Abstract
In both budding and fission yeast, a large number of ribonucleotides are incorporated into DNA during replication by the major replicative polymerases (Pols α, δ, and ε). They are subsequently removed by RNase H2-dependent repair, which if defective leads to replication stress and genome instability. To extend these studies to humans, where an RNase H2 defect results in an autoimmune disease, here we compare the ability of human and yeast Pol δ to incorporate, proofread, and bypass ribonucleotides during DNA synthesis. In reactions containing nucleotide concentrations estimated to be present in mammalian cells, human Pol δ stably incorporates one rNTP for approximately 2,000 dNTPs, a ratio similar to that for yeast Pol δ. This result predicts that human Pol δ may introduce more than a million ribonucleotides into the nuclear genome per replication cycle, an amount recently reported to be present in the genome of RNase H2-defective mouse cells. Consistent with such abundant stable incorporation, we show that the 3´-exonuclease activity of yeast and human Pol δ largely fails to edit ribonucleotides during polymerization. We also show that, like yeast Pol δ, human Pol δ pauses as it bypasses ribonucleotides in DNA templates, with four consecutive ribonucleotides in a DNA template being more problematic than single ribonucleotides. In conjunction with recent studies in yeast and mice, this ribonucleotide incorporation may be relevant to impaired development and disease when RNase H2 is defective in mammals. As one tool to investigate ribonucleotide incorporation by Pol δ in human cells, we show that human Pol δ containing a Leu606Met substitution in the polymerase active site incorporates 7-fold more ribonucleotides into DNA than does wild type Pol δ.
Keywords: DNA polymerase delta, ribonucleotide, incorporation, proofreading, bypass
1. INTRODUCTION
The presence of a ribonucleotide in DNA is potentially problematic because the 2´-oxygen on the ribose renders the DNA backbone susceptible to cleavage and potentially changes the conformation of the sugar pucker. Thus shortly after the discovery of DNA polymerases [1], it became of interest to examine how effectively DNA polymerases prevent ribonucleotide incorporation during DNA synthesis. Numerous studies since then (reviewed in [2, 3]) have revealed that discrimination against ribonucleotide incorporation can be high, but varies widely among DNA polymerases and is not absolute. The probability of ribonucleotide incorporation is increased by the fact that in both budding yeast [4] and in mammalian cells [5], ribonucleoside triphosphate (rNTP) concentrations are higher than deoxyribonucleoside triphosphates (dNTP) concentrations. These facts led us to examine the frequency of stable ribonucleotide incorporation into DNA by the three DNA polymerases that replicate the budding yeast nuclear genome, Pols α, δ, and ε. In reactions containing physiological concentrations of the rNTPs and dNTPs, these replicases incorporate a surprisingly large number of ribonucleotides into DNA [4]. The biological relevance of these ribonucleotides was revealed by deleting the budding yeast RNH201 gene that encodes the catalytic subunit of RNase H2, the enzyme that initiates removal of ribonucleotides from DNA (see [6, 7] and references therein). Budding yeast rnh201Δ strains accumulate large numbers of ribonucleotides in their genome and they have several phenotypes characteristic of replicative stress, including genome instability [6–9]. Large numbers of ribonucleotides are also incorporated by Pol ε into DNA in fission yeast, and these are also removed in a RNaseH2-dependent manner [10]. Moreover, Pol ε from budding yeast can proofread incorporated ribonucleotides, albeit not as efficiently as a misincorporated base [11].
The phenotypes of RNase H2-defective yeast may be relevant to the fact that defects in RNase H2 in humans result in Aicardi-Goutières syndrome, a recessive neuroinflammatory condition with similarities to autoimmune diseases [12]. It is therefore of interest to know whether the causes and consequences of ribonucleotide incorporation during DNA replication in yeast extend to higher eukaryotes. Of particular relevance here is a recent study demonstrating that more than a million ribonucleotides are present in the genome of RNase H2-defective mouse embryo cells [13]. These could result from failure to completely remove RNA primers from Okazaki fragments and/or from rNMPs incorporated by DNA polymerases during replication. As an initial step towards understanding the origins of ribonucleotides in the mammalian nuclear genome, and their possible relevance to human biology, we describe the ability to incorporate and proofread ribonucleotides during DNA synthesis in vitro by human Pol δ which has been inferred to be the primary lagging strand replicase [14] and which has high fidelity and can efficiently proofread base-base mismatches [15]. We also examine Pol δ bypass of single and multiple consecutive ribonucleotides in DNA templates. We observe pausing during bypass that may contribute to the replicative stress observed in RNase H2-defective cells. Finally, having previously showed that a Leu612Met variant of yeast Pol δ that has lower than normal fidelity [14] also incorporates increased numbers of ribonucleotides into DNA, here we demonstrate that a similar variant in human Pol δ incorporates large amounts of ribonucleotides into DNA. Overall, the results suggest that the biochemical properties of yeast Pol δ regarding ribonucleotide processing are conserved in human Pol δ, which has several implications that are discussed below.
