Abstract
Engineering functional protein scaffolds capable of carrying out chemical catalysis is a major challenge in enzyme design. Starting from a non-catalytic protein scaffold, we recently generated a novel RNA ligase by in vitro directed evolution. This artificial enzyme lost its original fold and adopted an entirely novel structure with dramatically enhanced conformational dynamics, demonstrating that a primordial fold with suitable flexibility is sufficient to carry out enzymatic function.
The known structures of naturally occurring proteins can be assigned to an apparently finite number of different fold families1,2. Starting from an existing fold, divergent evolution through a combination of gene duplication and mutations is a common path for proteins to acquire new functions while retaining their original fold3,4. However, the origin of those biological folds remains subject to debate5,6. Only a few examples have been described in which new function acquisition is accompanied by a simultaneous change in the protein fold. Those examples have largely been generated by rational design or involve protein binders7–13.
Recently, we created artificial RNA ligase enzymes by in vitro evolution14,15. These enzymes catalyze the joining of a 5′-triphosphorylated RNA to the 3′-hydroxyl group of a second RNA, a reaction for which no natural enzyme catalysts have been found. We began with a non-catalytic small protein domain consisting of two zinc finger motifs from the DNA binding domain of human retinoid-X-receptor (hRXRα)16 (Fig. 1). Two adjacent loops of this protein were randomized to generate a combinatorial library of mutants as input for the selection and evolution process17. Although zinc fingers are common structural motifs, they are not known to take part in catalysis in natural proteins. In contrast, we isolated from the zinc finger library active enzymes that exhibit rate accelerations of more than two-million-fold14.
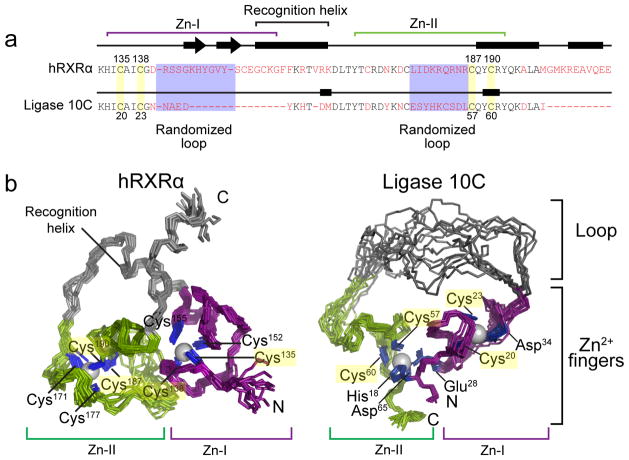
Figure 1. Changes in primary sequence and three-dimensional structure upon directed evolution of the hRXRα scaffold to the ligase enzyme 10C.
(a) Comparison of the primary sequences of hRXRα16 (residues 132–208) and the artificially evolved ligase (residues 17–68). The two zinc finger regions are highlighted with purple and green brackets. The red letters denote residues that are not conserved between the two sequences. (b) Three-dimensional structure of hRXRα16 and NMR ensemble of ligase 10C (for clarity, flexible termini are not shown). Although both proteins contain two zinc fingers, the overall structures are substantially different. Only two zinc-coordinating cysteines of each zinc finger in hRXRα are still coordinating zinc in ligase 10C (highlighted in yellow, see also Fig. 1a) while all other ligands differ in the two structures. In contrast to hRXRα zinc finger Zn-II in ligase 10C comprises residues of both N- and C-terminal sequence, imposing a cyclic structure to the enzyme. Residues involved in zinc coordination are labeled and shown in blue. Note that the new ligase lost both helical domains of the DNA binding domain (grey in left structure), replacing the recognition helix with a long unstructured loop (grey in right structure).
Sequence analysis of the artificial enzyme showed that several amino acids essential to maintaining zinc finger structure integrity were mutated or deleted, suggesting that the original scaffold may have been abandoned during the process of mutagenesis and evolution. The original hRXRα scaffold consisted of two loop-helix domains, each containing a zinc ion tetrahedrally coordinated by four cysteines16. However, during evolution of the ligase enzyme, only half of the zinc-coordinating cysteines had been conserved. In the starting scaffold, two helices were packed perpendicularly to form the globular fold and build the hydrophobic core and an additional helix was located at the C-terminus (Fig. 1b). In the ligase enzyme, seven residues of the former DNA recognition helix and ten residues of the C-terminal helix were deleted from the original hRXRα scaffold.
