dhiCD5+CD19+ B cell subset frequencies and numbers were determined. The data are presented as in (B, n=7). (D) B cell GFP expression in IL-10 reporter Tiger mice. Five d after infection, splenocytes were stimulated ex vivo with LPIM for 5 h before staining for cell surface CD19 and evaluating CD19+ B cell intracellular GFP expression by flow cytometry. Numbers indicate cell frequencies within the indicated gates. Values in bar graphs represent mean (± SEM, n=5) frequencies or numbers of CD19+GFP+ B cells 5 d post-infection. (E) Spleen and lymph node B10+B10pro cell frequencies and numbers following Listeria infection. B10+B10pro cells were enumerated after 48 h of stimulation with CD40 mAb, with LPIM added during the final 5 h of culture. Representative histograms show IL-10 expression by viable and single spleen CD19+ B cells, with bar graphs as in (B, n=10). (F–G) Listeria infection enhances B10 cell IL-10 production. Purified spleen (F) CD19+ B cells or (G) CD1dhiCD5+ and CD1dloCD5− B cell subsets were isolated from mice 5 d after infection and cultured for 72 h with or without LPS. Culture supernatant fluid IL-10 concentrations were measured by ELISA. Bar graphs indicate mean IL-10 concentrations (± SEM, n=3). (H) Treg cells do not expand following Listeria infection. Histograms show representative FoxP3 and CD25 expression by splenic CD4+ T cells 5 d post-infection. Values in the bar graphs represent mean (± SEM, n=5) frequencies or numbers of CD25+FoxP3+ CD4+ Treg cells, activated CD25+FoxP3− CD4+ T cells (Tact), or B10 cells. (A–H) Significant differences between sample means for mice given PBS or infected are indicated: *; p<0.05; **; p <0.01.
Following Listeria infection, spleen B10 cell numbers dropped by day 3, but expanded 2-fold by 5 d post-infection (p≤0.03) and remained elevated in the spleen and lymph nodes by d 8 when compared with uninfected control mice. The spleen CD1dhiCD5+ B cell subset was also significantly expanded by 5 d post-infection, while the CD1dloCD5+ B cell subset did not increase significantly (Fig. 1C). When B10 cell numbers were quantified using Tiger IL-10/GFP reporter mice (37), similar results were obtained. Listeria infection of Tiger mice did not induce detectable GFP expression, but GFP+ cell frequencies and numbers paralleled B10 cell cytoplasmic IL-10 expression in wild type mice following LPIM stimulation (Fig. 1B and D). Thus, B10 cell numbers increased significantly following infection.
Agonistic CD40 signals induce B10pro cell maturation, which renders these cells competent to express IL-10 following 5 h of LPIM stimulation (26, 30). Both B10 and B10pro cells (B10+B10pro) are quantified in this assay, as the B cells that acquire IL-10 competence in vitro (e.g., matured B10pro cells) cannot be differentiated from preexisting B10 cells that express cytoplasmic IL-10 after 5 h of LPIM stimulation. Spleen B10+B10pro cell numbers decreased by 87% (p=0.02) on d 3 post-infection, but returned to uninfected levels by 5–8 d post-infection (Fig. 1E). Infection did not significantly affect lymph node B10+B10pro cell numbers.
Listeria infection induced modest but measurable IL-10 secretion by purified spleen B cells cultured ex vivo without stimulation (Fig. 1F). LPS stimulation in vitro induced significant B cell IL-10 production, with more IL-10 produced by B cells isolated from Listeria-infected mice. The CD1dhiCD5+ B cell subset was enriched 10-fold for IL-10-secreting B cells relative to CD1dloCD5− B cells (Fig. 1G). Thus, Listeria infection was predominantly activating a subset of B cells within the CD1dhiCD5+ B10+B10pro cell pool.
Spleen FoxP3+CD25+CD4+ regulatory T cell (Treg) frequencies or numbers were not significantly changed following Listeria infection (d 5, Fig. 1H). However, the frequency and number of activated CD4+ T cells (FoxP3−CD25+) increased by five-fold. Thus, the B10 cell subset preferentially expanded following Listeria infection, while the B10pro and Treg cell subsets did not.
B10 cells inhibit L. monocytogenes clearance
The impact of B cells and B10 cells on Listeria infection was assessed using three different mouse models; CD19−/− mice, CD20 mAb-treated mice, and CD22 mAb-treated mice. Most B cell subsets develop normally in CD19−/− mice (53), but they are deficient in B10 cells (13, 26, 30, 32, 34, 35, 54, 55). Relative to wild type littermates, total spleen B cell numbers were 32% (p=0.006) lower in uninfected and day 5 infected CD19−/− mice (Fig. 2A). However, B10 cell numbers were reduced by 83% (p=0.001) in CD19−/− mice, and this did not change with infection. CD20 mAb treatment depletes all mature B cells and B10 cells (13, 20, 27, 56). Total B cell depletion by CD20 mAb reduced B10 cell and B cell numbers by >97% (p<0.0001) in both control and Listeria-infected mice (Fig. 2B). CD22 mAb treatment does not significantly reduce peripheral B cell numbers but depletes the majority of spleen CD1dhiCD5+ B cells, which results in significant B10 cell reductions (26, 31, 57). The term “depletion” as used in conjunction with CD20 and CD22 mAb treatments in this study designates significant reductions in the frequencies and numbers of tissue lymphocytes that either express a mature B cell phenotype (CD20 mAb), a CD1dhiCD5+ phenotype (CD22 mAb), or that are able to express IL-10 (CD20 or CD22 mAb) as published (13, 20, 26, 27, 31, 56, 57). CD22 mAb treatment reduced CD1dhiCD5+ (B10 cell-enriched) cell numbers by 60% (p=0.01) in uninfected mice and by 83% (p=0.003) in infected mice, but did not significantly affect total B cell numbers (Fig. 2C). B10 cell numbers were reduced by 80% (p=0.002) in uninfected mice and by 85% (p=0.03) in infected mice following CD22 mAb treatment. In contrast with CD20 and CD22 mAb-treated mice, B10 cell numbers increased 1.6-fold (p=0.02) in control mAb-treated mice by 5 d post-infection when compared with uninfected littermates (Fig. 2B–C). Thus, CD19-deficiency, total B cell depletion using CD20 mAb, and CD22 mAb treatment each significantly reduced B10 cell numbers following infection.
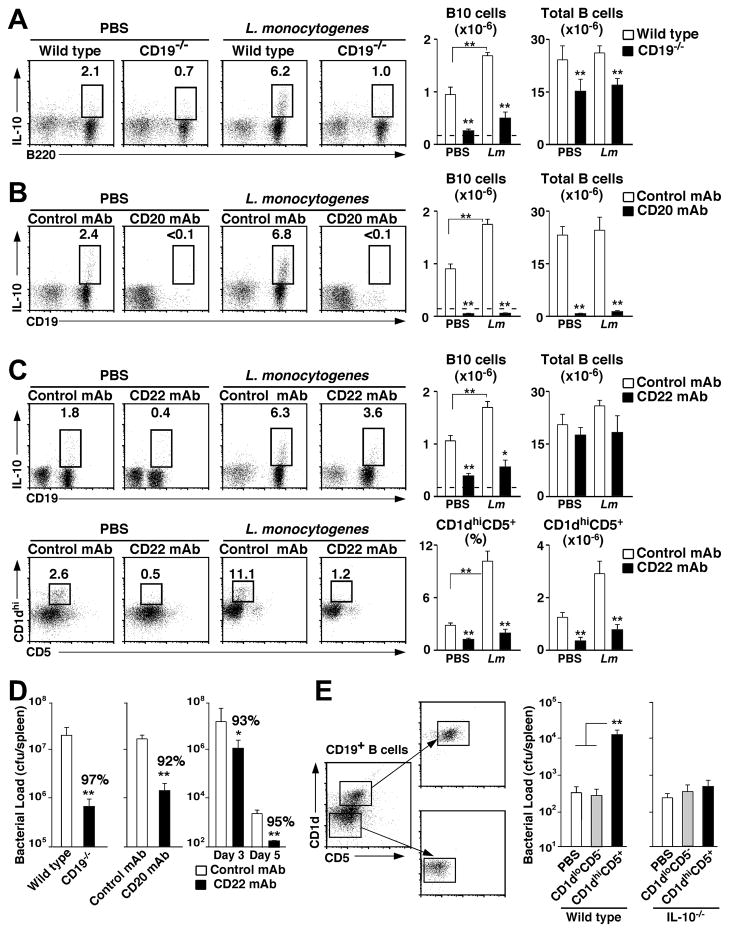
Figure 2. B10 cells inhibit Listeria clearance.