2. MATERIALS AND METHODS
2.1 Materials and reagents
Oligonucleotides were purchased from Oligos Etc., Inc. (Wilsonville, OR). dNTPs and rNTPs were purchased from GE-Healthcare. Yeast Pol δ was purified as described previously [16]. Yeast PCNA was purified as described previously [17]. Proofreading deficient human Pol δ D402A variants (Exo−) were constructed as previously decribed [18]. Proofreading proficient (Exo+) and proofreading deficient human Pol δ, Pol δ Leu606Met, and PCNA were purified as described previously [19].
2.2 Ribonucleotide incorporation assay
Stable incorporation of rNMPs by yeast and human Pol δ was analyzed using a substrate made by annealing a 40-mer 32P-labeled primer strand (5′-CCAGTGAATTTCTGCAGGTCGACTCCAAAGGTCAACCCGG) to a 70-mer template strand (5′-ATGACCATGATTACGAATTCCAGCTCGGTACCGGGTT GACCTTTGGAGTCGACCTGCAGAAATTCACTGG). Reaction mixtures contained 100 nM DNA substrate, and the reaction buffer contained 20 mM Tris (pH 7.8), 200 µg/mL BSA, 1 mM DTT, 90 mM NaCl, 8 mM Mg-acetate, and 400 nM human or yeast PCNA was added to the reaction mixture. Nucleotide substrates were added at cellular concentrations (Table 1) and contained all eight nucleotides. Reactions were initiated by adding 40 nM human or yeast Pol δ. Incubation was at 37°C for human Pol δ and at 30°C for yeast Pol δ. Reactions were terminated after 30 min by adding an equal volume of formamide loading dye, and the reaction products were separated in a denaturing 8% polyacrylamide gel. Full-length reaction products identified by exposing the gel to x-ray film were excised and purified as described [4]. Equivalent amounts of recovered products (as determined by scintillation counting) were treated with either 0.3 M KCl or 0.3 M KOH for 2 h at 55°C. Following addition of an equal volume of formamide loading dye, equivalent amounts of pre- and postexcision samples were analyzed by electrophoresis in a denaturing 8% polyacrylamide gel. Products were detected and quantified using a PhosphorImager and ImageQuaNT software (Molecular Dynamics).
Table 1.
| Yeast | Human | |||
|---|---|---|---|---|
| Base | dNTP | rNTP | dNTP | rNTP |
| A | 16 | 3,000 | 24 | 3,200 |
| C | 14 | 500 | 29 | 280 |
| G | 12 | 700 | 5.2 | 470 |
| T/U | 14 | 1,700 | 37 | 570 |
2.3 Bypass efficiency assay
Reaction mixtures contained 100 nM DNA substrate and the reaction buffer contained 20 mM Tris (pH 7.8), 200 µg/mL BSA, 1 mM DTT, 90 mM NaCl, 8 mM Mg-acetate and 400 nM human or yeast PCNA. The dGTP, dCTP, dATP and dTTP concentrations used are shown in Table 1. All components except the polymerase were mixed on ice and then incubated at 30°C for 2 minutes. 40 nM of the polymerase was added to initiate the reaction and aliquots were removed at 3, 6, and 9 minutes. Following addition of an equal volume of formamide loading dye, samples were analyzed by electrophoresis in a denaturing 12% polyacrylamide gel. Products were detected and quantified using a PhosphorImager and ImageQuaNT software (Molecular Dynamics).