NMR structural analyses of the ligase 10C (Online Methods), chosen for its superior solubility and thermostability, revealed that the evolved ligase lost the original zinc finger scaffold, adopting an entirely novel structure (Fig. 1b). This new three-dimensional structure still contained two zinc sites that constitute the folding core of the protein, however, the two Zn2+ ions were coordinated by several new ligands with a different register. The deletion of two N-terminal cysteines during directed evolution resulted in the concomitant rearrangement of the local geometry of the zinc-binding loop. Additionally, the short stretch of anti-parallel β-sheet within the first zinc finger (Zn-I) was also deleted. The C-terminal loop-helix domains and the recognition helix of hRXRα responsible for binding to the DNA groove18 were lost completely; the latter was replaced by an unstructured loop of twenty amino acids connecting the two new zinc fingers. The zinc fingers made up the most structured region, as demonstrated by the presence of short- and long-range nuclear Overhauser Effect (NOE) contacts. Moreover, several long-range NOEs indicated that the two metal-binding loops are in close proximity, while most of the protein presented only short range NOE contacts (Supplementary Results, Supplementary Fig. 1 and Supplementary Table 1). The conformational ensemble resulting from simulated annealing calculations showed two well-defined regions (residues 17–35 and 49–69) with root-mean-square-deviation from the average of less than 1 Å, while the large loop encompassing residues 36–48 was completely unstructured (RMSD greater than 6 Å). The three-dimensional structure of the enzyme was compounded by residual dipolar coupling measurements, which also helped to better define the local geometry around the zinc binding sites (Supplementary Fig. 2,3).
The two metal centers were responsible for the overall fold of the ligase. In the absence of Zn2+, the NMR fingerprint spectrum of the enzyme displayed broad and mostly unresolved resonances, typical of a molten globule. Titration of Zn2+ to the metal-free protein first saturated the C-terminal Zn2+ binding site (Zn-II) and induced a substantial structural rearrangement with sharper and more dispersed resonances (Supplementary Fig. 4,5). The transition between the unfolded and folded states of the ligase involved multiple intermediate species. For selected resonances, we could discern the presence of two distinct states in slow exchange in the NMR time scale. Complete saturation with Zn2+ funneled the enzyme into a more defined structure, with the complete resolution of fingerprint resonances showing only one population of peaks. Elemental analysis by inductively coupled plasma mass spectrometry revealed 2.74 ± 0.01 equivalents (± s.d.) of bound zinc per ligase molecule. We were able to fit the thermocalorimetry data using models with two or more Zn2+ binding sites, however, the fit does not improve significantly with n>2 (Supplementary Fig. 6 and Supplementary Table 2). Assigning two sites in accordance with the NMR titration data leads to one binding site Zn-II with higher affinity (Kd ~ 3μM), and a second binding site Zn-I with lower affinity for Zn2+ (Kd ~ 93 μM). These values were further supported by the zinc concentration dependence of the enzyme activity, showing a steep drop in activity at concentrations below 100 μM Zn2+ (Supplementary Fig. 7). Notably, the ligase affinity for Zn2+ was substantially lower than those reported for natural zinc-containing proteins which commonly have dissociations constants of 10−8–10−13 M19. Structure calculations were carried out in the absence of explicit Zn2+ ions to avoid conformational search bias and converged toward a structural ensemble with two distinct Zn2+ binding sites: the tetra coordinated N-terminal site (Zn-I) with weaker binding affinity, and the hexa coordinated C-terminal loop (Zn-II) with higher binding affinity (Supplementary Fig. 2,3). EXAFS data corroborated these results, showing that both Zn sites coordinated with two S(Cys) ligands with a Zn-S distance of 2.3 Å, and at least one site had four Zn-N/O ligands while the other site had two to four. These atom ligands can be either protein based or water molecules (Supplementary Fig. 8).