Spleen B10 cell numbers in (A) CD19−/− mice, (B) mice given either CD20 or isotype control mAb on d −7 or (C) mice given CD22 or isotype control mAb on d −7 and −1 before Listeria infection or PBS treatment on d 0. Five d later, splenocytes were stimulated ex vivo with LPIM for 5 h before staining for cell surface (A) B220, (B) CD19, or (C) CD19, CD1d, and CD5, and intracellular IL-10 expression. (D) CD19-deficiency, total B cell depletion, and CD22 mAb treatment enhance bacterial clearance. Mice treated as in (A–C) were infected with Listeria on day 0, and the number of bacteria present within their spleens was quantified 3 d (and 5 d, for CD22 mAb) later. Values represent mean (± SEM, n=8) bacterial loads in each set of mice. (E) B10 cells inhibit bacterial clearance through IL-10-dependent mechanisms. Representative fractionation of purified spleen CD19+ B cells from wild type or IL-10−/− mice into CD1dhiCD5+ (B10 cell enriched) or CD1dloCD5− B cell populations by cell sorting. The cells were then adoptively transferred into CD19−/− mice. One d later, the recipient mice were treated with PBS or infected with Listeria. Bar graphs represent mean (± SEM, n=7) spleen bacterial loads 3 d later. (A–E) Significant differences between means are indicated: *; p<0.05; **; p <0.01.
Spleen bacteria loads were reduced significantly (p≤0.01) by CD19-deficiency (97%), total B cell depletion using CD20 mAb (92%), or CD22 mAb treatment (93%, notice different scale on bar graph) as early as 3 d post-infection when compared with wild type or control mAb-treated littermates (Fig. 2D). Spleen bacteria loads also remained 95% lower 5 d post-infection in CD22 mAb-treated mice when compared with control mAb-treated littermates. Similar results were obtained for liver bacteria loads (data not shown). Thus, CD19-deficiency, total B cell depletion, and CD22 mAb treatment significantly accelerated Listeria clearance in vivo.
Adoptive transfer experiments were used to verify that bacterial clearance was accelerated due to the absence of regulatory B10 cells. Purified spleen CD1dhiCD5+ (B10 cell-enriched) and CD1dloCD5− (B10 cell-deficient) B cell subsets from wild type mice were adoptively transferred into B10 cell-deficient CD19−/− mice prior to Listeria infection (Fig. 2E). Spleen bacteria loads were 38-fold higher in CD19−/− mice given CD1dhiCD5+ B cells in comparison with littermates given PBS or CD1dloCD5− B cells 3 d post-infection (p=0.01). CD1dhiCD5+ and CD1dloCD5− B cells from IL-10−/− mice were also transferred into CD19−/− mice before infection. However, spleen bacteria levels were equivalent in mice that received PBS, IL-10−/− CD1dhiCD5+ B cells, or IL-10−/− CD1dloCD5− B cells (3 d post-infection; p=0.29). Thus, three independent mouse models where B cells or subsets of B cells were depleted and adoptive transfer experiments demonstrated that B10 cells inhibited Listeria clearance through IL-10-dependent mechanisms.
B10 cell depletion enhances macrophage cytokine production
The effect of B10 cell depletion on spleen macrophage and neutrophil numbers and activation was assessed ex vivo 1 d after Listeria infection. In wild type mice, CD22 mAb treatment did not significantly alter the frequencies or numbers of spleen CD11b+Ly6C+ macrophages or CD11b+Ly6G+ neutrophils (Fig. 3A). Similarly, CD22 mAb treatment did not result in spleen F4/80+CD11b+ macrophage activation as measured by ex vivo CD80 and CD86 expression (Fig. 3B). However, CD22 mAb treatment significantly enhanced ex vivo macrophage production of IFN-γ (2.6-fold, p=0.02) and TNF-α (40%, p=0.03) when compared with macrophages from control mAb-treated mice (Fig. 3C). By contrast, CD22 mAb treatment did not significantly affect IFN-γ production by CD11c+ (dendritic) or DX-5+ (natural killer) cells (data not shown). Thus, in vivo CD22 mAb treatment significantly enhanced macrophage cytokine production.
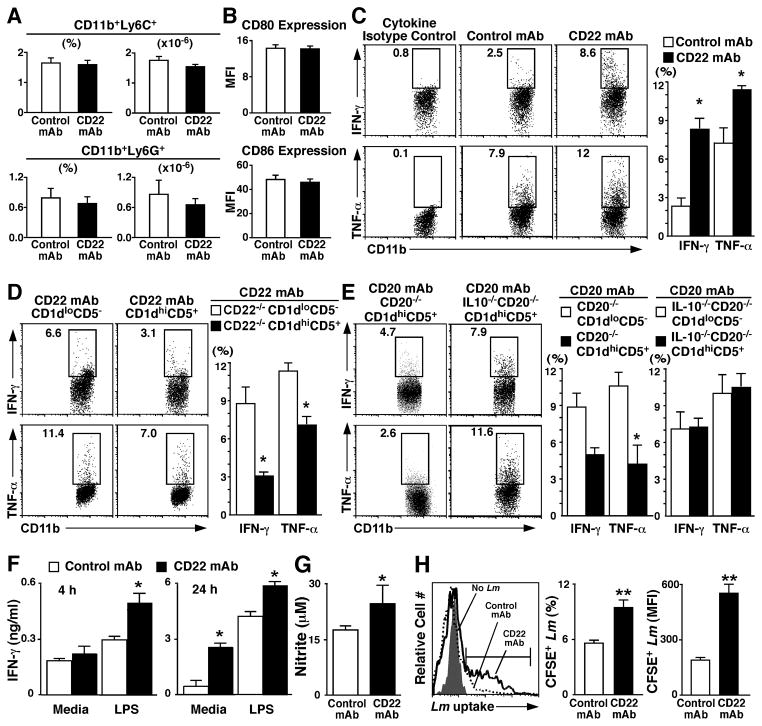
Figure 3. B10 cell depletion enhances macrophage cytokine production and phagocytosis.
