Abstract
We previously compared the expression of several human factor VIII (fVIII) transgene variants and demonstrated the superior expression properties of B domain deleted porcine fVIII. Subsequently, a hybrid human/porcine fVIII molecule (HP-fVIII) comprising 91% human amino acid sequence was engineered to maintain the high-expression characteristics of porcine fVIII. The bioengineered construct then was used effectively to treat knockout mice with hemophilia A. In the current study, we focused on optimizing self-inactivating (SIN) lentiviral vector systems by analyzing the efficacy of various lentiviral components in terms of virus production, transduction efficiency and transgene expression. Specifically, three parameters were evaluated: 1) the woodchuck hepatitis post-transcriptional regulatory element (WPRE), 2) HIV versus SIV viral vector systems, and 3) various internal promoters. The inclusion of a WPRE sequence had negligible effects on viral production and HP-fVIII expression. HIV and SIV vectors were compared and found to be similar with respect to transduction efficiency in both K562s and HEK-293T cells. However, there was an enhanced expression of HP-fVIII by the SIV system, which was evident in both K562 and BHK-M cell lines. To further compare expression of HP-fVIII from an SIV-based lentiviral system, we constructed expression vectors containing the high expression transgene and a human elongation factor-1 alpha (EF1α), cytomegalovirus (CMV) or phosphoglycerate kinase (PGK) promoter. Expression was significantly greater from the CMV promoter, which also yielded therapeutic levels of HP-fVIII in hemophilia A mice. Based on these studies, an optimized vector contains the HP-fVIII transgene driven by a CMV internal promoter within a SIV-based lentiviral backbone lacking a WPRE.
Keywords: hemophilia, hematopoietic stem cells, gene therapy, lentivirus
INTRODUCTION
Hemophilia A is a monogenic disease caused by mutations in the gene encoding fVIII, resulting in the inability to properly form a blood clot. Hemophilia A is a prime candidate for gene therapy in that only a moderate increase in fVIII (2–5% of normal equating to 2–5 ng/mL) is required to be therapeutically effective. In addition, the current treatment, consisting of repetitive prophylactic administration of recombinant fVIII as a means of protein replacement, is expensive, invasive and does not always result in patient compliance. Further complicating the current therapy, 30% of individuals with severe and moderately severe hemophilia develop an immunogenic response in the form of neutralizing antibodies against the administered fVIII.1 These inhibitors make managing a bleeding episode extremely complicated. Therefore, new therapeutic approaches are needed to treat hemophilia A. fVIII gene therapy attempts to rectify the presence of a mutant fVIII gene with either the addition of a functional gene or correction of the original gene. In the case of gene addition, delivery of the fVIII gene is not restricted to a certain cell type because, theoretically, any tissue with exposure to the vasculature is suitable as a cellular target. In addition, the therapeutic window is large, as fVIII levels as high as 150% of normal have not been associated with adverse effects such as thrombosis.2 As a result, a number of viral and non-viral delivery strategies have been postulated.
Despite promising preclinical data, hemophilia A gene therapy clinical trials have not progressed past phase 1 trials due to limited expression of fVIII.3–5 Although each trial was unique in regards to the cell type modified and the viral vector used, all trials yielded less than therapeutic levels of fVIII. To overcome low level transgene expression, we recently compared several high-expression fVIII transgene sequences and demonstrated enhanced expression of a B domain deleted porcine fVIII sequence, both in vitro and in vivo.6–8 Upon comparison of a series of hybrid human/porcine cDNAs, the domains responsible for the high expression characteristics of porcine fVIII were identified as the A1 and A3 domains.9 These findings resulted in the construction of a high expression human/porcine transgene. The bioengineered construct was then introduced via lentiviral vectors into hematopoietic stem cells (HSC) ex vivo and used to effectively treat fVIII knockout mice with hemophilia A, yielding therapeutic levels of fVIII.10 Further optimization resulted in the inclusion of the porcine C1 domain and three alanine substitutions in the A2 domain in order to reduce immunogenicity.11, 12 The final high expression B domain deleted fVIII transgene (HP-fVIII) contains human A2 and C2 domains in addition to porcine A1, A3, and C1 domains. This HP-fVIII has been shown to maintain the high expression characteristics of porcine fVIII.10
Lentiviral vectors are promising vectors for the delivery of the fVIII transgene because they provide stable integration and are able to transduce both dividing and non-dividing cells.13 Lentiviral vectors, unlike adeno-associated viral vectors, are less constrained by the size of the transgene. Vector size constraints are an issue for fVIII gene therapy since the B domain deleted fVIII cDNA is approximately 4.4kb (for review see Johnston et al., 2011).14 For these reasons, lentiviral vectors are reasonable for gene therapy applications aimed at the treatment of hemophilia A utilizing ex vivo modification of HSCs. Within lentiviral vectors, a woodchuck post-transcriptional regulatory element (WPRE) is routinely added to the 3’ end of the transgene. The inclusion of this sequence is for increased transgene expression, as it has been demonstrated that a 2- to 5- fold increase in expression is achieved with a WPRE sequence, which in part is due to an increased export of mRNA and possibly due to facilitating transcript processing.15–17 However, a recent report showed that enhanced transgene expression in the presence of a WPRE sequence was dependent on the promoter and cell line used, where in some instances the inclusion yielded no increase or decreased expression.17, 18 In addition, the WPRE codes for the first sixty amino acids of the hepadnavirus × protein, a protein that has been linked to oncogenesis.19 Therefore, the function of the WPRE appears to be more complex than originally assumed and may need to be evaluated in conjunction with individual transgenes, which has not been done for fVIII.
Previous studies have focused on optimizing the fVIII transgene for enhanced fVIII expression (for review see Doering and Spencer, 2009),20 but the components of the viral vector system have not been as well characterized. This report focuses on optimizing the lentiviral vector for virus production, transduction efficiency, and transgene expression with the use of a fVIII transgene that has been bioengineered for high level fVIII expression (HP-fVIII). The optimized lentiviral vector was utilized in ex vivo HSC transduction studies to determine in vivo HP-fVIII expression.