The directed evolution process that yielded the artificial enzyme was based only on product formation without structural constraints14. As a result, the ligase enzyme evolved into a new structure with substantially increased conformational dynamics compared to the original DNA-binding scaffold20. In fact, ligase 10C showed an overall increase in structural plasticity and malleability. While the two zinc fingers exhibited heteronuclear NOEs similar to the original scaffold, the region where the two helical domains were deleted displayed much higher flexibility, with heteronuclear NOEs below 0.5. These data indicate augmentation of conformational dynamics in the picosecond to nanosecond time scale supported by longitudinal (T1) and transverse (T2) nuclear spin-relaxation measurements as well as hydrogen/deuterium exchange data (Fig. 2a, Supplementary Fig. 9,10a).
Figure 2. Conformational dynamics of the ligase enzyme 10C.

(a) Mapping of the heteronuclear NOEs (proxy for fast dynamics on a picosecond-nanosecond time scale) on ligase 10C. Residues involved in zinc coordination are labeled. (b) Mapping of the exchange rates (Rex) obtained from relaxation dispersion measurements as a proxy for slow dynamics (microsecond-millisecond time scale). The color gradient and thickness of the backbone indicate that the ligase fast dynamics is located mostly in the unstructured loop, while the slow dynamics is located mostly in the region N-terminal to the Zn-II site (residues 46–53) and is potentially correlated to catalytic activity.
A distinct signature of the hRXRα structure is slow (microsecond-millisecond) conformational dynamics20 (Supplementary Fig. 10b), which may be correlated with the protein’s ability to optimize protein-DNA interactions. The in vitro evolution of hRXRα into the RNA ligase redistributed those conformational dynamics, particularly in the region N-terminal to the Zn2+ binding site Zn-II (residues 46–53, Fig. 2b).
To probe the substrate binding surface, we carried out an NMR titration with a pseudo-substrate that lacked the 2′-hydroxyl group, preventing enzyme turnover (Supplementary Fig. 11). Chemical shift perturbation mapping of the ligase structure (Fig. 3a and Supplementary Fig. 12) indicated that one of the highly perturbed regions in the substrate bound form (residues 46–53) corresponds to high values of chemical exchange (slow conformational dynamics) in the substrate-free form (Fig. 2b). Notably, most alanine mutations in this region decreased or completely obliterated the enzyme’s activity (Fig. 3b,c). Specifically, mutations E48A, Y50A and H51A abolished enzymatic activity, whereas C47A and C53A caused a 97% reduction in ligase function. These residues’ high conservation among evolved ligase variants further demonstrated their importance (Fig. 3d). The combined results suggest that this protein region (residues 46–53) is important for substrate recognition and binding and may contain the active site of the enzyme. Four of those five mutation-sensitive residues (C47, E48, H51, C53) are good potential metal ligands. Many natural enzymes, such as polymerases, that catalyze chemical reactions similar to the specific RNA ligation described here use a mechanism involving catalytic divalent metal ion cofactors, which are coordinated jointly by the nucleic acid substrates and active site residues of the enzyme21. One may speculate that upon forming the enzyme-substrate complex some of the mutation-sensitive residues in ligase 10C are involved in binding additional Zn2+ ions that facilitate catalysis, but are not bound by the protein alone. However, additional experiments studying the enzyme in complex with substrate are needed to elucidate the catalytic mechanism of our artificial enzyme.
Figure 3. Substrate binding surface of ligase 10C probed by NMR and alanine scanning.
(a) Mapping of NMR chemical shift perturbations (intensity changes ΔI) for the evolved ligase shows regions affected by formation of the complex with RNA substrate. The most perturbed regions are indicated by thicker lines and darker colors. Residues involved in zinc coordination are labeled. (b) Region of ten single alanine mutants that were tested individually is shown in red and mapped onto one NMR conformer. (c) Ligase activity assay of ligase 10C and alanine mutants by gel shift. The lower band corresponds to unligated 32P-labeled PPP-substrate, and the upper band corresponds to ligated product. Ligation activity is normalized to the activity of ligase 10C. Percentages shown represent the mean values from two independent experiments. Residues with activity below detection limit are shown in blue. (d) Sequence conservation analysis of region 45–54 for ligase enzymes generated by directed evolution14. The sequences were analyzed using the web based application WebLogo (V 2.8.2) from a dataset of 49 enzyme sequences. Residues E48, Y50, and H51 (blue) which completely lost activity during alanine scanning (see above) were conserved among all sequences. The two residues with 97% reduced activity of their alanine mutant were either conserved (C53) or had one alternative amino acid (C47). None of the other residues in this region were conserved.