In all experiments, wild type mice were given CD22 or isotype control mAb on d −7 and −1 and infected with Listeria on d 0. (A) CD22 mAb-treatment does not alter macrophage or neutrophil numbers in vivo. Frequencies and numbers of viable spleen CD11b+Ly6C+ macrophages and CD11b+Ly6G+ neutrophils were quantified ex vivo 1 d post-infection with Listeria by immunofluorescence staining with flow cytometry analysis. Values represent mean (± SEM, n=3) cell frequencies and numbers among spleen mononuclear cells. (B) CD22 mAb-treatment does not induce macrophage activation in vivo. Spleen F4/80+CD11b+ macrophage CD80 and CD86 mean fluorescence staining intensities (MFI) were quantified ex vivo 1 d post-infection with Listeria by immunofluorescence staining with flow cytometry analysis. Values represent mean (± SEM, n=3) MFI values with isotype control mAb background staining subtracted. (C) CD22 mAb-treatment enhances macrophage cytokine production. Representative histograms show intracellular IFN-γ and TNF-α expression by viable, single F4/80+CD11b+ spleen macrophages directly ex vivo 1 d post-infection with Listeria as assessed by immunofluorescence staining. Background staining was determined using isotype-matched control antibodies. (D) Adoptively transferred CD1dhiCD5+ B10 cells inhibit macrophage cytokine production in CD22 mAb-treated mice. In parallel with the experiments in (C), mice were given either control or CD22 mAb. Subsequently, the mice were given purified spleen CD1dhiCD5+ or CD1dloCD5−CD19+ B cells from CD22−/−CD154Tg mice. One day later, the recipient mice were infected with Listeria, with spleen macrophage IFN-γ and TNF-α expression quantified as in (C). (C,D) Bar graphs show mean (± SEM, n=5) frequencies of cytokine expressing cells within the indicated gates. (E) Spleen CD1dhiCD5+ B cells inhibit macrophage cytokine production through IL-10 production. Wild type mice were given control or CD20 mAb as in figure 2B–C. Subsequently, the mice were given either purified spleen CD1dhiCD5+ or CD1dloCD5− CD19+ B cells from CD20−/− or IL-10−/−CD20−/− mice. One day later, the recipient mice were infected with Listeria, with spleen macrophage IFN-γ and TNF-α expression assessed as in (C). (F–G) CD22 mAb-treatment in vivo regulates spleen macrophage cytokine and nitric oxide production ex vivo. Adherent macrophages isolated from CD22 or control mAb-treated Listeria-infected mice generated as in (C) were stimulated with LPS. Mean (± SEM) supernatant fluid (F) IFN-γ (4 or 24 h cultures, n=6) or (G) nitrate (24 h culture, n=6) concentrations were quantified by ELISA. (H) CD22 mAb-treatment enhances macrophage phagocytosis of CFSE-labeled Listeria. Mice were treated with CD22 or isotype-matched control mAb on d −7 and −1. Spleen CD11b+ macrophages were isolated on d 0 and cultured with CFSE-labeled Listeria for 2 h. The histogram shows representative intracellular CFSE fluorescence of macrophages from CD22 or control mAb-treated mice cultured without (No Lm) or with CFSE-labeled Listeria. The horizontal line indicates the flow cytometry gate used for quantifying CFSE-positive cells. Bar graphs represent mean (± SEM, n=8) frequencies of CFSE+ macrophages or mean fluorescent intensities (MFI) of CD11b+CFSE+ cells. (A–H) Significant differences between the sample means are indicated: *; p<0.05; **; p <0.01.
Adoptive transfer experiments were carried out in parallel with the experiments described above to verify that the absence of regulatory B10 cells following CD22 mAb treatment enhanced infection-induced macrophage cytokine production. First, spleen CD1dhiCD5+ and CD1dloCD5− B cells were purified from CD22−/−CD154Tg mice, in which the majority of CD1dhi B cells are B10 cells (31). These B cell subsets were then adoptively transferred into wild type mice that were given CD22 mAb prior to Listeria infection. Since the adoptively transferred B cells are CD22-deficient, they are not affected by CD22 mAb treatment (57). IFN-γ and TNF-α production by macrophages from CD22 mAb-treated mice (Fig. 3C) were reduced by 67% and 54% (p=0.01), respectively, by the adoptive transfer of CD1dhiCD5+ CD22−/− B cells but were not affected by the adoptive transfer of CD1dloCD5− CD22−/− B cells (Fig. 3D). In fact, IFN-γ and TNF-α production were virtually identical in macrophages from CD22 mAb-treated mice given CD1dhiCD5+ CD22−/− B cells and untreated control mice. Thus, the CD22 mAb-induced effects were directly attributable to CD1dhiCD5+ B cell depletion.
A second set of adoptive transfer experiments were carried out to confirm that CD1dhiCD5+ B cell regulation of macrophage cytokine production was IL-10 dependent. Spleen CD1dhiCD5+ and CD1dloCD5− B cells were purified from CD20−/− or IL-10−/−CD20−/− mice and adoptively transferred into wild type mice that were given CD20 mAb prior to Listeria infection. Macrophage IFN-γ and TNF-α production were elevated similarly in mice given either CD20 or CD22 mAb relative to macrophages isolated from control mAb-treated Listeria-infected mice (Fig. 3C–E). However, the adoptive transfer of CD1dhiCD5+ B cells from CD20−/− mice reduced IFN-γ and TNF-α production in mice given CD20 mAb by 43% and 60%, respectively, in comparison with CD1dloCD5− CD20−/− B cells (Fig. 3E). Neither CD1dhiCD5+ nor CD1dloCD5− B cells from IL-10−/−CD20−/− mice affected IFN-γ and TNF-α production by macrophages from CD20 mAb-treated mice (p=0.8). Thus, endogenous B10 cells found within the CD1dhiCD5+ B cell subpopulation negatively regulated macrophage cytokine production through IL-10-dependent pathways.
To verify that the ex vivo changes in macrophage IFN-γ secretion were durable, macrophages purified from CD22 or control mAb-treated mice were cultured with LPS in vitro. Following 4 h of LPS stimulation, macrophages from CD22 mAb-treated mice secreted 62% (p=0.03) more IFN-γ than macrophages from control mAb-treated mice (Fig. 3F). After 24 h of culture without LPS stimulation, macrophages from CD22 mAb-treated mice secreted 9.2-fold (p=0.01) more IFN-γ than macrophages from control mAb-treated mice. After LPS stimulation, macrophages from CD22 mAb-treated mice secreted 30% (p=0.03) more IFN-γ than macrophages from control mice. Nitric oxide production by macrophages from CD22 mAb-treated mice was also 40% (p=0.04) higher than for macrophages from control mAb-treated mice (Fig. 3G). Thereby, B10 cell negative regulation of macrophage cytokine and nitric oxide production in vivo was persistent and not reversed by ex vivo stimulation.
B10 cell depletion enhances macrophage phagocytosis of Listeria
Since macrophages play an important role in host defense during Listeria infections, the ability of spleen CD11b+ macrophages from mice treated with either CD22 or control mAb for 7 d were analyzed for their ability to phagocytose CFSE-labeled Listeria ex vivo in the absence of extrinsic stimulation. Trypan blue was used to quench CFSE fluorescence from cell surface-associated bacteria during flow cytometry analysis as described (51). Within two hours, twice as many macrophages from CD22 mAb-treated mice (p=0.01) had internalized bacteria when compared with macrophages from control mAb-treated mice (Fig. 3H). Furthermore, macrophages from CD22 mAb-treated mice that had internalized bacteria contained twice as many (p=0.01) bacteria as macrophages from control mAb-treated mice. Thus, in vivo B10 cell depletion following CD22 mAb treatment significantly enhanced the ex vivo phagocytic capacity of spleen macrophages.
B10 cell depletion reduces Ag-specific CD4+ T cell activation
The effect of CD22 mAb-induced B10 cell depletion on cellular immune responses was quantified in wild type mice. CD22 mAb treatment did not significantly affect spleen CD4+ or CD8+ T cells in either uninfected or Listeria-infected mice (Fig. 4A). However, as shown in Figure 1A, Listeria infection results in a transient reduction in CD4+ T cell numbers, and this effect was blocked in mice given CD22 mAb but not control mAb. Therefore, B10 cell depletion reduced the effect of Listeria infection on spleen CD4+ T cell frequencies.