3. RESULTS AND DISCUSSION
3.1 Abundant ribonucleotide incorporation by human Pol δ
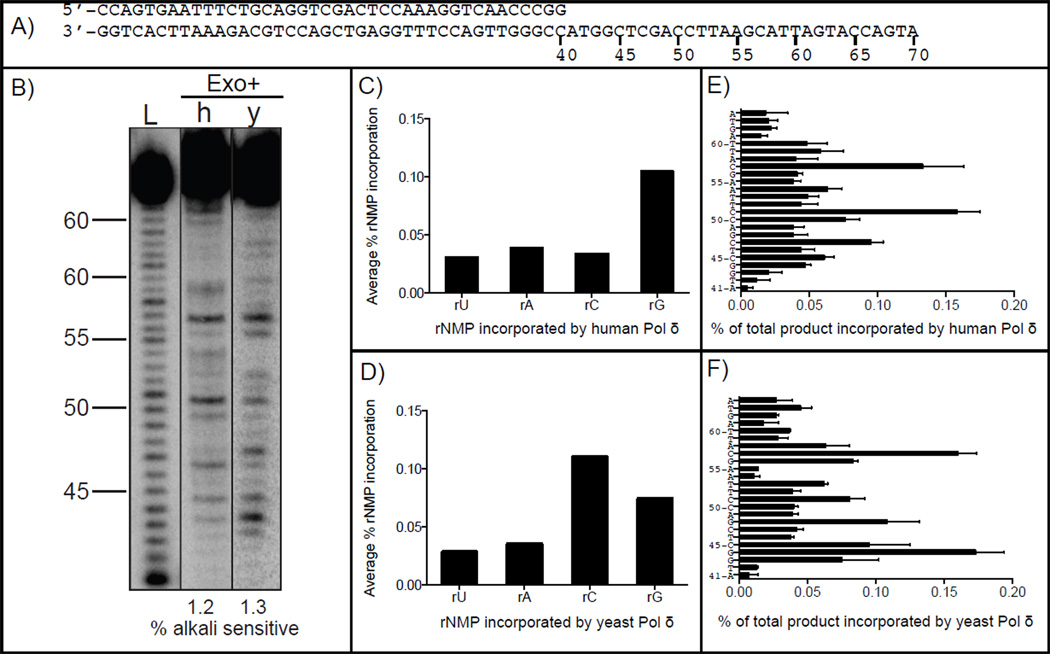
Human Pol δ is a heterotetramer comprised of four subunits: the catalytic subunit (p125) and three accesory subunits (p68, p50 and p12). The frequency with which four subunit human Pol δ stably incorporates ribonucleotides into DNA was examined in DNA synthesis reaction mixtures containing human PCNA and the rNTP and dNTP concentrations estimated to be present in mammalian cells (Table 1, from [5]). Yeast Pol δ is a heterotrimer comprised of three subunits: the catalytic subunit (pol3), and two accesory subunits (pol31 and pol32). A parallel reaction was performed using three subunit yeast Pol δ and yeast PCNA, in this case using the rNTP and dNTP concentrations in yeast (Table 1, from [4]). After extending a 5´-32P-end-labeled 40-mer primer hybridized to a 70-mer template (Fig. 1A), full-length products were gel-purified and treated with 0.3 M KCl (control) or with 0.3M KOH to hydrolyze the DNA backbone at locations where rNMPs are present. The DNA products were separated by electrophoresis in a denaturing polyacrylamide gel and quantified by phosphorimaging. The results (Fig. 1B) show that 1.2±0.1% of the reaction products generated by human Pol δ (lane 2) were sensitive to alkaline hydrolysis. When divided by the 24 positions quantified here (nucleotides 41 through 65, Fig. 1A), the average value is one incorporated ribonucleotide per 2,000 deoxyribonucleotides. This ratio is remarkably similar to that for yeast Pol δ (lane 3, 1.3±0.2% total, 1:1,800). We conclude that the ability of Pol δ to incorporate ribonucleotides into DNA during synthesis in the presence of physiological nucleotide pools is conserved between budding yeast and humans. In a model wherein human Pol δ performs about 90% of lagging strand replication of the six billion nucleotide nuclear genome, the results in Figure 1B predict the incorporation of 3,000,000 ribonucleotides by Pol δ into the nascent lagging strand per round of DNA replication. Given the simplicity of the analysis in vitro compared to the complexity of replication in vivo, it is amazing that Reijns et. al. [13] recently reported the presence of >1,000,000 single or di-ribonucleotides per cell in mouse embryos deficient in RNase H2, the enzyme that initiates removal of ribonucleotides incorporated into DNA during replication in yeast [6]. While the study in mice did not identify the source of the ribonucleotides in the genome, i.e., whether incorporated by RNA primase, a replicase or during repair synthesis [20, 21], our data imply that large numbers of ribonucleotides may be incorporated during replication in human cells by Pol δ.
Figure 1.
Stable incorporation of ribonucleotides into DNA by proofreading-proficient human (h) and yeast (y) Pol δ. (A) Primer-template sequences. (B) Alkali cleavage products of polymerase reactions with all eight NTPs at cellular concentrations by human and yeast Pol δ in the presence PCNA. The relative amount of ribonucleotides incorporated into the primer strand is indicated below each lane. L indicates the ladder. (C) Average frequency of ribonucleotide incorporation by human Pol δ according to the incorporated ribonucleotide. (D) Average frequency of ribonucleotide incorporation by yeast Pol δ according to the incorporated ribonucleotide. (E) Frequency of ribonucleotide incorporation by human Pol δ at each of 24 template positions. (F) Frequency of ribonucleotide incorporation by yeast Pol δ at each of 24 template positions. Results are from at least two independent experiments.
Variations from the average ribonucleotide incorporation are seen in the gel images (Fig. 1B). These variations depend on the identity of the nucleotide (Fig. 1, panels C and D) and its position in the DNA sequence (Fig. 1, panels E and F), with the latter variations sometimes exceeding 10-fold. These observations predict that ribonucleotide incorporation into the human genome will not be random. This could be relevant to the non-random distribution of short deletions in repetitive sequences observed in yeast [6, 9] and the increased presence of micronuclei and chromosomal rearrangements in mice [13] that are associated with unrepaired ribonucleotides in RNase H2-defective cells. Theoretically, the non-random presence of ribonucleotides in DNA could be relevant to any signaling functions that ribonucleotides in DNA might have (discussed in [4]), such as signaling for mismatch repair. For these reasons, it will be of interest in the future to map ribonucleotides in yeast and mammalian genomes, preferably at single-nucleotide resolution.