The increased flexibility of the new ligase structure relative to the parent hRXRα could potentially originate from their different functional roles. hRXRα is a DNA binder and has been proposed to work through an induced-fit mechanism16. In contrast, the RNA ligase has evolved to function as a catalyst. This role requires additional flexibility to optimize interactions with a target molecule and carry out chemical catalysis using transient interactions that occur in excited conformational states rather than through a stable, low energy complex22–24. This argument is supported by an independent directed evolution experiment in which the same hRXRα library yielded proteins that bind ATP and maintain the original, non-catalytic DNA binding scaffold, but have no catalytic function17. In contrast, the evolution of the ligase enzyme resulted in a different structure and increased dynamics.
Compared to natural enzymes which evolved over billions of years, the laboratory-evolved ligase enzyme contained substantially fewer secondary structure elements such as α-helices and β-strands, and instead exhibited increased flexibility. The complete reorganization of the starting scaffold during in vitro evolution may have led to the loss of these structural elements. This novel structure has not been subjected to extensive selection pressure which shaped contemporary enzymes during their natural evolution and can therefore be considered an early or primordial catalytic fold. Further evolution of this enzyme in vitro or inside a cell will explore if incremental mutations lead to structural and dynamic properties more similar to natural enzymes. While flexibility has been suggested to increase the probability of developing new functions5, it also reduces overall protein stability; a trade-off which enzymes must balance during evolution.
This report describes the first new protein structure emerging simultaneously with a novel enzymatic function. This ligase evolved in the absence of selection pressure to maintain the protein’s original function (DNA binding). Would proteins evolving in nature also more readily adopt new folds and functions if they were freed from maintaining their original function? While the search for such examples in nature is still ongoing, the simplified environment of in vitro evolution enables us to generate precedents and study basic principles of complex natural evolution. Finally, in vitro directed evolution has the potential to produce novel biocatalysts for a wide range of applications. The unique structure of the artificial ligase enzyme demonstrates that this approach can successfully generate novel enzymes without being limited to known biological folds25.
Online methods
All chemical compounds used in this study were purchased from Sigma-Aldrich unless noted otherwise and were of Molecular Biology Grade, and certified for the absence of ribonucleases when used for ligation reactions.
Sequence of RNA ligase 10C
MGAPVPYPDPLEPRGGKHICAICGNNAEDYKHTDMDLTYTDRDYKNCESYHKCSDLCQYCRYQKDLAIHHQHHHGGSMGMSGSGTGY
All ligase protein preparations consisted of the sequence above except for point mutations in the case of ligase mutants. Note that the sequence HHQHHH functions similarly to a 6xHis-tag.
Expression and purification of 15N-labeled ligase protein for NMR studies
Ligase samples were expressed in E. coli BL21-DE3 Rosetta strain cells (Novagen). Cells were grown in LB with 36 μg/mL kanamycin overnight at 37 °C. This culture was then used to inoculate 1 L of LB medium containing 36 μg/mL kanamycin. The cultures were grown to an OD600 of 0.6–0.8 at 37 °C, spun down, and resuspended in M9 minimal medium (50 mM Na2PHO4, 22 mM KH2PHO4, 8.5 mM NaCl, 2 mM MgSO4, 1 mg/L thiamine, 1 mg/L biotin, 60 μM ZnSO4, 10 g/L dextrose, 1 g/L 15NH4Cl, and 36 μg/mL kanamycin, pH = 7.3). Cultures were shaken for 1 h at 37 °C, induced with 1 mM IPTG, and shaken overnight at room temperature before being spun down and stored at −20 °C.