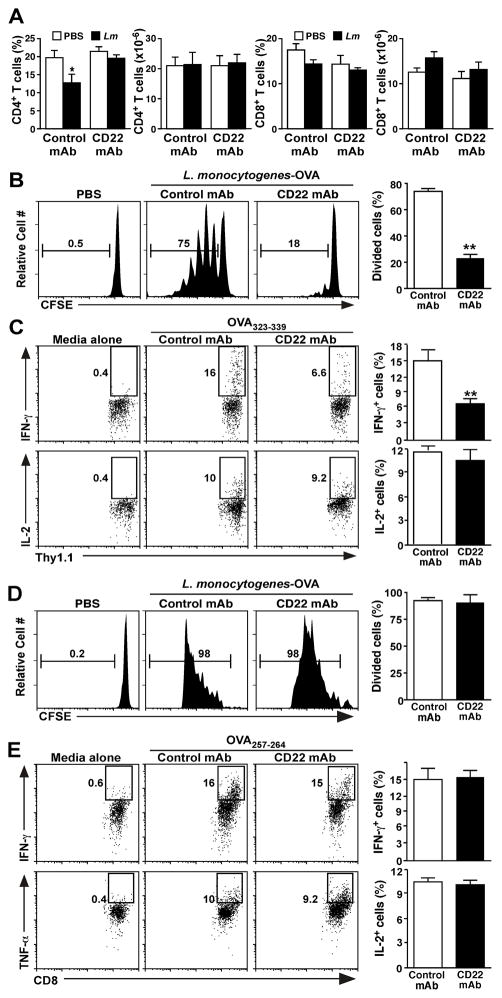
Figure 4. B10 cells regulate Ag-specific CD4+ T cell activation.
Wild type mice were given CD22 or control mAb on day −7 and −1 and were infected with Listeria-OVA on day 0. (A) CD22 mAb treatment normalizes spleen CD4+ T cell numbers following Listeria infection. Values represent mean (± SEM, n=6) frequencies and numbers of total spleen CD4+ and CD8+ T cells 5 d post-infection or treatment with PBS. (B) CD22 mAb-treatment reduces Ag-specific spleen CD4+ T cell proliferation. CD22 or control mAb-treated and infected mice received CFSE-labeled spleen Thy1.1+OT-II CD4+ T cells on d −1. The in vivo proliferation of adoptively transferred CFSE-labeled Thy1.1+OT-II T cells was assessed by CFSE dilution 3 d following infection. Representative histograms demonstrate CFSE staining for uninfected mice (PBS) versus infected mice treated with control or CD22 mAb, with the percentages of dividing cells indicated. Bar graph values represent mean (± SEM, n=6) frequencies of spleen Thy1.1+OT-II T cells with diluted CFSE in infected mice. (C) CD22 mAb-treatment reduces Ag-specific CD4+ T cell cytokine production. Splenocytes from control or CD22 mAb treated mice given Thy1.1+ OT-II T cells as in (B) were harvested 3 d post-Listeria infection and stimulated ex vivo with media alone or OVA323-339 peptide for 4 h in the presence of brefeldin A. IFN-γ, IL-2, and TNF-α production were assessed by intracellular staining with flow cytometry analysis. Representative dot plots show relative frequencies of cytokine expressing spleen Thy1.1+ T cells within the indicated gates. Bar graphs represent mean (± SEM, n=6) frequencies of Thy1.1+ OT-II T cells expressing each cytokine. (D) B10 cell depletion does not reduce Ag-specific spleen CD8+ T cell proliferation. CD22 or control mAb-treated and infected mice received CFSE-labeled spleen Thy1.1+ OT-I CD8+ T cells on d −1. The in vivo proliferation of adoptively transferred CFSE-labeled spleen Thy1.1+ OT-I T cells was assessed as in (B; n=6). (E) B10 cell depletion does not reduce spleen Ag-specific CD8+ T cell cytokine production. Cytokine production by adoptively transferred Thy1.1+ OT-I T cells was assessed as in (C) using OVA257-264 peptide (n=4). (A–E) Significant differences between the indicated sample means among control mAb and CD22 mAb treated mice are indicated: *; p<0.05; **; p <0.01.
The involvement of B10 cells during Ag-specific CD4+ T cell proliferation and function was quantified in vivo using Thy1.1+ congenic mice that express an OVA-specific T cell receptor transgene (40). CD4+ T cells purified from OT-II mice were CFSE-labeled and adoptively transferred into CD22 or control mAb-treated wild type mice 1 d prior to Listeria infection. Uninfected control mice were given PBS. Spleen Thy1.1+ OT-II T cell proliferation was assessed 3 d later by measuring CFSE dilution. Relatively few OT-II T cells from CD22 mAb-treated mice diluted CFSE (p<0.001, Fig. 4B), while OT-II T cells from infected mice given control mAb proliferated vigorously, with 75% of the T cells diluting CFSE. Consistent with these results, half as many OT-II T cells isolated from CD22 mAb-treated mice were induced to produce IFN-γ when stimulated ex vivo with OVA323-339 peptide in comparison with OT-II T cells from infected mice given control mAb (p=0.005; Fig. 4C). CD4+ OT-II T cell production of IL-2 was equivalent in both CD22 and control mAb-treated mice after infection. Thus, CD4+ T cell proliferation and cytokine production were reduced in mice given CD22 mAb to deplete CD1dhiCD5+ B10 cells.
The effects of B10 cell depletion using CD22 mAb on Ag-specific CD8+ T cell proliferation and function was also quantified in vivo using mice that express an OVA-specific T cell receptor transgene (41). CD8+ T cells purified from OT-I mice were CFSE-labeled and adoptively transferred into CD22 or control mAb-treated wild type mice 1 d prior to Listeria infection, with spleen OT-I T cell proliferation assessed 3 d later. OT-I T cells proliferated vigorously in infected mice given control or CD22 mAb, with >95% of the T cells diluting CFSE (p=0.4; Fig. 4D). Ag-specific OT-I T cells from B10 cell-depleted or control-treated mice also produced similar amounts of IFN-γ when stimulated ex vivo with OVA257-264 peptide (p=0.7; Fig. 4E). Thus, B10 cell depletion using CD22 mAb reduced Ag-specific CD4+ but not CD8+ T cell proliferation and cytokine production in vivo.
B10 cell regulation is dependent on cognate interactions with CD4+ T cells
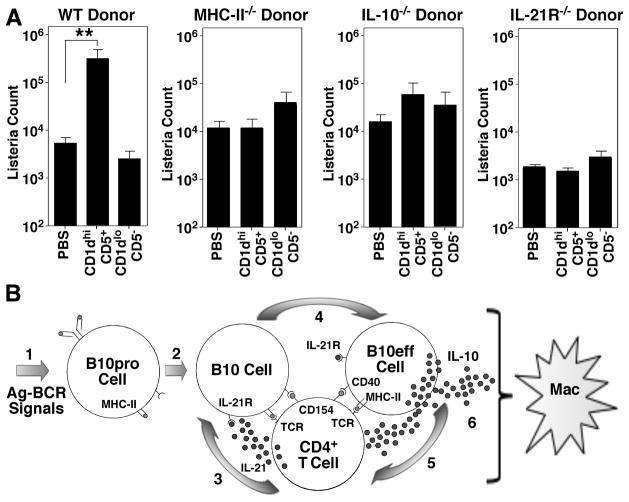
B10 cell regulation of T cell-mediated autoimmunity requires cognate interactions with CD4+ T cells through MHC-class II and IL-21 dependent pathways to induce B10 cell effector function and IL-10 production (32). Whether inflammatory signals or bacterial products can induce regulatory B10 cell function independent of cognate T cell interactions was therefore examined using CD19−/− mice infected with Listeria. Remarkably, the adoptive transfer of wild type CD1dhiCD5+ B cells resulted in a subsequent 58-fold (p≤0.01) increase in spleen bacteria loads following Listeria infection when compared to littermates given PBS or CD1dloCD5− B cells (Fig. 5A). By contrast, the adoptive transfer of either CD1dhiCD5+ or CD1dloCD5− B cells from MHC-II−/− mice, IL-21 receptor deficient (IL-21R−/−) mice, or IL-10−/− mice did not significantly affect bacterial clearance. Thus, IL-10, IL-21, and MHC-II expression were required to elicit regulatory B10 cell function during innate immune responses to this bacterial pathogen.
Figure 5.