3.2 Lack of ribonucleotide proofreading by human and yeast Pol δ
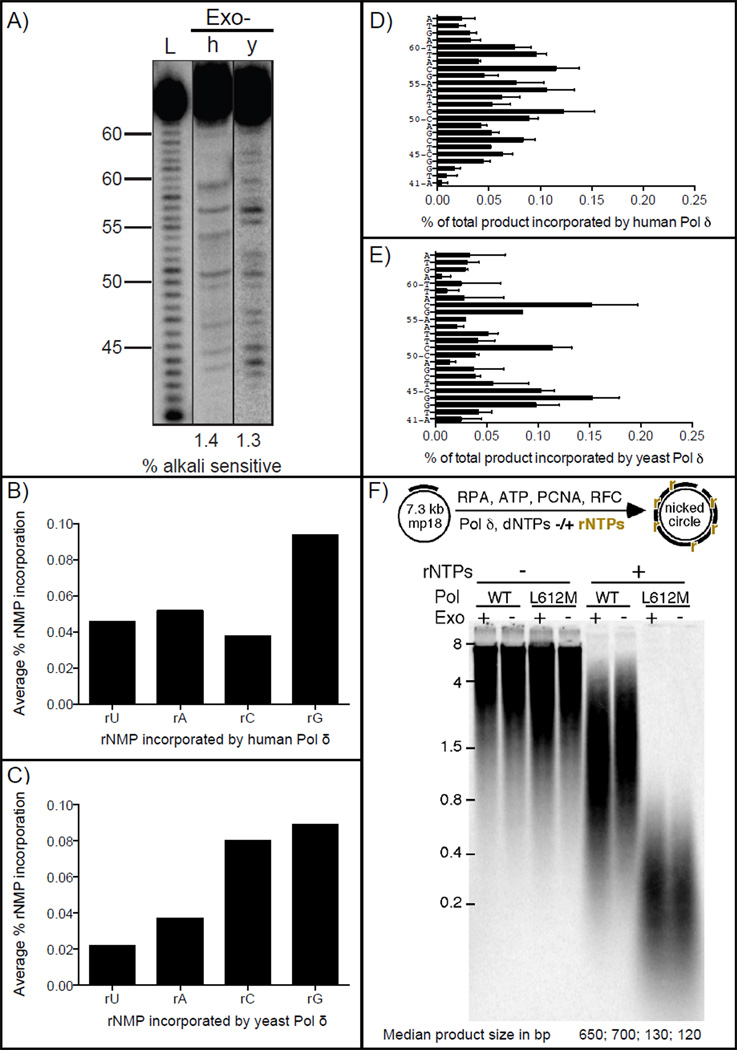
We recently reported that the 3´ exonuclease activity of yeast Pol δ can weakly proofread newly inserted ribonucleotides [11]. To determine if this is also the case for yeast and human Pol δ, ribonucleotide incorporation experiments were performed using exonuclease-deficient Pol δ derivatives. The results (Fig. 2) were compared to those described above for the exonuclease-proficient enzymes (Fig. 1). The proportion of ribonucleotides incorporated into DNA by the exonuclease-deficient polymerases (Fig. 2A) was similar to that seen with wild type Pol δ, for both human (lane 2, 1.4±0.2%) and yeast (lane 3, 1.3±0.0%) Pol δ. Likewise, the proportion of rU, rA, rC and rG incorporated (Fig. 2, panels B and C) and the site-specific distributions of newly incorporated ribonucleotides (panels D and E) were similar, albeit not identical. These data indicate that proofreading of ribonucleotides by Pol δ is very inefficient. This suggests that, unlike the efficient proofreading of base-base mismatches created during replication, the editing function of the major lagging strand replicase does not protect the nuclear genome against ribonucleotide incorporation during lagging strand replication, either in budding yeast or in human cells.
Figure 2.
Stable incorporation of ribonucleotides into DNA by proofreading-deficient human (h) and yeast (y) Pol δ. (A) Alkali cleavage products of reactions with all eight NTPs at cellular concentrations by human and yeast Pol δ in the presence of PCNA. The relative amount of ribonucleotides incorporated into the primer strand is indicated below each lane. L indicates the ladder. (B) Average frequency of ribonucleotide incorporation by human Pol δ according to the incorporated ribonucleotide. (C) Average frequency of ribonucleotide incorporation by yeast Pol δ according to the incorporated ribonucleotide. (D) Frequency of ribonucleotide incorporation by human Pol δ at each of 24 template positions. (E) Frequency of ribonucleotide incorporation by yeast Pol δ at each of 24 template positions. (F) Primed RPA-coated SS-M13mp18 DNA was fully replicated by proofreading proficient and deficient yeast Pol δ or the Leu612Met mutant forms under standard conditions with or without rNTPs, as described before (6). Reactions were treated with 0.3 M NaOH. The products were recovered by ethanol precipitation and separated on a 1% alkaline agarose gel. Median product sizes are indicated below the gel image. Results are from at least two independent experiments.