RESULTS
Assessment of the requirement for a WPRE sequence for the production of virus containing an HP-fVIII transgene
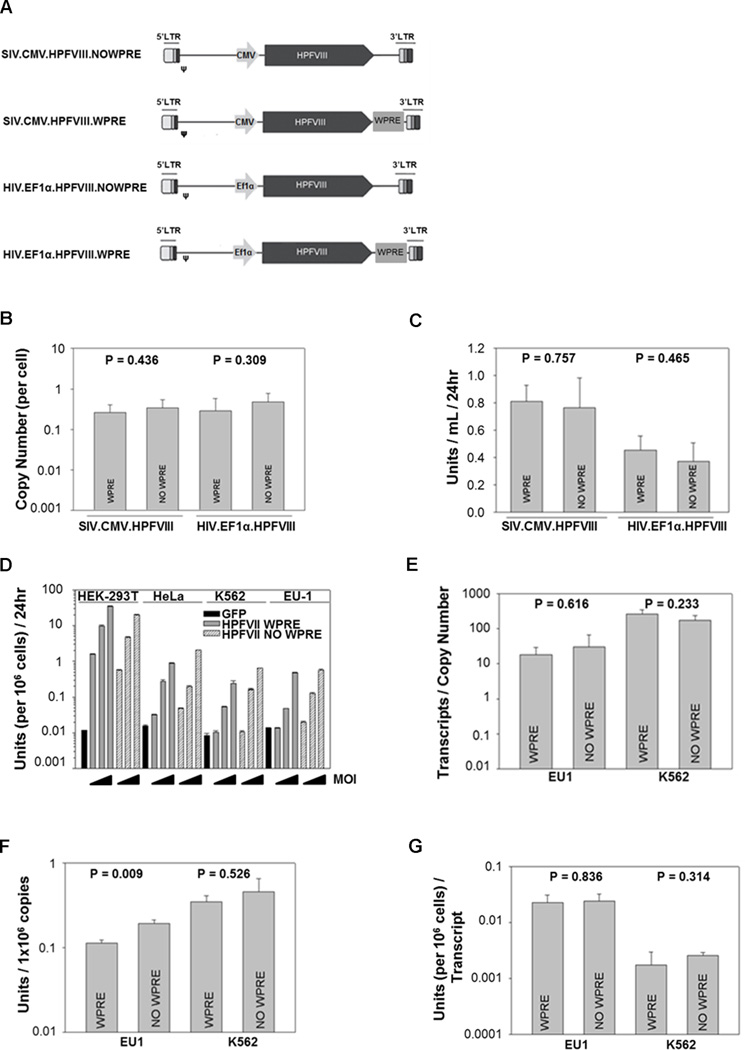
A recent report showed that enhanced transgene expression as a result of a WPRE is dependent on both the promoter and cell line used, and should thus be assessed for individual transgene scenarios.18 The WPRE was evaluated in the context of an optimized HP-fVIII transgene. Both SIV and HIV expression plasmids were constructed with and without a WPRE sequence (Figure 1A) and used to produce recombinant viral vector from HEK-293T cells. Virus, with and without a WPRE, was generated in triplicate under identical conditions. Unconcentrated viral supernatant then was used to transduce HEK-293T cells to assess viral titer. In this manner, viral production was analyzed in the context of a highly transducible cell line for comparisons. DNA was extracted from the transduced cells and analyzed for viral copy number by qPCR. For both lentiviral vectors, assuming equal transduction efficiency, the absence of a WPRE did not affect viral production (p = 0.436 for SIV and p = 0.309 for HIV) (Figure 1B).
Figure 1.
Evaluation of the WPRE sequence in SIV and HIV vector systems. (A) Schematic representation of SIV and HIV-based lentiviral vectors. (B) Recombinant virus was produced under identical conditions (n=3). Unconcentrated viral supernatant (0.5mL) was used to transduce HEK-293T cells. Seventy-two hours later, genomic DNA was isolated from the transduced cells and used to assess viral copy number by qPCR. (C) To assess HP-fVIII activity, the expression plasmids were transiently transfected into HEK-293T cells. HP-fVIII activity was detected by an APTT-reagent based one-stage coagulation assay 48 hours post transfection. Twenty-four hours prior to the APTT-reagent based one-stage coagulation assay, conditioned media was replaced with AIM-V (serum-free). Each bar represents the mean ± standard deviation of 6 wells measured in duplicate. (D) HP-fVIII activity was measured following viral transduction of HEK-293T, HeLa, K562 and EU1 cells with an HIV based lentivirus virus at increasing MOIs (MOI 5, 25, and 125). As a negative control, the cells were transduced with an HIV based lentivirus encoding the GFP transgene (MOI 125). (E) An SIV-based lentivirus also was used to assess HP-fVIII expression in EU1 and K562 cells (MOI 1). Seventy-two hours post- transduction, genomic DNA and total RNA were isolated from the cells. HP-fVIII transcripts and HP-fVIII copy number was assessed by qRT-PCR and qPCR, respectively. (F) HP-fVIII activity was assessed 24 hr after exchange of conditioned media with serum-free AIM-V media by an APTT-reagent based one-stage coagulation assay. (G) HP-fVIII activity was normalized to transcript levels. Each bar represents the mean ± standard deviation of 3 wells measured in duplicate. P values were derived from a student’s t test.
Evaluation of the expression of the HP-fVIII transgene from lentiviral vectors containing a WPRE element
Two independent internal promoters and vector systems then were used to evaluate the effects of a WPRE on expression, an EF1α promoter in an HIV lentiviral system and a CMV promoter in a SIV lentiviral system. The HP-fVIII expression constructs were transiently transfected into HEK-293T cells. Forty-eight hours later, HP-fVIII expression was quantified using an activated partial thromboplastin (APTT) reagent-based one-stage coagulation assay. Comparison of HP-fVIII expression showed that the WPRE did not enhance expression of the transgene driven from either an EF1α (p = 0.465) or a CMV (p = 0.757) internal promoter (Figure 1C). In order to ensure that this effect was not specific to expression after transient transfection of HEK-293T cells, HeLa cells, two hematopoietic cell lines (EU1 and K562 cells) and HEK-293T cells were transduced with the HIV-based lentiviral vector system and assessed for HP-fVIII activity. No difference in HP-fVIII expression was observed in any cell line when comparing cells transduced with vectors containing a WPRE to those without (Figure 1D). The lack of effect was further analyzed in a hematopoietic context in which two hematopoietic cell lines (EU1 and K562s) were transduced with the SIV-based lentiviral vector system. HP-fVIII expression was assessed at both the transcript and protein level. To correct for transduction efficiency, transcript and fVIII activity levels were normalized to copy number for each viral preparation. No enhancement in HP-fVIII transcripts was observed when a WPRE was included in the lentiviral vector (Figure 1E). As shown in Figure 1F, a similar observation was noted for the activity of expressed HP-VIII from transduced K562 cells when normalized to copy number. However, significantly more activity was observed from EU1 cells that were transduced with a SIV viral vector devoid of a WPRE sequence than those that were transduced with a SIV virus containing a WPRE sequence (p = 0.009) (Figure 1F). This relative enhancement was not observed upon normalization of HP-fVIII activity to transcripts (Figure 1G). We have previously demonstrated a correlation between fVIII transcript number and fVIII activity.10 However, less correlation has been documented between copy number and fVIII expression and would account for the lack of enhancement seen as a result of HP-fVIII activity normalization to transcripts.21 In addition, no statistically significant difference was found in the amount of transcriptional read-through when a WPRE sequence was present as compared to when it was removed (p = 0.905) (data not shown). Taken together, these data demonstrate that the WPRE does not enhance viral titer, transduction, or HP-fVIII expression, and led to the removal of the WPRE element from both the HIV-based and SIV-based lentiviral vectors in subsequent studies.