B10 cell regulation of innate immune responses to Listeria requires cognate interactions with CD4+ T cells. (A) B10 cell MHC class II, IL-10, and IL-21R expression are required for regulation of innate immune responses. CD19−/− mice were either given PBS or purified spleen CD1dhiCD5+ or CD1dloCD5− B cells from naïve wild type (WT), MHC-II−/−, IL-10−/−, or IL-21R−/− mice 1 day before Listeria infection. Values represent mean (±SEM) spleen bacterial counts from ≥4 mice in each group 3 days after infection. Significant differences between the indicated sample means are shown: **, p<0.01. (B) Model for pathogen (Ag)-specific B10 cell function. B cells capture Ags that trigger appropriate BCR signals (step 1) and promote IL-10-competent B10pro cell development. During immune responses (step 2), B10pro cells present peptides to Ag-specific CD4+ T cells through cognate interactions that induce T cell activation and CD40/CD154 interactions. Activated T cells may produce IL-21 locally, which binds to proximal B10 cell IL-21R (step 3). IL-21R signals induce B10 cell IL-10 production and effector function (B10eff, step 4), which may negatively regulate Ag-specific T cell function (step 5). Indirectly, B10 effector cell production of IL-10 in the local environment negatively regulates macrophage activation and effector function.
DISCUSSION
The current studies demonstrate that Listeria manipulates early immune responses through a strategy of immune evasion that involves preferential B10 cell expansion. Specifically, spleen and lymph node B10 cell frequencies and numbers were acutely expanded by day 5 of infection (Fig. 1), with B10 cell production of IL-10 dramatically reducing bacteria clearance (Fig. 2). Enhanced bacteria clearance in the absence of B10 cells or following their in vivo depletion paralleled significantly improved macrophage phagocytosis of bacteria (Fig. 3H) and their significantly increased production of IFN-γ, TNF-α and nitric oxide ex vivo (Fig. 3C–G). Remarkably, B10 cell negative regulation of macrophage cytokine production persisted even when the macrophages were restimulated ex vivo with LPS (Fig. 3F). Enhanced bacteria clearance following B10 cell depletion using CD22 mAb also significantly reduced Ag-specific CD4+ T cell expansion in response to Listeria infection, but did not reduce Ag-specific CD8+ T cell expansion (Fig. 4). Thereby, B10 cell expansion normally inhibits Listeria clearance in vivo by reducing macrophage activation, which promotes bacteria persistence and the generation of foreign Ags that drive CD4+ T cell expansion. B10 cell expansion thus shapes the qualitative and quantitative nature of both innate and cellular immune responses during acute Listeria infections.
In contrast with B10 cell expansion, B10 cell depletion significantly reduced spleen and liver bacteria loads by 82–97% (Fig. 2, data not shown). This was demonstrated in three ways. First, enhanced bacteria clearance was observed in CD19−/− mice that have dramatically reduced numbers of B10 and B10pro cells, but otherwise have near normal B cell numbers (Fig. 2C, ref. 13). Enhanced clearance of Listeria in the absence of B10 cells was similar to the rapid clearance of Listeria in IL-10−/− mice (7). Second, Listeria clearance was also dramatically accelerated in mice depleted of all mature B cells using a potent CD20 mAb (Fig. 2B). This result is consistent with enhanced bacteria clearance in B cell-deficient μMT mice (58). Third, Listeria loads were significantly reduced in mice given a ligand-blocking CD22 mAb that deleted most CD1dhiCD5+ B cells and 60–80% of spleen B10 cells (Fig. 2C, refs. 31, 57). However, the vast majority of spleen B cells remain intact after CD22 mAb treatment. The elimination of CD1dhiCD5+ cells and B10 cells by select CD22 mAbs is not a consequence of the initiation of CD22 negative regulation or CD22 removal from the cell surface during B cell Ag receptor signaling (unpublished observations), but likely reflects the diversity of effects elicited by CD22 binding to appropriate in vivo ligands (59). Remarkably, CD20 mAb and the ligand-blocking CD22 mAb used in these studies deplete different percentages of B cells and utilize distinct mechanisms of action to deplete B10 cells (43, 57), but their effects on bacteria clearance were identical to those observed in CD19−/− mice without B10 cells. Moreover, B10 cell depletion by CD20 mAb and this ligand-blocking CD22 mAb can also exacerbate cellular immunity and autoimmune disease symptoms similarly (26, 31, 57). Thereby, B cell and particularly B10 cell manipulation has significant effects on bacteria clearance and innate immune responses.
Listeria initially infects macrophages, with the subsequent elicitation of Ag-specific adaptive immunity (60). Macrophages also serve as the primary in vivo mediators of Listeria clearance (61). Thereby, enhanced Listeria clearance in the absence of B10 cells is likely to result from the enhanced ability of macrophages from B10 cell-depleted mice to phagocytose bacteria (Fig. 3H) and express cytokines (Fig. 3C–E) and nitric oxide (Fig. 3G) in vivo. That B cell regulation of bacteria clearance by monocytes was B10 cell and IL-10-dependent was confirmed by adoptive transfer experiments. First, the adoptive transfer of wild type CD1dhiCD5+ B cells that are enriched for B10+B10pro cells (13, 32) reduced Listeria clearance in CD19−/− mice to levels seen in wild type mice (Fig. 2E). By contrast, the adoptive transfer of IL-10−/− CD1dhiCD5+ B cells or CD1dloCD5− B cells was without effect. Second, the adoptive transfer of CD1dhiCD5+ B cells from CD22−/−CD154Tg mice, where the majority of CD1dhiCD5+ B cells are B10 cells (31), normalized in vivo macrophage TNF-α and IFN-γ production in CD22 mAb-treated mice, while CD22−/−CD1dloCD5− B cells were without effect (Fig. 3D). Third, the adoptive transfer of CD1dhiCD5+ B cells from CD20−/− mice normalized in vivo macrophage TNF-α and IFN-γ production in CD20 mAb-treated mice, while CD20−/−CD1dhiCD5+ B cells were without effect (Fig. 3E). Fourth, the adoptive transfer of CD1dhiCD5+ B cells from IL-10−/−CD20−/− mice were unable to normalize in vivo macrophage TNF-α and IFN-γ production in CD20 mAb-treated mice. Consistent with these findings, small numbers of adoptively transferred B10 cells inhibit macrophage activation through IL-10-dependent mechanisms and thereby dramatically suppress antibody-mediated lymphoma depletion in vivo, as well as cytokine production in vivo and in vitro (56). B10 cells also negatively regulate human macrophage activation and phagocytic activity (33). Exogenous IL-10 also down-regulates macrophage cytokine production in vitro, their generation of reactive oxygen intermediates, and cell surface expression of MHC class II and other costimulatory molecules (62–64). B10 cell inhibition of Listeria clearance is also consistent with the proposed role for B1 cell-derived IL-10 in the regulation of B. hermsii clearance from the blood (15). These collective studies thereby demonstrate that IL-10 production by the B10 cell subset has substantial regulatory effects on macrophage function, including the clearance of Listeria and potentially other pathogens.
Acute expansion of the B10 cell subset after Listeria infection (Fig. 1B–D) was similar to the rapid increase in B10 cells that occurs during inflammation (13, 20, 26, 56) and the regulatory B cell increases observed with other inflammatory conditions (15–18). By contrast, there was only modest expansion of the FoxP3+CD4+ regulatory T cell subset by day 5 following infection (Fig. 1H). B10 cell expansion at early time-points in the absence of regulatory T cell expansion also occurs during experimental autoimmune encephalomyelitis in mice (26). Thereby, acute Listeria infection may preferentially expand B10 cells but not regulatory T cells to manipulate IL-10 levels and increase bacterial loads that may be normally required to drive early Treg and CD4+ T cell expansion. Rapid Listeria clearance in the absence of B10 cells following CD22 mAb-treatment may thereby limit the availability of foreign Ags and explain the dramatic reductions in CD4+ T cell proliferation and cytokine production in the current studies (Fig. 4B–C). Alternatively, the absence of B10 cells may facilitate the rapid development of potent CD8+ effector T cells that facilitate bacteria clearance. Reduced bacteria loads with augmented T cell responses have been observed in μMT mice after Listeria infection (65). Acute Listeria-driven expansion of the B10 cell subset thereby accelerates the accumulation of stimulatory foreign Ags that determine the magnitude of CD4+ but not CD8+ T cell proliferation and cytokine production.