We also examined proofreading by yeast Pol δ during synthesis to copy circular M13mp18 DNA in the presence of RFC, PCNA, and RPA (Fig. 2F, lane 5 and 6), i.e., a reaction that more closely mimics lagging strand DNA replication in cells. The median size of alkali-generated products for this reaction were similar for the wild type and exonuclease-deficient polymerases (650 ± 100 vs. 700 ± 100 bp) (Fig. 2F). This confirms the lack of ribonucleotide proofreading by yeast Pol δ, and it further supports an earlier study showing that high-density ribonucleotide incorporation occurs with proofreading-proficient Pol δ even in the presence of the accessory proteins involved in lagging strand replication in vivo [7].
3.3 Ribonucleotide bypass by human Pol δ
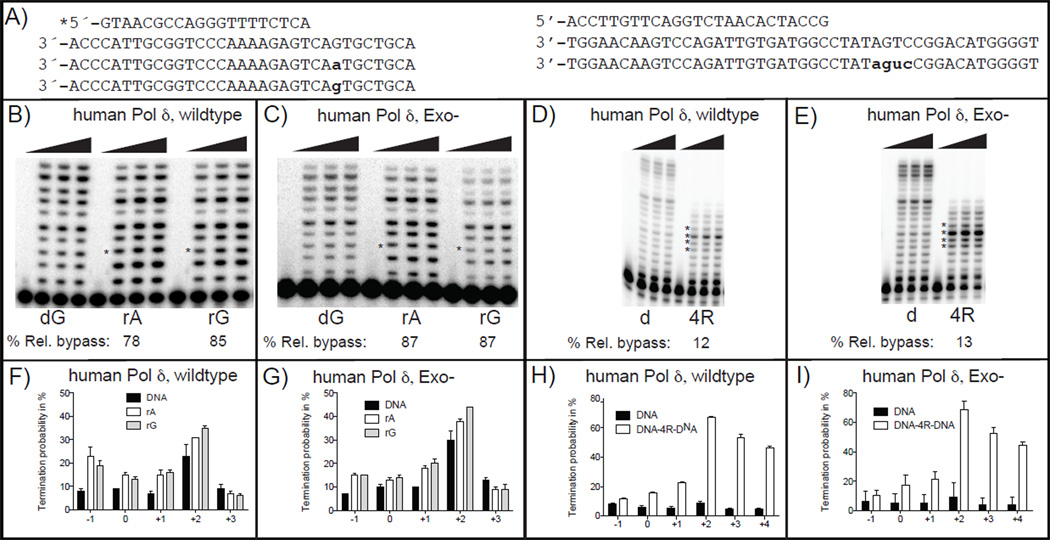
In yeast cells defective in RNase H2, unrepaired ribonucleotides incorporated into DNA will be in the template strand used for the next round of replication. Having previously reported on the ribonucleotide bypass capacity of yeast Pol δ [22, 23], here we performed similar experiments with human Pol δ. Bypass was examined using DNA templates containing either rA, rG, or four consecutive ribonucleotides (Fig. 3A). The ribonucleotides in all three templates were bypassed by wild type Pol δ (Fig. 3B/D). Relative to the all DNA (control) template, the single ribonucleotide bypass efficiency of wild type Pol δ was 78% for rA and 85% for rG (Fig. 3B). Examination of termination probabilities during processive synthesis (Fig. 3F) indicated that reduced bypass is a consequence of a slight reduction in incorporation efficiency for a stretch of four consecutive nucleotides as bypass proceeds, i.e., dNTP insertion opposite the nucleotide before the template ribonucleotide (designated −1) through the template nucleotide two bases beyond the ribonucleotide (designated +2). Bypass of four consecutive ribonucleotides in the template was more problematic, occurring with a relative bypass efficiency of only 10% (Fig. 3D). With this substrate, insertion becomes increasingly difficult as synthesis proceeds (Fig. 3H), i.e., as the number of ribonucleotide-containing base pairs within the duplex DNA upstream of the polymerase active site increases. When reactions were performed in parallel with exonuclease-deficient human Pol δ, ribonucleotide bypass parameters were only slightly affected (Fig. 3C/G/E/I), indicating that the dominant determinant of ribonucleotide bypass is the polymerase, not the exonuclease.
Figure 3.