Frozen cell pellets were resuspended in lysis buffer (20 mM HEPES, 400 mM NaCl, 100 μM ZnCl2, 100 mg/L Triton X-100, 5 mM β-mercaptoethanol, pH = 7.4) and lysed using a S-450D Digital Sonifier (Branson). Cell debris was removed by centrifugation and the His-tagged ligase protein was purified by affinity chromatography using Ni-NTA Superflow resin (QIAGEN). The protein was eluted with acidic elution buffer (20 mM NaOAc, 400 mM NaCl, 0.1 mM ZnCl2, 100 mg/L Triton X-100, 5 mM β-mercaptoethanol, pH = 4.5) into 1 M HEPES at pH = 7.5, and immediately mixed to adjust the pH. Protein purification was evaluated by SDS-PAGE on Ready Gel precast gels (Bio-Rad). Elution fractions containing ligase protein were concentrated under high pressure in a stirred-cell concentrator unit with a 5,000 MWCO Ultracel Ultrafiltration cellulose membrane (Millipore) and dialyzed into FPLC buffer (20 mM HEPES, 150 mM NaCl, 0.1 mM ZnCl2, and 0.5 mM β-mercaptoethanol, pH = 7.5).
Monomer ligase protein was isolated by size-exclusion chromatography using the AKTA FPLC system (GE Healthcare) equipped with a 10 mm × 300 mm column (Tricorn) and Superdex 75 resin (GE Healthcare). The separation was carried out in FPLC buffer. Fractions containing monomer protein were pooled and concentrated using 10,000 MWCO Ultra-4 Centrifugal Filter units (Millipore).Purity was assessed by SDS PAGE gel (Supplementary Figure 13).
Expression and purification of 15N/13C-labeled ligase samples for NMR studies
Ligase samples were expressed in E. coli BL21-DE3 Rosetta strain cells (Novagen). Cells were grown in LB with 36 μg/mL kanamycin overnight at 37 °C, spun down, and resuspended in M9 minimal medium (contents as described above, except with 2 g/L 13C-dextrose). The resuspended cells were used to inoculate 100 mL of M9 minimal medium and were grown to an OD600 of 0.6 at 37 °C, at which time the culture was used to inoculate 900 mL of M9 minimal medium. The 1 L culture was grown to an OD600 of 1.0 at 37 °C, induced with 1 mM IPTG, and shaken overnight at 37 °C before being spun down and stored at −20 °C. The 15N/13C-labeled protein was purified in the same manner as the 15N-labeled protein samples.
Expression of selectively labeled ligase protein for NMR studies
Ligase samples were expressed in E. coli BL21-DE3 Rosetta strain cells (Novagen). Cells were grown in LB with 36 μg/mL kanamycin overnight at 37 °C, spun down, and used to inoculate 1 L of selectively labeled M9 medium (40 mM Na2PHO4, 22 mM KH2PHO4, 8.5 mM NaCl, 1 mM MgSO4, 50 μM CaCl2, essential vitamins and minerals, and 36 μg/mL kanamycin, pH 7.0). To the medium was also added 250 mg of a single 15N-labeled amino acid (Cys, Leu, Lys or Tyr), 600 mg of the remaining 19 unlabeled amino acids and, except for labeling 15N Cys, one of the following additional amino acid supplements: 900 mg Gln, Asn and Arg when labeling 15N Lys; 900 mg Val, and Ile when labeling 15N Leu; and 900 mg Phe, Trp, Ala, Ser, Gly, and Cys when labeling 15N Tyr. Cultures were grown to an OD600 of 1.0 at 37 °C, induced with 1 mM IPTG, and shaken for 6 h at 37 °C before being spun down and stored at −20 °C. Selectively labeled protein was purified in the same manner as the 15N-labeled protein samples.