B10 cell function appears Ag-specific in models of inflammation, and Ag receptor specificity and signaling are critical components for B10 cell function in vivo (30, 32). For example, B10 cells fail to develop in CD19−/− mice where B cell Ag receptor signaling is muted (Fig. 2A). While B cell Ag receptor signaling appears to direct B10 cell development, cognate interactions with Ag-specific T cells producing IL-21 appear critical for the development of IL-10-producing B10 effector cells (32), which may then negatively regulate cognate CD4+ T cell partners (Fig. 5B). B10 cell expansion may thereby normally limit Ag-specific CD4+ T cell proliferation and cytokine production during acute Listeria infections, or may alternatively reshape the repertoire of CD4+ T cells that expand in response to infection. Consistent with this, regulatory B10 cell inhibition of bacteria clearance required that B10 cells express IL-10, MHC class II molecules, and CD21-R (Fig. 5A), arguing that B10 cells require cognate interactions with CD4+ T cells to induce their regulatory effector functions during infections (Fig. 5B) as occurs during T cell-mediated autoimmunity (32). Given this, macrophage function is likely to be regulated downstream of B10 cell interactions with CD4+ T cells. Under these conditions, B10 cell negative regulation of monocyte function would remain Ag-specific and may only affect macrophages within the local microenvironment of ongoing immune responses.
In addition to the current studies with B cells, IL-10 produced by other immune cells is known to modulate host immune responses to diverse pathogens including Listeria, Bordetella pertussis, lymphocytic choriomeningitis virus, and vaccinia virus (6, 9, 11, 66). Gram-negative bacteria also induce B cells to express IL-10 through a MyD88-dependent pathway, which inhibits immunity (14). It is therefore possible that pathogens also utilize TLR-signaling pathways to induce B10 cell IL-10 production. Consistent with this, inflammatory signals and bacterial products are potent stimulators of B10 cell maturation and IL-10 production (30). However, B10 cells develop normally in both MyD88-deficient and pathogen-deficient mice, and B10 cell function remains Ag-specific in models of inflammation, which suggests that Ag receptor specificity and signaling are also critical components for B10 cell function in vivo (30). Given the negative regulatory functions of B10 cells and their capacity to modulate both innate and adaptive immune responses, it is likely that pathogens will utilize multiple diverse mechanisms to manipulate B10 cell function and biology to modulate immune responses to their advantage. Alterations in the numbers and/or proportions of B10 cells during acute infections may thus have major ramifications for quantitatively and qualitatively programming subsequent innate and cellular immune responses that might be manipulated for the development of vaccines to a variety of pathogens.
Acknowledgments
We thank Dr. Damian Maseda for help with these experiments and their analysis.
These studies were supported by grants from the NIH, AI56363 and Southeastern Regional Center of Excellence for Emerging Infections and Biodefense (U54 AI057157), the Lymphoma Research Foundation, and the Division of Intramural Research, National Heart, Lung, and Blood Institute, NIH.
Footnotes
Author contributions: M.H., E.T.W and T.F.T. designed the research; M.H., E.T.W., G.M.V. and R.S. performed the experiments; R.S. and W.J.L provided essential reagents and input; M.H., E.T.W., D.J.D., M.T.H. and T.F.T. analyzed the data and wrote the paper.
Conflict of Interest: TFT is a paid consultant and shareholder for Angelica Therapeutics, Inc. All other authors declare no conflict of interest.
References
- 1.Portnoy DA. Manipulation of innate immunity by bacterial pathogens. Curr Opin Immunol. 2005;17:25–28. doi: 10.1016/j.coi.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 2.Majlessi L, Lo-Man R, Leclerc C. Regulatory B and T cells in infections. Microbes Infect. 2008;10:1030–1035. doi: 10.1016/j.micinf.2008.07.017. [DOI] [PubMed] [Google Scholar]
- 3.Portnoy DA, Auerbuch V, Glomski IJ. The cell biology of Listeria monocytogenes infection: the intersection of bacterial pathogenesis and cell-mediated immunity. J Cell Biol. 2002;158:409–414. doi: 10.1083/jcb.200205009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore KW, de Waal Malefyt R, Coffman RL, O’Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 5.Brooks DG, Trifilo MJ, Edelmann KH, Teyton L, McGavern DB, Oldstone MB. Interleukin-10 determines viral clearance or persistence in vivo. Nat Med. 2006;12:1301–1309. doi: 10.1038/nm1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks DG, Walsh KB, Elsaesser H, Oldstone MBA. IL-10 directly suppresses CD4 but not CD8 T cell effector and memory responses following acute viral infection. Proc Natl Acad Sci USA. 2010;107:3018–3023. doi: 10.1073/pnas.0914500107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai WJ, Kohler G, Brombacher F. Both innate and acquired immunity to Listeria monocytogenes infection are increased in IL-10-deficient mice. J Immunol. 1997;158:2259–2267. [PubMed] [Google Scholar]
- 8.Ronet C, Hauyon-La Torre Y, Revaz-Breton M, Mastelic B, Tacchini-Cottier F, Louis J, Launois P. Regulatory B cells shape the development of Th2 immune responses in BALB/c mice infected with Leishmania major through IL-10 production. J Immunol. 2010;184:886–894. doi: 10.4049/jimmunol.0901114. [DOI] [PubMed] [Google Scholar]
- 9.Wolfe DN, Karanikas AT, Hester SE, Kennett MJ, Harvill ET. IL-10 induction by Bordetella parapertussis limits a protective IFN-γ response. J Immunol. 2010;184:1392–1400. doi: 10.4049/jimmunol.0803045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagenbaugh A, Sharma S, Dubinett SM, Wei SH, Aranda R, Cheroutre H, Fowell DJ, Binder S, Tsao B, Locksley RM, Moore KW, Kronenberg M. Altered immune responses in interleukin 10 transgenic mice. J Exp Med. 1997;185:2101–2110. doi: 10.1084/jem.185.12.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blackburn SD, Wherry EJ. IL-10, T cell exhaustion and viral persistence. Trends Microbiol. 2007;15:143–146. doi: 10.1016/j.tim.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Maynard CL, Harrington LE, Janowski KM, Oliver JR, Zindl CL, Rudensky AY, Weaver CT. Regulatory T cells expressing interleukin 10 develop from Foxp3+ and Foxp3- precursor cells in the absence of interleukin 10. Nat Immunol. 2007;8:931–941. doi: 10.1038/ni1504. [DOI] [PubMed] [Google Scholar]