Evaluation of a codon optimized HP-fVIII transgene
To test the usefulness of codon optimization of HP-fVIII, the transgene was codon optimized for codon usage, the removal of mRNA secondary structures, as well as an increase in GC-content from approximately 48% to 52%. In addition, a Kozak sequence was included for increased translation initiation. The optimized high expression fVIII cDNA has 76% sequence identity at the nucleotide level to the original non-optimized sequence. HIV-based lentiviral expression plasmids containing either the non-optimized or the codon optimized HP-fVIII transgene regulated by the EF1α internal promoter without a WPRE sequence were transiently transfected into HEK-293T cells. No enhancement in HP-fVIII activity was noted from these cells as assessed by an APTT-reagent based one-stage coagulation assay forty-eight hours post transfection (p = 0.07) (data not shown). In addition, HEK-293T and K562 cells were transduced at an multiplicity of infection (MOI) of 1, 5, or 25 and assessed for HP-fVIII activity one week later. No significant enhancement was noted in HP-fVIII activity with the use of a codon optimized HP-fVIII transgene within HEK-293T cells (Figure 2A). However, greater expression of HP-fVIII was achieved at the highest MOI tested in K562 cells transduced with a HIV-based lentivirus containing the non-optimized HP-fVIII transgene (Figure 2A). In addition, BHK-M cells were serially transduced with an HIV-based lentivirus containing either the non-optimized or the codon optimized high expression fVIII transgene. No enhancement in HP-fVIII activity was noted with the use of a codon optimized HP-fVIII transgene (Figure 2B). Together, this data shows that the codon optimized transgene did not enhance expression.
Figure 2.
Evaluation of a codon optimized high expression fVIII transgene. HEK-293T and K562 cells were transduced with an HIV-based lentiviral vector at increasing MOIs (1, 5, and 25). Mock transductions were performed (denoted as Neg) with an HIV-based lentiviral vector containing an eGFP transgene at an MOI of 25. Twenty-four hours prior to assessment, conditioned media was exchanged for serum-free AIM-V. HP-FVIII activity was assessed by an APTT reagent-based one-stage coagulation assay a week post transduction (n=2). (A) BHK-M cells were serially transduced at an MOI of 30 for a total of 6 times. Seventy-two hours after transduction, HP-FVIII activity was assessed (n=2). (B) Black bars represent a non-optimized HP-FVIII transgene. White bars represent a codon optimized HP-FVIII transgene. Abbreviations: NonCO HP-FVIII (non-optimized HP-FVIII transgene) and CO HP-FVIII (codon optimized HP-FVIII transgene).
Comparison of the transduction efficiency and HP-fVIII expression of HIV and SIV-based lentiviral vectors containing the HP-fVIII transgene
To determine if there are inherent differences in HP-fVIII expression between HIV and SIV gene transfer systems, HIV and SIV-based expression plasmids were produced in which the only differences were derived from the vector system itself (Figure 3A). The HP-fVIII transgene was expressed in both lentiviral vectors from the EF1α internal promoter. Expression plasmids were used to produce virus from HEK-293T cells under identical conditions three separate times. Virus was quantified by assessing viral titer using 1mL unconcentrated viral supernatant, which was added to the highly transducible HEK-293T cell line as a baseline for comparison. Viral copy number was determined utilizing qPCR with primers specific for the HP-fVIII transgene and compared between HIV transduced and SIV transduced cells. Unconcentrated viral vector titers were significantly higher with the SIV vector system (p=0.014) (data not shown). Recombinant HIV and SIV then was used to transduce BHK-M, K562 and HEK-293T cells at a MOI of 5. Seventy two hours after viral addition, transduction efficiency was measured by analyzing the copy number of HP-fVIII by qPCR. As previously observed (Figure 1D), BHK-M and K562 cells are transduced less efficiently with lentiviral vectors than HEK-293T cells, as noted by lower HP-fVIII copy numbers (Figure 3B). Comparison of the two vector systems within cell lines, on the other hand, did not show any difference with respect to the transduction efficiency in both K562 cells (p = 0.837) and HEK-293T cells (p = 0.120) (Figure 3B). Identical copy numbers in HEK-293T cells were observed and expected since these cells were used to calculate viral titer (Figure 3B). However, a slight difference was seen with BHK-M cells. Greater copy numbers were noted when a SIV-based vector was used to transduce BHK-M cells compared to the HIV-based vector (p=0.045).
Figure 3.
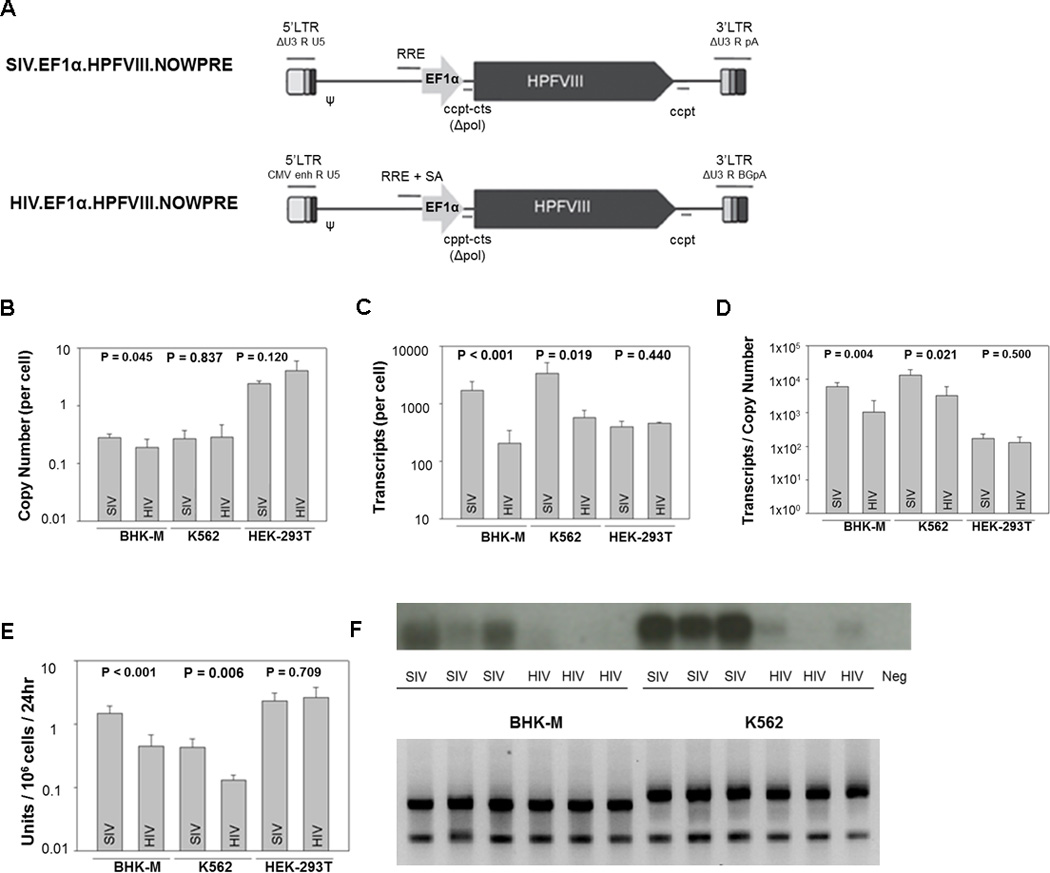
Comparison of SIV and HIV-vector systems encoding HP-fVIII. SIV and HIV expression plasmids were generated to encode HP-fVIII as denoted in (A). Recombinant virus was produced under identical conditions (n=3). Each viral preparation was utilized to transduce two wells of BHK-M, K562, and HEK-293T cells at an MOI of 5. Seventy-two hours post-transduction, total DNA and genomic DNA were isolated and (B) copy number and (C) transcript levels were quantified by qRT-PCR and qPCR, respectively. (D) Transcript levels were normalized to copy number. (E) Twenty-four hours prior to isolation, the conditioned media was exchanged for serum-free AIM-V in order to assess the HP-fVIII activity by an APTT reagent-based one-stage coagulation assay. Bars represent the mean ± the standard deviation of three sets of virus each added to two wells, while each well was measured in duplicate. (F) Enhanced transcript levels were confirmed by Northern Blot analysis. P values were derived from a student’s t test. Equivalent amounts of ribosomal RNA were apparent among each sample (lower panel).