PAGE phosphorimages of bypass of a single or four consecutive ribonucleotides by proofreading proficient (Exo+) and deficient human Pol δ (Exo−). (A) Primer-template sequences, letters in bold face and small caps indicates positions of a ribonucleotide. (B) Proofreading proficient (Exo+) human Pol δ bypassing a single rA or rG. (C) Proofreading deficient Pol δ (Exo−) bypassing rA or rG. (D) Proofreading proficient human Pol δ (Exo+) bypassing four consecutive ribonucleotides. (E) Proofreading deficient Pol δ (Exo−) bypassing four consecutive ribonucleotides. Gel images of reaction products shown in Figures 3B–E were quantified as described in Methods and bar graph of termination probabilities at each incorporation is shown in Figures 3F–I, respectively. Position “0” corresponds to the location of the ribonucleotide, “−1” indicates the preceding incorporation, and “+1” and “+3” indicate sequential incorporations after insertion at “0”. Error bars represent the standard deviations. Results are from at least two independent experiments.
The ribonucleotide bypass capacity of human Pol δ is remarkably similar to that of yeast Pol δ [22, 23], i.e., this biochemical property is conserved between budding yeast and humans. Pausing during ribonucleotide bypass by Pol δ may be relevant to the replication stress observed in RNase H-defective yeast [6] and mouse cells [13]. Even slight pausing due to single ribonucleotide bypass may be biologically relevant given the sheer number of ribonucleotides present in RNase H2-defective yeast and mouse cells. Also of interest is the difficulty with which Pol δ bypasses four consecutive ribonucleotides (Fig. 3, panels D and H). RNA-DNA hybrid substrates containing four consecutive ribonucleotides in one strand are subject to cleavage by both RNase H1 and RNase H2 [24]. This is interesting because when grown in the presence of the replication inhibitor hydroxyurea, survival of yeast cells lacking both RNases H depends on two lesion tolerance pathways, Rad5-dependent template switching and Rev3-dependent translesion synthesis by Pol δ [22]. We previously showed that yeast Pol δ efficiently bypasses four consecutive ribonucleotides in a DNA template, whereas yeast Pol δ has difficulty doing so [22]. The fact that human Pol δ also has difficulty bypassing multiple consecutive ribonucleotides in DNA suggests that perhaps in humans as in yeast, certain ribonucleotide-containing templates may stall replication fork progression to the degree that Pol δ -dependent TLS is invoked. Precedents for such replication fork stalling at ribonucleotides are studies of mating type switching in fission yeast, where two consecutive ribonucleotides at a specific location in the genome are reported to stall replication to initiate switching [25].
3.4 Ribonucleotide incorporation by human L606M Pol δ
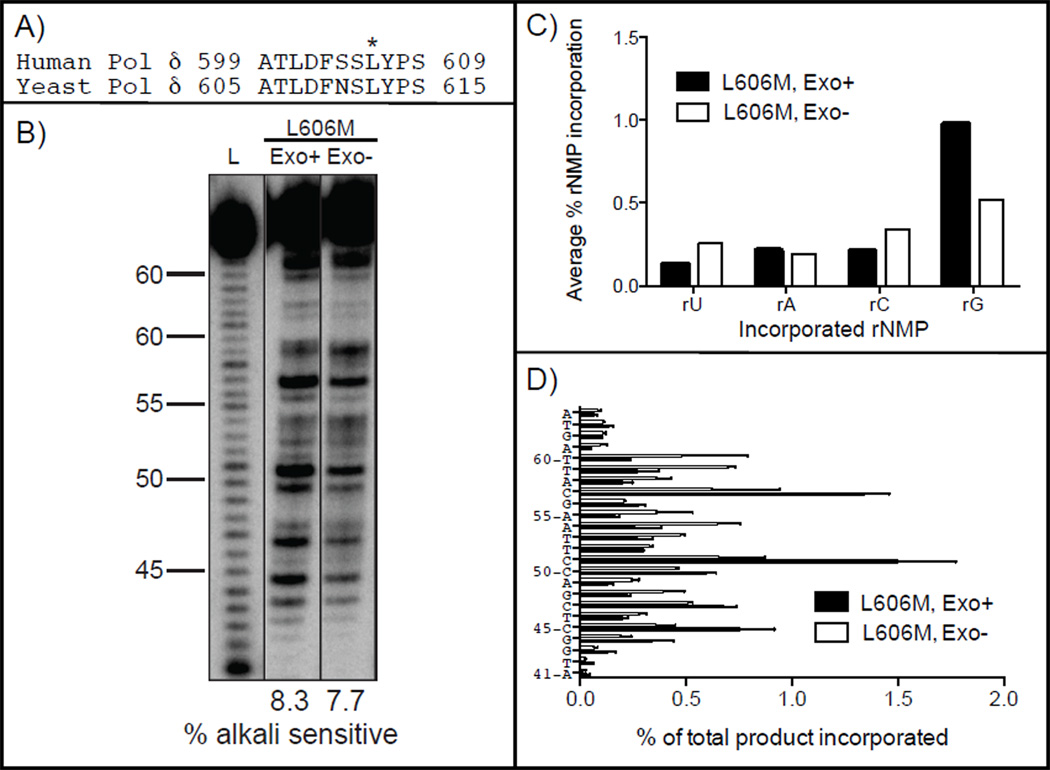
In budding and fission yeasts, investigating of ribonucleotide incorporation and its consequences have been facilitated by the availability of variants of Pol δ and Pol ε that have an altered propensity to incorporate ribonucleotides during DNA synthesis [6, 10]. These variants differ from their wild type parent replicases by a single amino acid change at the active site adjacent to a tyrosine that acts as a steric gate to prevent ribonucleotide incorporation. Increased ribonucleotide incorporation and lack of proofreading were also observed with the Leu612Met variant of yeast Pol δ (Fig. 2F, lane 7 and 8). The median product size was 130 ± 50 bp for the yeast Pol δ Leu612Met variant and 120 ± 50 bp for the proofreading deficient variant. When a Leu612Met variant of Pol δ is present in RNase H2-defective yeast it results in an increased number of ribonucleotides that are preferentially present in the nascent lagging strand (manuscript in preparation). In order to facilitate future studies of ribonucleotide incorporation during replication in human cells, we examined the ability of the homologous Leu606Met variant of human Pol δ to incorporate ribonucleotides in reactions performed