- 13.Yanaba K, Bouaziz J-D, Haas KM, Poe JC, Fujimoto M, Tedder TF. A regulatory B cell subset with a unique CD1dhiCD5+ phenotype controls T cell-dependent inflammatory responses. Immunity. 2008;28:639–650. doi: 10.1016/j.immuni.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 14.Neves P, Lampropoulou V, Calderon-Gomez E, Roch T, Stervbo U, Shen P, Kuhl AA, Loddenkemper C, Haury M, Nedospasov SA, Kaufmann SH, Steinhoff U, Calado DP, Fillatreau S. Signaling via the MyD88 adaptor protein in B cells suppresses protective immunity during Salmonella typhimurium infection. Immunity. 2010;33:777–790. doi: 10.1016/j.immuni.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 15.Sindhava V, Woodman ME, Stevenson B, Bondada S. Interleukin-10 mediated autoregulation of murine B-1 B-cells and its role in Borrelia hermsii infection. PLoS ONE. 2010;5:e11445. doi: 10.1371/journal.pone.0011445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gillan V, Lawrence RA, Devaney E. B cells play a regulatory role in mice infected with the L3 of Brugia pahangi. Int Immunol. 2005;17:373–382. doi: 10.1093/intimm/dxh217. [DOI] [PubMed] [Google Scholar]
- 17.Correale J, Farez M, Razzitte G. Helminth infections associated with multiple sclerosis induce regulatory B cells. Ann Neurol. 2008;64:187–199. doi: 10.1002/ana.21438. [DOI] [PubMed] [Google Scholar]
- 18.Mangan NE, Fallon RE, Smith P, van Rooijen N, McKenzie AN, Fallon PG. Helminth infection protects mice from anaphylaxis via IL-10-producing B cells. J Immunol. 2004;173:6346–6356. doi: 10.4049/jimmunol.173.10.6346. [DOI] [PubMed] [Google Scholar]
- 19.DiLillo DJ, Horikawa M, Tedder TF. B-lymphocyte effector functions in health and disease. Immunol Res. 2011;49:281–292. doi: 10.1007/s12026-010-8189-3. [DOI] [PubMed] [Google Scholar]
- 20.Matsushita T, Yanaba K, Bouaziz JD, Fujimoto M, Tedder TF. Regulatory B cells inhibit EAE initiation in mice while other B cells promote disease progression. J Clin Invest. 2008;118:3420–3430. doi: 10.1172/JCI36030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DiLillo DJ, Matsushita T, Tedder TF. B10 cells and regulatory B cells balance immune responses during inflammation, autoimmunity, and cancer. Ann N Y Acad Sci. 2010;1183:38–57. doi: 10.1111/j.1749-6632.2009.05137.x. [DOI] [PubMed] [Google Scholar]
- 22.Ray A, Basu S, Williams CB, Salzman NH, Dittel BN. A novel IL-10-independent regulatory role for B cells in suppressing autoimmunity by maintenance of regulatory T cells via GITR ligand. J Immunol. 2012;188:3188–3198. doi: 10.4049/jimmunol.1103354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans JG, Chavez-Rueda KA, Eddaoudi A, Meyer-Bahlburg A, Rawlings DJ, Ehrenstein MR, Mauri C. Novel suppressive function of transitional 2 B cells in experimental arthritis. J Immunol. 2007;178:7868–7878. doi: 10.4049/jimmunol.178.12.7868. [DOI] [PubMed] [Google Scholar]
- 24.Madan R, Demircik F, Surianarayanan S, Allen JL, Divanovic S, Trompette A, Yogev N, Gu Y, Khodoun M, Hildeman D, Boespflug N, Fogolin MB, Grobe L, Greweling M, Finkelman FD, Cardin R, Mohrs M, Muller W, Waisman A, Roers A, Karp CL. Nonredundant roles for B cell-derived IL-10 in immune counter-regulation. J Immunol. 2009;183:2312–2320. doi: 10.4049/jimmunol.0900185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouaziz JD, Yanaba K, Tedder TF. Regulatory B cells as inhibitors of immune responses and inflammation. Immunol Rev. 2008;224:201–214. doi: 10.1111/j.1600-065X.2008.00661.x. [DOI] [PubMed] [Google Scholar]
- 26.Matsushita T, Horikawa M, Iwata Y, Tedder TF. Regulatory B cells (B10 cells) and regulatory T cells have independent roles in controlling EAE initiation and late-phase immunopathogenesis. J Immunol. 2010;185:2240–2252. doi: 10.4049/jimmunol.1001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haas KM, Watanabe R, Matsushita T, Nakashima H, Ishiura N, Okochi H, Fujimoto M, Tedder TF. Protective and pathogenic roles for B cells during systemic autoimmunity in NZB/W F1 mice. J Immunol. 2010;184:4789–4800. doi: 10.4049/jimmunol.0902391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brummel R, Lenert P. Activation of marginal zone B cells from lupus mice with type A(D) CpG-oligodeoxynucleotides. J Immunol. 2005;174:2429–2434. doi: 10.4049/jimmunol.174.4.2429. [DOI] [PubMed] [Google Scholar]
- 29.Spencer NF, Daynes RA. IL-12 directly stimulates expression of IL-10 by CD5+ B cells and IL-6 by both CD5+ and CD5− B cells: possible involvement in age-associated cytokine dysregulation. Int Immunol. 1997;9:745–754. doi: 10.1093/intimm/9.5.745. [DOI] [PubMed] [Google Scholar]
- 30.Yanaba K, Bouaziz JD, Matsushita T, Tsubata T, Tedder TF. The development and function of regulatory B cells expressing IL-10 (B10 cells) requires antigen receptor diversity and TLR signals. J Immunol. 2009;182:7459–7472. doi: 10.4049/jimmunol.0900270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poe JC, Smith SH, Haas KM, Yanaba K, Tsubata T, Matsushita T, Tedder TF. Amplified B lymphocyte CD40 signaling drives regulatory B10 cell expansion in mice. PLoS ONE. 2011;6:e22464. doi: 10.1371/journal.pone.0022464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshizaki A, Miyagaki T, DiLillo DJ, Matsushita T, Horikawa M, Kountikov EI, Spolski R, Poe JC, Leonard WJ, Tedder TF. Regulatory B cells control T cell autoimmunity through IL-21-dependent cognate interactions. Nature. 2012 doi: 10.1038/nature11501. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwata Y, Matsushita T, Horikawa M, DiLillo DJ, Yanaba K, Venturi GM, Szabolcs PM, Bernstein SH, Magro CM, Williams AD, Hall RP, St Clair EW, Tedder TF. Characterization of a rare IL-10-competent B cell subset in humans that parallels mouse regulatory B10 cells. Blood. 2011;117:530–541. doi: 10.1182/blood-2010-07-294249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe R, Ishiura N, Nakashima H, Kuwano Y, Okochi H, Tamaki K, Sato S, Tedder TF, Fujimoto M. Regulatory B cells (B10 cells) have a suppressive role in murine lupus: CD19 and B10 cell deficiency exacerbates systemic autoimmunity. J Immunol. 2010;184:4801–4809. doi: 10.4049/jimmunol.0902385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanaba K, Yoshizaki A, Asano Y, Kadono T, Tedder TF, Sato S. IL-10-producing regulatory B10 cells inhibit intestinal injury in a mouse model. Am J Pathol. 2011;178:735–743. doi: 10.1016/j.ajpath.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maseda D, Smith SH, DiLillo DJ, Bryant JM, Candando KM, Weaver CT, Tedder TF. Regulatory B10 cells differentiate into antibody-secreting cells after transient IL-10 production in vivo. J Immunol. 2012;188:1036–1048. doi: 10.4049/jimmunol.1102500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamanaka M, Kim ST, Wan YY, Sutterwala FS, Lara-Tejero M, Galan JE, Harhaj E, Flavell RA. Expression of interleukin-10 in intestinal lymphocytes detected by an interleukin-10 reporter knockin tiger mouse. Immunity. 2006;25:941–952. doi: 10.1016/j.immuni.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 38.Grusby MJ, Auchincloss H, Jr, Lee R, Johnson RS, Spencer JP, Zijlstra M, Jaenisch R, Papaioannou VE, Glimcher LH. Mice lacking major histocompatibility complex class I and class II molecules. Proc Natl Acad Sci U S A. 1993;90:3913–3917. doi: 10.1073/pnas.90.9.3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engel P, Zhou LJ, Ord DC, Sato S, Koller B, Tedder TF. Abnormal B lymphocyte development, activation and differentiation in mice that lack or overexpress the CD19 signal transduction molecule. Immunity. 1995;3:39–50. doi: 10.1016/1074-7613(95)90157-4. [DOI] [PubMed] [Google Scholar]