RNA was extracted and fVIII transcripts were quantified using qPCR from the BHK-M, K562 and HEK-293T cells transduced with either HIV or SIV-based vectors. BHK-M cells were included in this analysis as a high-expressing fVIII cell line. It was expected that the expression of the transgene would be similar between the two viral vectors, since similar MOIs were used and similar copy numbers were determined. However, there was enhanced HP-fVIII RNA expression by the SIV system, which was evident in both BHK-M (p < 0.001) and K562 (p = 0.019) cell lines (Figure 3C and 3D). Enhanced HP-fVIII RNA levels were confirmed by Northern blot analysis (Figure 3F upper panel). Supernatants from the transduced cells were used to assess HP-fVIII activity by an APTT reagent-based one-stage coagulation assay. HP-fVIII activity was significantly increased for K562 (p=0.006) and BHK-M (p < 0.001) cells transduced with SIV (Figure 3E). In K562 and BHK-M cells, greater RNA levels led to greater protein production. This is consistent with previous findings that showed a strong correlation between RNA and fVIII activity.10 Based on this set of data, the SIV-based vector system was selected for further studies.
Comparison of three internal promoters for the expression of HP-fVIII
SIN vectors require internal promoters for transgene expression due to the inactivation of the viral 5’ long terminal repeat (LTR) upon transfer of the U3 deletion during integration. A balance between adequate transgene expression and the elimination of transactivation of nearby genes must be maintained by the internal promoter. Three ubiquitous heterologous promoters, varying in enhancer activity, were evaluated in transduced HEK-293T cells. The human EF1α promoter, the CMV promoter, and the yeast PGK promoter were incorporated into the SIV expression plasmid and used to produce virus under identical conditions (Figure 4A). Viral titer was determined for each vector by qPCR. Vector then was added to HEK-293T cells at an MOI of 3. HEK-293T cells, although unable to accurately depict the transcription profile of HP-fVIII from sca-1+ cells, were chosen due to the inherent superior transduction capabilities as compared to the hematopoietic EU1 and K562 cell lines. Transgene expression was greatest when driven by the CMV promoter and least by the PGK promoter as evaluated by the level of HP-fVIII transcripts from transduced cells (Figure 4B). In addition, HP-fVIII activity normalized to copy number showed that HP-fVIII activity is greatest when expression is driven by the CMV promoter and least by the PGK promoter (Figure 4C).
Figure 4.
Effects of various internal promoters on HP-fVIII activity in vitro. (A) Schematic representation of SIN SIV-based expression vectors constructed with the PGK, EF1α or CMV internal promoter to make recombinant lentivirus. (B) HEK-293T cells were transduced at an MOI of 3. Seventy-two hours post-transduction, total RNA and genomic DNA were isolated from the cells and quantified by qPCR for transcript levels. (C) Twenty-four hours prior to isolation, the conditioned media was exchanged for serum-free AIM-V in order to assess HP-fVIII activity which was normalized to copy number to correct for transduction efficiency. Each bar represents the mean ± the standard deviation of 3 wells. P values were determined by a one way ANOVA.
Assessment of an optimized lentiviral vector expressing HP-fVIII in hemophilia A mice
Based on the above analyses, an SIV-based lentiviral vector containing the HP-fVIII transgene expressed from a CMV promoter without the inclusion of a WPRE was predicted to be optimal for HP-fVIII expression in vitro. This construct then was evaluated in vivo in hemophilia A mice. Sca-1+ cells were isolated from CD45.1 mice and transduced with the optimized SIV vector encoding HP-fVIII. CD45.1 sca-1+ cells were transduced twice, 24 hr between transductions, at an MOI of 15 each time, which resulted in 30–60% transduction efficiency with similar eGFP encoding vectors. However, with fVIII containing vectors only 3–10% is expected since previous findings showed a ten-fold lower transduction efficiency when using a vector encoding fVIII as compared to eGFP (data not shown, Doering et al., 2009 10). Transduction protocols were designed to ensure that transduced cells would only contain, on average, one or fewer copies of the vector. The transduced sca-1+ cells then were transplanted into lethally irradiated (11 Gy split dose total body irradiation) hemophilia A mice. Three months after transplantation, donor cell engraftment was measured by flow cytometry. Average engraftment in the peripheral blood and spleen was shown to be approximately 90% (Figure 5A). Every two weeks HP-fVIII levels were quantified by an ELISA assay. HP-fVIII levels persisted for the duration of the study (four months) with a range between 2 and 20 ng/mL (Figure 5B). A Coatest activity assay showed a similar HP-fVIII expression profile (Figure 5C). Copy numbers in six mice remained below the detectable level of five percent gene-modified cells by qPCR with one each having 0.07, 0.10, 0.11 and 0.14 vector copies per genome. No correlation between copy number and HP-fVIII expression was observed in these mice.
Figure 5.
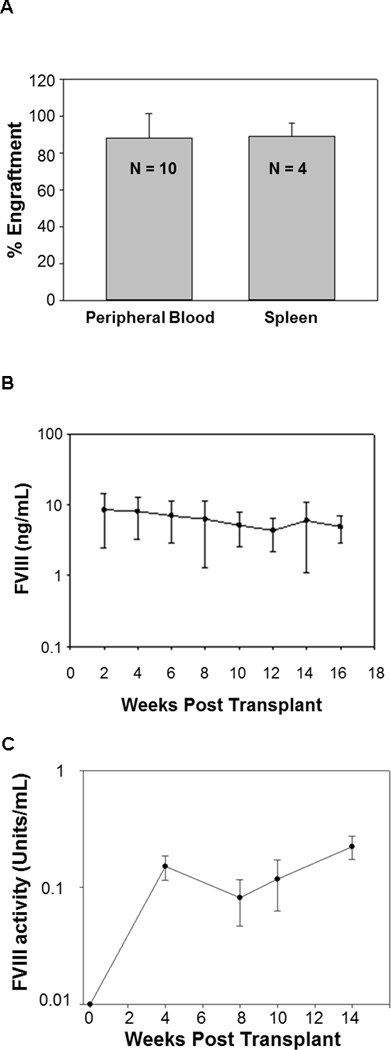
In vivo expression of the optimized vector in transplanted hemophilia A mice. Sca-1+ cells were isolated from CD45.1 mice and transduced with the optimized vector with an MOI of 15 (X2). Approximately 1×106 cells were transplanted into Hemophilia A mice (CD45.2) (n=14). (A) Engraftment was measured from peripheral blood and splenocytes of three mice and was assessed three months after transplant by flow cytometry. (B) HP-fVIII expression levels were assessed within the mice every two weeks following transplantation by an ELISA. (C) A chromogenic test (Coatest assay) was used to assess HP-fVIII activity in a subset of mice (n=3).