as described above. The proportion of ribonucleotides incorporated by Leu606Met Pol δ was 8.3±0.5% (Fig. 4B, lane 2), which is 7-fold higher than for wild type Pol δ (1.2%, Fig. 1B). The priority order for ribonucleotide incorporation remained similar to wild type, with rG being the highest (Fig. 4C), as did the site-specific variations in ribonucleotide incorporation at various positions (Fig. 4D). Similar results were observed with exonuclease-deficient Leu606Met Pol δ (Fig. 4B, lane 3, 7.7±0.0%), indicating that, as for wild type human Pol δ, newly incorporated ribonucleotides are not efficiently proofread during synthesis by Leu606Met Pol δ. These results indicate that this variant of Pol δ will be a useful tool to study ribonucleotide incorporation in human cells and the consequences of failure to repair these non-canonical nucleotides in DNA. Ribonucleotides incorporated by this variant could potentially be high-density physical biomarkers of Pol δ action during lagging strand replication and possibly during recombination and DNA synthesis associated with excision repair processes.
Figure 4.
Stable incorporation of ribonucleotides into DNA by proofreading proficient (Exo+) and deficient (Exo−) Leu606Met human Pol δ. (A) An alignment of motif A for human and yeast Pol δ is shown and the asterisk indicates the position of the Leu606Met mutation for human Pol δ. (B) Alkali cleavage products of reactions with all eight NTPs at cellular concentrations Leu606Met human Pol δ in the presence of PCNA. The relative amount of ribonucleotides incorporated into the primer strand is indicated below each lane. L indicates the ladder. (C) Average frequency of ribonucleotide incorporation by human Pol δ according to the incorporated ribonucleotide. (D) Frequency of ribonucleotide incorporation by human Pol δ at each of 24 template positions. Results are from at least two independent experiments.
Highlights.
The ribonucleotide incorporation properties of yeast Pol δ are conserved in humans
Pol δ does not efficiently proofread newly inserted ribonucleotides
Consecutive ribonucleotides in a DNA template impede synthesis by human Pol δ
A variant of human Pol δ incorporates more ribonucleotides than does wild type Pol δ
ACKNOWLEDGEMENTS
We thank Roel M. Schaaper and Jessica S. Williams for helpful comments on the manuscript. This work was supported by Project Z01 ES065070 to T.A.K., Project GM032431 to P.M.B. and Project 5RO1 GM031973 to M.Y.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Kornberg A, Baker TA. DNA Replication. Herndon, Virginia: University Science Books; 2005. [Google Scholar]
- 2.Brown JA, Suo ZC. Unlocking the Sugar "Steric Gate" of DNA Polymerases. Biochemistry. 2011;50:1135–1142. doi: 10.1021/bi101915z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joyce CM. Choosing the right sugar: How polymerases select a nucleotide substrate. Proceedings of the National Academy of Sciences. 1997;94:1619–1622. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McElhinny SAN, Watts BE, Kumar D, Watt DL, Lundstrom EB, Burgers PMJ, Johansson E, Chabes A, Kunkel TA. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Traut TW. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 6.McElhinny SAN, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, Johansson E, Chabes A, Kunkel TA. Genome instability due to ribonucleotide incorporation into DNA. Nat. Chem. Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sparks JL, Chon H, Cerritelli SM, Kunkel TA, Johansson E, Crouch RJ, Burgers PM. RNase H2-Initiated Ribonucleotide Excision Repair. Molecular Cell. 2012;47:980–986. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark AB, Lujan SA, Kissling GE, Kunkel TA. Mismatch repair-independent tandem repeat sequence instability resulting from ribonucleotide incorporation by DNA polymerase epsilon. DNA Repair. 2011;10:476–482. doi: 10.1016/j.dnarep.