- 40.Foulds KE, Zenewicz LA, Shedlock DJ, Jiang J, Troy AE, Shen H. Cutting edge: CD4 and CD8 T cells are intrinsically different in their proliferative responses. J Immunol. 2002;168:1528–1532. doi: 10.4049/jimmunol.168.4.1528. [DOI] [PubMed] [Google Scholar]
- 41.Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 42.Ozaki K, Spolski R, Feng CG, Qi CF, Cheng J, Sher A, Morse HC, 3rd, Liu C, Schwartzberg PL, Leonard WJ. A critical role for IL-21 in regulating immunoglobulin production. Science. 2002;298:1630–1634. doi: 10.1126/science.1077002. [DOI] [PubMed] [Google Scholar]
- 43.Uchida J, Hamaguchi Y, Oliver JA, Ravetch JV, Poe JC, Haas KM, Tedder TF. The innate mononuclear phagocyte network depletes B lymphocytes through Fc receptor-dependent mechanisms during anti-CD20 antibody immunotherapy. J Exp Med. 2004;199:1659–1669. doi: 10.1084/jem.20040119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou LJ, Smith HM, Waldschmidt TJ, Schwarting R, Daley J, Tedder TF. Tissue-specific expression of the human CD19 gene in transgenic mice inhibits antigen-independent B lymphocyte development. Mol Cell Biol. 1994;14:3884–3894. doi: 10.1128/mcb.14.6.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsushita T, Tedder TF. Identifying regulatory B cells (B10 cells) that produce IL-10. Methods Mol Biol. 2011;677:99–111. doi: 10.1007/978-1-60761-869-0_7. [DOI] [PubMed] [Google Scholar]
- 46.Pope C, Kim SK, Marzo A, Williams K, Jiang J, Shen H, Lefrancois L. Organ-specific regulation of the CD8 T cell response to Listeria monocytogenes infection. J Immunol. 2001;166:3402–3409. doi: 10.4049/jimmunol.166.5.3402. [DOI] [PubMed] [Google Scholar]
- 47.Bouaziz JD, Yanaba K, Venturi GM, Wang Y, Tisch RM, Poe JC, Tedder TF. Therapeutic B cell depletion impairs adaptive and autoreactive CD4+ T cell activation in mice. Proc Natl Acad Sci USA. 2007;104:20882–20887. doi: 10.1073/pnas.0709205105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen Y, Kawamura I, Nomura T, Tsuchiya K, Hara H, Dewamitta SR, Sakai S, Qu H, Daim S, Yamamoto T, Mitsuyama M. Toll-like receptor 2- and MyD88-dependent phosphatidylinositol 3-kinase and Rac1 activation facilitates the phagocytosis of Listeria monocytogenes by murine macrophages. Infect Immun. 2010;78:2857–2867. doi: 10.1128/IAI.01138-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Westcott MM, Henry CJ, Cook AS, Grant KW, Hiltbold EM. Differential susceptibility of bone marrow-derived dendritic cells and macrophages to productive infection with Listeria monocytogenes. Cell Microbiol. 2007;9:1397–1411. doi: 10.1111/j.1462-5822.2006.00880.x. [DOI] [PubMed] [Google Scholar]
- 50.Westcott MM, Henry CJ, Amis JE, Hiltbold EM. Dendritic cells inhibit the progression of Listeria monocytogenes intracellular infection by retaining bacteria in major histocompatibility complex class II-rich phagosomes and by limiting cytosolic growth. Infect Immun. 2010;78:2956–2965. doi: 10.1128/IAI.01027-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pils S, Schmitter T, Neske F, Hauck CR. Quantification of bacterial invasion into adherent cells by flow cytometry. J Microbiol Met. 2006;65:301–310. doi: 10.1016/j.mimet.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 52.Mandel TE, Cheers C. Resistance and susceptibility of mice to bacterial infection: histopathology of listeriosis in resistant and susceptible strains. Infect Immun. 1980;30:851–861. doi: 10.1128/iai.30.3.851-861.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haas KM, Poe JC, Steeber DA, Tedder TF. B-1a and B-1b cells exhibit distinct developmental requirements and have unique functional roles in innate and adaptive immunity to S. pneumoniae. Immunity. 2005;23:7–18. doi: 10.1016/j.immuni.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 54.Matsushita T, Fujimoto M, Hasegawa M, Komura K, Takehara K, Tedder TF, Sato S. Inhibitory role of CD19 in the progression of experimental autoimmune encephalomyelitis by regulating cytokine response. Am J Pathol. 2006;168:812–821. doi: 10.2353/ajpath.2006.050923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe R, Fujimoto M, Ishiura N, Kuwano Y, Nakashima H, Yazawa N, Okochi H, Sato S, Tedder TF, Tamaki K. CD19 expression in B cells is important for suppression of contact hypersensitivity. Am J Pathol. 2007;171:560–570. doi: 10.2353/ajpath.2007.061279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horikawa M, Minard-Colin V, Matsushita T, Tedder TF. Regulatory B cell production of IL-10 inhibits lymphoma depletion during CD20 immunotherapy in mice. J Clin Invest. 2011;121:4268–4280. doi: 10.1172/JCI59266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haas KM, Sen S, Sanford IG, Miller AS, Poe JC, Tedder TF. CD22 ligand binding regulates normal and malignant B lymphocyte survival in vivo. J Immunol. 2006;177:3063–3073. doi: 10.4049/jimmunol.177.5.3063. [DOI] [PubMed] [Google Scholar]
- 58.Kelly-Scumpia KM, Scumpia PO, Weinstein JS, Delano MJ, Cuenca AG, Nacionales DC, Wynn JL, Lee PY, Kumagai Y, Efron PA, Akira S, Wasserfall C, Atkinson MA, Moldawer LL. B cells enhance early innate immune responses during bacterial sepsis. J Exp Med. 2011;208:1673–1682. doi: 10.1084/jem.20101715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poe JC, Tedder TF. CD22 and Siglec-G/10 in B cell function and tolerance. Trends Immunol. 2012;33:413–420. doi: 10.1016/j.it.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamon M, Bierne H, Cossart P. Listeria monocytogenes: a multifaceted model. Nat Rev Microbiol. 2006;4:423–434. doi: 10.1038/nrmicro1413. [DOI] [PubMed] [Google Scholar]
- 61.Portnoy DA. Innate immunity to a facultative intracellular bacterial pathogen. Curr Opin Immunol. 1992;4:20–24. doi: 10.1016/0952-7915(92)90118-x. [DOI] [PubMed] [Google Scholar]
- 62.Fiorentino DF, Zlotnik A, Mosmann TR, Howard M, O’Garra A. IL-10 inhibits cytokine production by activated macrophages. J Immunol. 1991;147:3815–3822. [PubMed] [Google Scholar]
- 63.Fleming SD, Campbell PA. Some macrophages kill Listeria monocytogenes while others do not. Immunol Rev. 1997;158:69–77. doi: 10.1111/j.1600-065x.1997.tb00993.x. [DOI] [PubMed] [Google Scholar]
- 64.O’Farrell AM, Liu Y, Moore KW, Mui AL. IL-10 inhibits macrophage activation and proliferation by distinct signaling mechanisms: evidence for Stat3-dependent and -independent pathways. EMBO J. 1998;17:1006–1018. doi: 10.1093/emboj/17.4.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matsuzaki G, Vordermeier HM, Hashimoto A, Nomoto K, Ivanyi J. The role of B cells in the establishment of T cell response in mice infected with an intracellular bacteria, Listeria monocytogenes. Cell Immunol. 1999;194:178–185. doi: 10.1006/cimm.1999.1503. [DOI] [PubMed] [Google Scholar]
- 66.Nagamatsu K, Kuwae A, Konaka T, Nagai S, Yoshida S, Eguchi M, Watanabe M, Mimuro H, Koyasu S, Abe A. Bordetella evades the host immune system by inducing IL-10 through a type III effector, BopN. J Exp Med. 2009;206:3073–3088. doi: 10.1084/jem.20090494. [DOI] [PMC free article] [PubMed] [Google Scholar]