DISCUSSION
To date, three clinical trials have been initiated for gene therapy applications to treat hemophilia A.3–5 However, each trial failed to yield sustained therapeutic levels of human fVIII. To overcome the obstacle of limited expression, a human/porcine fVIII hybrid has been constructed exhibiting expression 19-fold higher than human B domain deleted fVIII.10 Further optimization of the transgene resulted in a theoretically less immunogenic high expression fVIII transgene. With the production of an extensively optimized transgene, efforts can now be directed toward optimizating the viral vector for transgene delivery and expression.
A number of viral vectors have been considered for the modification of cells including both nonintegrating and integrating vectors. Nonintegrating vectors, such as the adenoviral and adeno-associated viral vectors exist within cells extrachromosomally and are limited by potential vector genome loss. Yet, adenoviral vectors and adeno-associated viral vectors are appealing in that they efficiently transduce both dividing and non-dividing cells. With respect to the treatment of hemophilia A, adenoviral vectors have been used in neonates to produce tolerance22, 23 but currently are less clinically desirable due to toxic side effects experienced in clinical trials24. However, adeno-associated viral vectors are being extensively evaluated for use in gene therapy, especially for hemophilia B. Unfortunately, adeno-associated viral vectors are limited by their genetic carrying capacity. The vector cassette associated with the 4.4 kb of the B-domain deleted fVIII cDNA including regulatory elements is at and above the carrying capacity of an adeno-associated viral vector for the treatment of hemophilia A. Several groups are attempting to overcome this limitation by separating the genetic payload, i.e. the heavy chain and light chain of fVIII, into two different vectors.23, 25, 26 Other groups are attempting to minimize the regulatory elements by utilizing smaller internal promoters.27–31
Several groups have focused on the use of integrating viral vectors such as lentiviral vectors. Lentiviral vectors are suitable vectors for hemophilia A gene therapy applications in that they (1) stably integrate within the host genome, (2) are able to transduce quiescent cells and (3) can encapsulate large transgenes such as HP-fVIII. Although insertional mutagenesis was observed clinically with the use of integrating γ-retroviral vectors,32, 33 to date similar issues have not been observed with lentiviral vectors.34–36 This may be due to integration site preferences between the two vectors. In addition, clinically used lentiviral vectors contain safety measures that are now routinely incorporated, including a 133bp deletion within the U3 region of the 3’ LTR that self-inactivates the vector, creating a replication incompetent or SIN vector37, 38 (for review see Pauwels et al., 2009).39 Comparative studies have shown that SIN lentiviral vectors are less oncogenic than γ-retroviruses.40, 41
A WPRE often is incorporated into lentiviral vectors since the demonstration of 2- to 5-fold enhanced GFP and luciferase expression.15–17 However, a recent report showed that the enhanced expression was dependent on the promoter and cell line used, and in some scenarios, the presence of a WPRE resulted in a decrease in transgene expression.18 It is apparent that the WPRE is more complex than originally assumed and requires individual transgene evaluation. With the HP-fVIII transgene, the WPRE was found to be negligible in regards to viral production, transgene expression and transcriptional read-through. Therefore, for our high expression construct, there appears to be no benefit to include a WPRE. However, it may be useful for lower expressing fVIII constructs and should be tested in conjunction with each.
Codon usage bias within species can result in ribosomal stalling at rare codons during translation. For this reason, algorithms have been designed to optimize the translation process in order to ensure efficient protein production. A variety of other features are included in the codon optimization process including the elimination of mRNA secondary structures and the inclusion of translational performance sequences. Codon optimization of our high expressing hybrid fVIII transgene did not enhance HP-fVIII expression. The lack of enhancement was noted with both HEK-293T, K562 and BHK-M cells. It is of note that although a less robust EF1α internal promoter was utilized still no enhancement was observed in this analysis. This is contrary to previous observations that showed increased activity after codon optimization of a human fVIII transgene.42 The ineffectiveness of the codon optimization process to enhance the expression of HP-fVIII may be due to the innate high expressing characteristics of HP-fVIII. Besides potentially eliminating ribosomal stalling, another possible mechanism for the enhanced levels of fVIII from a codon optimized fVIII transgene could be due to the removal of transcriptional silencers and/or inhibitory sequences found within the human fVIII cDNA and not within the porcine fVIII domains.43–46
The HIV and SIV-based vector systems also were analyzed in this study in regards to HP-fVIII expression. The production of HIV and SIV lentiviral vectors were optimized separately. Using standard production conditions, higher viral titers routinely were achieved with the SIV system compared to HIV. After each viral preparation was quantified with respect to viral titer, transduction at identical MOIs yielded integration events that were not statistically different in K562 and HEK-293T cells. However, BHK-M and K562 cells transduced with SIV expressed HP-fVIII more efficiently than those cells transduced with identical amounts of HIV. This suggests that although SIV and HIV integrated at similar levels, the integration events of SIV may be within regions of the genome that promoted greater expression than those regions where HIV integrated. This potential difference requires further study to test this hypothesis. It is interesting that this same phenomenon was not observed with a GFP transgene (data not shown) and that the enhanced expression of HP-fVIII also was not seen with HEK-293T cells. This suggests a potential transgene and cell type specificity of SIV enhancement. This data also suggests that SIV is, at a minimum, as efficient at gene transfer and expression of HP-fVIII as HIV. SIV-based vectors may be a safer option for gene therapy applications to combat hemophilia A since a number of adults with hemophilia are HIV-1 positive after receiving plasma-derived fVIII infusion products before HIV testing became routine. The use of HIV vectors can raise safety concerns due to possible recombination events of the vector and wild-type HIV. SIV, on the other hand, is less likely to recombine due to sequence differences between HIV and SIV.
Different internal promoters also were tested as a component of the expression vector for HP-fVIII expression. SIN-lentiviral vectors require internal promoters to direct expression of the transgene due to the inactivation of the 5’ LTR during integration. This safety feature removes the enhancer element of the LTRs preventing transactivation of nearby genes. Therefore, the internal promoter must be sufficiently strong to provide adequate expression of the transgene without having the capacity to transactivate genes nearby. Not surprisingly the CMV promoter was superior to the EF1α and PGK promoters at expressing HP-fVIII in vitro using the SIV-based vector system. The enhanced expression from the CMV promoter in vitro is not unexpected since the CMV promoter contains the strongest enhancer among the three internal promoters. However, with the stronger enhancer activity the transactivation of nearby genes is a concern with the use of CMV as well as the possibility of methylation-induced inactivation of the promoter. Within hemophilia A mice under limiting transduction protocols using an SIV vector without a WPRE, the CMV promoter directed expression of HP-fVIII at therapeutically relevant levels.