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim N, Huang SYN, Williams JS, Li YC, Clark AB, Cho JE, Kunkel TA, Pommier Y, Jinks-Robertson S. Mutagenic Processing of Ribonucleotides in DNA by Yeast Topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyabe I, Kunkel TA, Carr AM. The Major Roles of DNA Polymerases Epsilon and Delta at the Eukaryotic Replication Fork Are Evolutionarily Conserved. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams JS, Clausen AR, McElhinny SAN, Watts BE, Johansson E, Kunkel TA. Proofreading of ribonucleotides inserted into DNA by yeast DNA polymerase epsilon. DNA Repair. 2012;11:649–656. doi: 10.1016/j.dnarep.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, Baumann C, Baxter P, Bertini E, Chandler KE, Chitayat D, Cau D, Dery C, Fazzi E, Goizet C, King MD, Klepper J, Lacombe D, Lanzi G, Lyall H, Martinez-Frias ML, Mathieu M, McKeown C, Monier A, Oade Y, Quarrell OW, Rittey CD, Rogers RC, Sanchis A, Stephenson JBP, Tacke U, Till M, Tolmie JL, Tomlin P, Voit T, Weschke B, Woods CG, Lebon P, Bonthron DT, Ponting CP, Jackson AP. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutiéres syndrome and mimic congenital viral brain infection. Nature Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 13.Reijns MAM, Rabe B, Rigby RE, Mill P, Astell KR, Lettice LA, Boyle S, Leitch A, Keighren M, Kilanowski F, Devenney PS, Sexton D, Grimes G, Holt IJ, Hill RE, Taylor MS, Lawson KA, Dorin JR, Jackson AP. Enzymatic Removal of Ribonucleotides from DNA Is Essential for Mammalian Genome Integrity and Development. Cell. 2012;149:1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McElhinny SAN, Gordenin DA, Stith CM, Burgers PMJ, Kunkel TA. Division of labor at the eukaryotic replication fork. Molecular Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fortune JM, Pavlov YI, Welch CM, Johansson E, Burgers PMJ, Kunkel TA. Saccharomyces cerevisiae DNA polymerase delta - High fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J. Biol. Chem. 2005;280:29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- 16.Burgers PMJ, Gerik KJ. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 1998;273:19756–19762. doi: 10.1074/jbc.273.31.19756. [DOI] [PubMed] [Google Scholar]
- 17.Eissenberg JC, Ayyagari R, Gomes XV, Burgers PMJ. Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase delta and DNA polymerase epsilon. Mol. Cell. Biol. 1997;17:6367–6378. doi: 10.1128/mcb.17.11.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fazlieva R, Spittle CS, Morrissey D, Hayashi H, Yan H, Matsumoto Y. Proofreading exonuclease activity of human DNA polymerase and its effects on lesion-bypass DNA synthesis. Nucleic Acids Res. 2009;37:2854–2866. doi: 10.1093/nar/gkp155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie B, Mazloum N, Liu L, Rahmeh A, Li H, Lee M. Reconstitution and characterization of the human DNA polymerase delta four-subunit holoenzyme. Biochemistry. 2002;41:13133–13142. doi: 10.1021/bi0262707. [DOI] [PubMed] [Google Scholar]
- 20.McElhinny SAN, Ramsden DA. Polymerase Mu is a DNA-directed DNA/RNA polymerase. Mol. Cell. Biol. 2003;23:2309–2315. doi: 10.1128/MCB.23.7.2309-2315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen Y, Koh KD, Weiss B, Storici F. Mispaired rNMPs in DNA are mutagenic and are targets of mismatch repair and RNases H. Nat. Struct. Mol. Biol. 2012;19:98–104. doi: 10.1038/nsmb.2176. [DOI] [PubMed] [Google Scholar]
- 22.Lazzaro F, Novarina D, Amara F, Watt DL, Stone JE, Costanzo V, Burgers PM, Kunkel TA, Plevani P, Muzi-Falconi M. RNase H and Postreplication Repair Protect Cells from Ribonucleotides Incorporated in DNA. Molecular Cell. 2012;45:99–110. doi: 10.1016/j.molcel.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watt DL, Johansson E, Burgers PM, Kunkel TA. Replication of ribonucleotide-containing DNA templates by yeast replicative polymerases. DNA Repair. 2011;10:897–902. doi: 10.1016/j.dnarep.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. Febs J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vengrova S, Dalgaard JZ. The wild-type Schizosaccharomyces pombe mat1 imprint consists of two ribonucleotides. EMBO Rep. 2006;7:59–65. doi: 10.1038/sj.embor.7400576. [DOI] [PMC free article] [PubMed] [Google Scholar]