Several fVIII transgenes have been studied extensively for use in gene therapy applications of hemophilia A. HP-fVIII overcomes low level expression obstacles while theoretically reducing the immunogenicity. Although the transgene has been fairly well described the expression vector has not been as well characterized. In this study, a lentiviral vector was optimized for HP-fVIII expression. Under the conditions tested, the SIV-based lentiviral backbone was found to be more effective for HP-fVIII expression than an HIV-based lentiviral backbone. Within the SIV-based lentiviral backbone, the CMV internal promoter was shown to drive HP-fVIII expression efficiently, but it is realized that this promoter may have complications due to possible transactivation and promoter inactivation. The WPRE was found to be unnecessary and was removed from the lentivector. In vivo data shows that the optimized vector provides sustained HP-fVIII expression in hemophilia A mice, supporting its further development for hemophilia A.
MATERIALS AND METHODS
Reagents
Dulbecco’s Modified Eagle’s medium (DMEM)/F-12, Aim V medium and StemPro-34 serum free medium were purchased from Invitrogen life technologies (Carlsbad, CA). Heat-inactivated fetal bovine serum (FBS) was purchased from Atlanta Biologicals (Atlanta, GA). Penicillin-streptomycin solution was purchased from Mediatech (Manassas, VA). Cell transfections were performed with polyethyleneimine purchased from Fisher Scientific (Pittsburg, PA). Plasmids utilized for viral preparation were isolated from bacterial stocks utilizing QIAGEN Hispeed midiprep plasmid kits (Valencia, CA). Nucleic acid isolation kits were purchased from QIAGEN (Valencia, CA). Integration events were analyzed using a qPCR SYBR Green Low Rox master mix from Thermo Fisher Scientific (Waltham, MA) an ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA) and oligonucleotide primers synthesized by Integrated DNA Technologies (Coralville, IA). HP-fVIII RNA was quantifed utilizing fVIII RNA standards generated with an mMessage mMachine kit (Ambion, Austin, TX). Northern blots were performed with the digoxigenin (DIG) nonradioactive nucleic acid-labeling and detection system (Roche, Indianapolis, IN). Human fVIII-deficient plasma and normal pooled human plasma (FACT) were purchased from George King Biomedical (Overland Park, KS). Automated activated partial thromboplastin reagent was purchased from BioMérieux (Durham, NC). Clotting times were measured using an STart Coagulation Instrument (Diagnostica Stago, Asnieres, France). Anti-pfVIII and anti-hfVIII monoclonal antibodies were a kind gift of Dr. Pete Lollar (Aflac Cancer Center and Blood Disorder Services, Emory University, Atlanta, GA). Sca-1+ cells were isolated using magnetic separation columns purchased from Miltenyi Biotec (Auburn, CA). Exon 16-disrupted hemophilia A mice have been previously described.47 All antibodies for flow cytometry were purchased from BD Pharmingen (San Diego, CA).
Vector production
Expression plasmids containing the HP-fVIII gene along with the necessary packaging plasmids were transiently transfected into HEK-293T cells utilizing polyethylenimine (6µg PEI / 1µg DNA). A 2:1:1 ratio of expression plasmid to packaging plasmids (expression plasmid:psPAX2:pVSVG) was used to manufacture research-grade HIV-based lentiviral vectors within the LentiMax production system. Research-grade SIV-based lentiviral vectors were manufactured using a 1.3:1:1:1.6 ratio of expression plasmid to packaging plasmids (expression plasmid:pCAG4:pVSVG:pSIV). One day after transfection, the media was replaced with DMEM-F12 containing 10 % FBS and 1 % penicillin / streptomycin. Conditioned medium from the HEK-293T viral producing cells was collected for the following three days, passed through 0.45µmol/l filter and stored at −80°C until concentration. Virus was concentrated by velocity sedimentation upon centrifugation at 10,000 × g (4°C) overnight. Viral pellets were resuspended in 1/100th of the original volume of StemPro media, and filtered through a 0.22 micron filter. Viral titer was assessed on HEK-293T cells with increasing vector volumes by qPCR seventy-two hours after viral addition. Unconcentrated viral supernatant had titers ranging from 5×105 to 2×106. Concentrated virus ranged from 5×107 to 1×108. Virus was stored in 1mL aliquots at −80°C.
Measurement of HP-fVIII transgene copy number
Total genomic DNA was isolated using the DNeasy® Blood & Tissue Kit following the manufacture’s protocol for cultured cells. DNA was quantified with a spectrophotometer at an absorbance of 260nm. To determine transgene copy number, 50ng of each sample was added to a 25µl real-time quantitative PCR reaction containing 1× SYBR green PCR master mix (Thermo Fisher Scientific, Surrey UK) and 0.01 µM forward and reverse primers. HP-fVIII specific primers annealing to the A1 porcine domain were utilitzed: forward primer, 5’- CAG GAG CTC TTC CGT TGG -3’ at position 164 and reverse primer, 5’- CTG GGC CTG GCA ACG C -3’ at position 239. Ct values for each sample were compared to Ct values produced from plasmid standards of known copy quantities. The equivalent copy number was then divided by 8333, the predicted number of genome equivalents in 50ng of DNA.
Measurement of HP-fVIII transcript expression
Total RNA was isolated using the RNeasy® Mini Kit following the manufacture’s protocol for animal cells seventy-two hours post transduction. RNA was then quantified spectrophotometrically at an absorbance of 260nm. HP-fVIII transcripts were measured by quantitative RT-PCR utilizing a porcine fVIII RNA standard as previously described.48 qPCR reactions were carried out in 25µl containing 1× SYBR green PCR master mix, 300µM forward and reverse primers, 12.5 units MultiScribe, 10 units RNase inhibitor, and 5ng of sample RNA. Reactions containing the porcine fVIII RNA standard also included 5ng of yeast tRNA, mimicking the RNA environment of the sample RNA. The oligonucleotide primers utilized for the qPCR reaction annealed within the A2 domain of the the fVIII cDNA sequence at positions 1897–1917 for the forward primer (5’-ATGCACAGCATCAATGGCTAT-3’) and at positions 2044–2063 for the reverse primer (5’-GTGAGTGTGTCTTCATAGAC-3’). One-step real-time RT-PCR was performed by incubation at 48 °C for 30 min for reverse transcription followed by one cycle at 95 °C for 10 min and 40 amplification cycles of 90 °C for 15 sec and then at 60 °C for 1 min. Postreaction dissociation was performed to confirm single-product amplification. Ct values for each sample were compared to Ct values produced from the porcine fVIII standards having known transcript quantities.
Measurement of HP-fVIII activity from cell lines
fVIII activity was measured from the supernatant of cells cultured in AIM V media for 24 hours before the assay as previously described.48 In short, the activated partial thromboplastin reagent-based one stage coagulation assay was performed in duplicate for each supernatant on a ST art Coagulation Instrument within human fVIII-deficient plasma. The clot time for each sample was compared to a standard curve based on dilutions of FACT.
Animals
Exon-16 deleted hemophilia A mice were obtained from Dr. Leon Hoyer (Holland Laboratories, American Red Cross, Rockville, MD). B6.SJL (CD45.1) mice were acquired from Dr. David Archer (Emory University, Atlanta, GA). Both strains were maintained at the animal care facility of Emory University. All procedures were approved by the Institutional Animal Care Committee at Emory University.
Isolation and transduction of murine stem cell antigen-1+ cells
Whole bone marrow was flushed from the femurs and tibias of 8 to 10 week old CD45.1 mice and then subjected to positive immunomagnetic bead selection. The isolated sca-1+ cells were cultured at a density of 106 cells per mL in StemPro media supplemented with L-glutamine (29µg/mL). For three days prior to transduction the cells were stimulated with murine stem cell factor (100ng/mL), murine interleukin-3 (20ng/mL), human interleukin-11 (100ng/mL), and human Flt-3 ligand (100ng/mL). Cells were transduced twice (MOI 15), 8 hours apart, and transplanted via tail-vein injection the following day into lethally irradiated (11Gy TBI using a Gammacell 40 Exactor) 8 to 10 week old recipient hemophilia A mice (CD45.2). Blood was collected from the transplanted mice every two weeks retro-orbitally and fVIII was measured using an ELISA specific for HP-fVIII. The ELISA only detects properly folded HP-fVIII in that the primary antibody detects the heavy chain (human A2 domain) and the secondary antibody detects the light chain (porcine A3 domain). In addition, HP-fVIII was detected from the plasma using a commercially available chromogenic substrate assay (COATEST SP FVIIII) as previously described.7
ACKNOWLEDGEMENTS
We would like to thank Arthur Nienhuis (St. Jude University, Memphis, TN) for the SIV vector system. This work was supported by grants from the National Heart, Lung and Blood Institute of the National Institutes of Health (5R01HL092179).
Footnotes
CONFLICT OF INTEREST STATEMENT
HTS and CBD have equity interest in Expression Therapeutics, LLC, which owns the bioengineered high expression technology. GD is an employee of Expression Therapeutics, LLC.
REFERENCES
- 1.Ehrenforth S, Kreuz W, Scharrer I, Kornhuber B. Factor VIII inhibitors in haemophiliacs. Lancet. 1992;340:253. doi: 10.1016/0140-6736(92)90530-g. [DOI] [PubMed] [Google Scholar]
- 2.VandenDriessche T, Collen D, Chuah MK. Viral vector-mediated gene therapy for hemophilia. Curr Gene Ther. 2001;1:301–315. doi: 10.2174/1566523013348508. [DOI] [PubMed] [Google Scholar]
- 3.Roth DA, Tawa NE, Jr, O'Brien JM, Treco DA, Selden RF. Nonviral transfer of the gene encoding coagulation factor VIII in patients with severe hemophilia A. N Engl J Med. 2001;344:1735–1742. doi: 10.1056/NEJM200106073442301. [DOI] [PubMed] [Google Scholar]
- 4.Powell JS, Ragni MV, White GC, 2nd, Lusher JM, Hillman-Wiseman C, Moon TE, et al. Phase 1 trial of FVIII gene transfer for severe hemophilia A using a retroviral construct administered by peripheral intravenous infusion. Blood. 2003;102:2038–2045. doi: 10.1182/blood-2003-01-0167. [DOI] [PubMed] [Google Scholar]
- 5.Berlfein J. Clinical trial update. Hemaware. 2003;8:52–53. [Google Scholar]
- 6.Ide LM, Gangadharan B, Chiang KY, Doering CB, Spencer HT. Hematopoietic stem-cell gene therapy of hemophilia A incorporating a porcine factor VIII transgene and nonmyeloablative conditioning regimens. Blood. 2007;110:2855–2863. doi: 10.1182/blood-2007-04-082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doering CB, Gangadharan B, Dukart HZ, Spencer HT. Hematopoietic stem cells encoding porcine factor VIII induce pro-coagulant activity in hemophilia A mice with pre-existing factor VIII immunity. Mol Ther. 2007;15:1093–1099. doi: 10.1038/sj.mt.6300146. [DOI] [PubMed] [Google Scholar]
- 8.Dooriss KL, Denning G, Gangadharan B, Javazon EH, McCarty DA, Spencer HT, et al. Comparison of factor VIII transgenes bioengineered for improved expression in gene therapy of hemophilia A. Hum Gene Ther. 2009;20:465–478. doi: 10.1089/hum.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doering CB, Healey JF, Parker ET, Barrow RT, Lollar P. Identification of porcine coagulation factor VIII domains responsible for high level expression via enhanced secretion. J Biol Chem. 2004;279:6546–6552. doi: 10.1074/jbc.M312451200. [DOI] [PubMed] [Google Scholar]
- 10.Doering CB, Denning G, Dooriss K, Gangadharan B, Johnston JM, Kerstann KW, et al. Directed engineering of a high-expression chimeric transgene as a strategy for gene therapy of hemophilia A. Mol Ther. 2009;17:1145–1154. doi: 10.1038/mt.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Healey JF, Parker ET, Barrow RT, Langley TJ, Church WR, Lollar P. The comparative immunogenicity of human and porcine factor VIII in haemophilia A mice. Thromb Haemost. 2009;102:35–41. doi: 10.1160/TH08-12-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lubin IM, Healey JF, Barrow RT, Scandella D, Lollar P. Analysis of the human factor VIII A2 inhibitor epitope by alanine scanning mutagenesis. J Biol Chem. 1997;272:30191–30195. doi: 10.1074/jbc.272.48.30191. [DOI] [PubMed] [Google Scholar]
- 13.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 14.Johnston J, Doering CB, Spencer HT. Gene Therapy Strategies Incorporating Large Transgenes. In: Chunsheng Kang., editor. Gene Therapy - Developments and Future Perspectives. Croatia: InTech; 2011. pp. 121–142. [Google Scholar]
- 15.Zufferey R, Donello JE, Trono D, Hope TJ. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J Virol. 1999;73:2886–2892. doi: 10.1128/jvi.73.4.2886-2892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brun S, Faucon-Biguet N, Mallet J. Optimization of transgene expression at the posttranscriptional level in neural cells: implications for gene therapy. Mol Ther. 2003;7:782–789. doi: 10.1016/s1525-0016(03)00097-2. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez-Murillo A, Lozano ML, Alvarez L, Jacome A, Almarza E, Navarro S, et al. Development of lentiviral vectors with optimized transcriptional activity for the gene therapy of patients with Fanconi anemia. Hum Gene Ther. 2010;21:623–630. doi: 10.1089/hum.2009.141. [DOI] [PubMed] [Google Scholar]
- 18.Klein R, Ruttkowski B, Knapp E, Salmons B, Gunzburg WH, Hohenadl C. WPRE-mediated enhancement of gene expression is promoter and cell line specific. Gene. 2006;372:153–161. doi: 10.1016/j.gene.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 19.Kingsman SM, Mitrophanous K, Olsen JC. Potential oncogene activity of the woodchuck hepatitis post-transcriptional regulatory element (WPRE) Gene Therapy. 2005;12:3–4. doi: 10.1038/sj.gt.3302417. [DOI] [PubMed] [Google Scholar]
- 20.Doering CB, Spencer HT. Advancements in gene transfer-based therapy for hemophilia A. Expert Rev Hematol. 2009;2:673–683. doi: 10.1586/EHM.09.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spencer HT, et al. Lentiviral vector platform for production of bioengineered recombinant coagulation factor VIII. Mol Ther. 2011;19:302–309. doi: 10.1038/mt.2010.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu C, Cela RG, Suzuki M, Lee B, Lipshutz GS. Neonatal helper-dependent adenoviral vector gene therapy mediates correction of hemophilia A and tolerance to human factor VIII. Proc Natl Acad Sci. 2011;108:2082–2087. doi: 10.1073/pnas.1015571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu C, Lipshutz GS. AAV-based neonatal gene therapy for hemophilia A: long-term correction and avoidance of immune responses in mice. Gene Therapy. 2012 doi: 10.1038/gt.2011.200. e-pub ahead of print 12 January, 2012. doi: 10.1038/gt.2011.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aruda V. Toward gene therapy for hemophilia A with novel adenoviral vectors: success and limitations in canine models. J Thromb Haemost. 2006;4:1215–1217. doi: 10.1111/j.1538-7836.2006.01964.x. [DOI] [PubMed] [Google Scholar]
- 25.Scallan CD, Liu T, Parker AE, Patarroyo-White SL, Chen H, Jiang H, et al. Phenotypic correction of a mouse model of hemophilia A using AAV2 vectors encoding the heavy and light chains of FVIII. Blood. 2003;102:3919–3926. doi: 10.1182/blood-2003-01-0222. [DOI] [PubMed] [Google Scholar]
- 26.Sarkar R, Mucci M, Addya S, Tetreault R, Bellinger DA, Nichols TC, et al. Long-term efficacy of adeno-associated virus serotypes 8 and 9 in hemophilia a dogs and mice. Hum Gene Ther. 2006;17:427–439. doi: 10.1089/hum.2006.17.427. [DOI] [PubMed] [Google Scholar]
- 27.Scallan CD, Lillicrap D, Jiang H, Qian X, Patarroyo-White SL, Parker AE, et al. Sustained phenotypic correction of canine hemophilia A using an adeno-associated viral vector. Blood. 2003;102:2031–2037. doi: 10.1182/blood-2003-01-0292. [DOI] [PubMed] [Google Scholar]
- 28.Sarkar R, Tetreault R, Gao G, Wang L, Bell P, Chandler R, et al. Total correction of hemophilia A mice with canine FVIII using an AAV 8 serotype. Blood. 2004;103:1253–1260. doi: 10.1182/blood-2003-08-2954. [DOI] [PubMed] [Google Scholar]
- 29.Jiang H, Lillicrap D, Patarroyo-White S, Liu T, Qian X, Scallan CD, et al. Multiyear therapeutic benefit of AAV serotypes 2, 6, and 8 delivering factor VIII to hemophilia A mice and dogs. Blood. 2006;108:107–115. doi: 10.1182/blood-2005-12-5115. [DOI] [PubMed] [Google Scholar]
- 30.Lu H, Chen L, Wang J, Huack B, Sarkar R, Zhou S, et al. Complete correction of hemophilia A with adeno-associated viral vectors containing a full-size expression cassette. Hum Gene Ther. 2008;19:648–654. doi: 10.1089/hum.2007.0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabatino DE, Lange AM, Altynova ES, Sarkar R, Zhou S, Merricks EP, et al. Efficacy and safety of long-term prophylaxis in severe hemophilia A dogs following liver gene therapy using AAV vectors. Mol Ther. 2011;19:442–449. doi: 10.1038/mt.2010.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 33.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 34.Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, Kutschera I, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 35.Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F, et al. Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biffi A, Bartolomae CC, Cesana D, Cartier N, Aubourg P, Ranzani M, et al. Lentiviral vector common integration sites in preclinical models and a clinical trial reflect a benign integration bias and not oncogenic selection. Blood. 2011;117:5332–5339. doi: 10.1182/blood-2010-09-306761. [DOI] [PubMed] [Google Scholar]
- 37.Miyoshi H, Blomer U, Takahashi M, Gage FH, Verma IM. Development of a self-inactivating lentivirus vector. J Virol. 1998;72(10):8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwakuma T, Cui Y, Chang LJ. Self-inactivating lentiviral vectors with U3 and U5 modifications. Virology. 1999;261:120–132. doi: 10.1006/viro.1999.9850. [DOI] [PubMed] [Google Scholar]
- 39.Pauwels K, Gijsbers R, Toelen J, Schambach A, Willard-Gallo K, Verheust C, et al. State-of-the-art lentiviral vectors for research use: risk assessment and biosafety recommendations. Curr Gene Ther. 2009;9:459–474. doi: 10.2174/156652309790031120. [DOI] [PubMed] [Google Scholar]
- 40.Montini E, Cesana D, Schmidt M, Sanvito F, Ponzoni M, Bartholomae C, et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat Biotechnol. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- 41.Montini E, Cesana D, Schmidt M, Sanvito F, Bartholomae CC, Ranzani M, et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J Clin Invest. 2009;119:964–975. doi: 10.1172/JCI37630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward NJ, Buckley SM, Waddington SN, Vandendriessche T, Chuah MK, Nathwani AC, et al. Codon optimization of human factor VIII cDNAs leads to high-level expression. Blood. 2011;117:798–807. doi: 10.1182/blood-2010-05-282707. [DOI] [PubMed] [Google Scholar]
- 43.Lynch CM, Israel DI, Kaufman RJ, Miller AD. Sequences in the coding region of clotting factor VIII act as dominant inhibitors of RNA accumulation and protein production. Hum Gene Ther. 1993;4:259–272. doi: 10.1089/hum.1993.4.3-259. [DOI] [PubMed] [Google Scholar]
- 44.Hoeben RC, Fallaux FJ, Cramer SJ, van den Wollenberg DJ, van Ormondt H, Briet E, et al. Expression of the blood-clotting factor-VIII cDNA is repressed by a transcriptional silencer located in its coding region. Blood. 1995;85:2447–2454. [PubMed] [Google Scholar]
- 45.Koeberl DD, Halbert CL, Krumm A, Miller AD. Sequences within the coding regions of clotting factor VIII and CFTR block transcriptional elongation. Hum Gene Ther. 1995;6:469–479. doi: 10.1089/hum.1995.6.4-469. [DOI] [PubMed] [Google Scholar]
- 46.Fallaux FJ, Hoeben RC, Cramer SJ, van den Wollenberg DJ, Briet E, van Ormondt H, et al. The human clotting factor VIII cDNA contains an autonomously replicating sequence consensus- and matrix attachment region-like sequence that binds a nuclear factor, represses heterologous gene expression, and mediates the transcriptional effects of sodium butyrate. MCB. 1996;16:4264–4272. doi: 10.1128/mcb.16.8.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bi L, Lawler AM, Antonarakis SE, High KA, Gearhart JD, Kazazian HH., Jr Targeted disruption of the mouse factor VIII gene produces a model of haemophilia A. Nat Genet. 1995;10:119–121. doi: 10.1038/ng0595-119. [DOI] [PubMed] [Google Scholar]
- 48.Doering CB, Healey JF, Parker ET, Barrow RT, Lollar P. High level expression of recombinant porcine coagulation factor VIII. J Biol Chem. 2002;277:38345–38349. doi: 10.1074/jbc.M206959200. [DOI] [PubMed] [Google Scholar]