Generation of ligase mutants
Ligase mutants were obtained by site-directed mutagenesis (QuikChange Lightning, Agilent). Plasmid DNA was purified using the QIA prep Spin Miniprep kit (QIAGEN). The ligase mutants were verified by DNA sequencing. The primer sequences used to generate the indicated mutations in the ligase were designed in accordance with the Quik Change Primer Design tool (Agilent) and were as follows:
| K45A F: | 5′-CTACACCGATCGAGACTACGCGAATTGTGAGAGCTACC |
| K45A R: | 5′GGTAGCTCTCACAATTCGCGTAGTCTCGATCGGTGTAG |
| N46A F: | 5′CCGATCGAGACTACAAGGCTTGTGAGAGCTACCATAAGTG |
| N46A R: | 5′CACTTATGGTAGCTCTCACAAGCCTTGTAGTCTCGATCGG |
| C47A F: | 5′CCGATCGAGACTACAAGAATGCTGAGAGCTACCATAA |
| C47A R: | 5′TTATGGTAGCTCTCAGCATTCTTGTAGTCTCGATCGG |
| E48A F: | 5′GACTACAAGAATTGTGCGAGCTACCATAAGTGCTCGG |
| E48A R: | 5′CCGAGCACTTATGGTAGCTCGCACAATTCTTGTAGTC |
| S49A F: | 5′AGACTACAAGAATTGTGAGGCCTACCATAAGTGCTCGGAC |
| S49A R: | 5′GTCCGAGCACTTATGGTAGGCCTCACAATTCTTGTAGTCT |
| Y50A F: | 5′CTACAAGAATTGTGAGAGCGCCCATAAGTGCTCGGACTTGTG |
| Y50A R: | 5′CACAAGTCCGAGCACTTATGGGCGCTCTCACAATTCTTGTAG |
| H51A F: | 5′CTACAAGAATTGTGAGAGCTACGCTAAGTGCTCGGACTTGTG |
| H51A R: | 5′CACAAGTCCGAGCACTTAGCGTAGCTCTCACAATTCTTGTAG |
| K52A F: | 5′ACAAGAATTGTGAGAGCTACCATGCGTGCTCGGACTTGTGC |
| K52A R: | 5′GCACAAGTCCGAGCACGCATGGTAGCTCTCACAATTCTTGT |
| C53A F: | 5′GTGAGAGCTACCATAAGGCCTCGGACTTGTGCCAGT |
| C53A R: | 5′ACTGGCACAAGTCCGAGGCCTTATGGTAGCTCTCAC |
| S54A F: | 5′GTGAGAGCTACCATAAGTGCGCGGACTTGTG |
| S54A R: | 5′CACAAGTCCGCGCACTTATGGTAGCTCTCAC |
Expression and purification of ligase mutants
Ligase mutants were expressed in E. coli BL21-DE3 Rosetta cells (Novagen). Cells were cultured in 1 L of LB medium with 36 μg/mL kanamycin to an OD600 of 0.8–1.0 at 37 °C. Cultures induced with 1 mM IPTG and shaken for 6 h at 37 °C before being spun down and stored at −20 °C. Ligase mutant proteins were purified by Ni-NTA affinity chromatography in the same manner as described for the 15N-labeled protein samples.
Analysis of metal content by ICP-MS
Ligase 10C was purified as described for the 15N-labeled protein samples and then dialyzed three times against buffer (100 mM NaCl, 10 mM β-mercaptoethanol, 20 mM TrisHCl at pH 7.5; pre-treated with Chelex 100 beads (Bio-Rad) for 2 h and filtered) at a ratio of 1/1,000. The metal content of 14 μM protein was measured by ICP MS (Thermo Scientific XSERIES 2 ICP-MS w/ESI PC3 Peltier cooled spray chamber, Department of Earth Sciences at the University of Minnesota).
Ligase activity assay for zinc dependence
Ligase 10C was purified as reported previously14. Zinc was removed from ligase 10C by treatment with ion exchange resin (Chelex 100, Bio-Rad). 5 μM Ligase 10C was incubated with 20 μM HO-substrate, 10 μM 32P-labeled PPP-substrate/splint, 20 mM HEPES (pH 7.5), 150 mM NaCl, 500 μM β-mercaptoethanol, and the indicated concentrations of ZnCl2 for 6 h at room temperature. The ligation reactions were quenched with 20 mM EDTA/8 M urea, heated to 95°C for 4 min, and separated by denaturing PAGE gel. The gel was analyzed using a GE Healthcare (Amersham Bioscience) Phosphorimager and ImageQuant software (Amersham Bioscience).
Ligase activity assay of 10C and alanine mutants
5 μM Ligase 10C (or alanine mutant) was incubated for 6 h at room temperature in the presence of 20 μM HO-substrate, 10 μM 32P-labeled PPP-substrate/splint, 24 mM HEPES (pH 7.5), 130 mM NaCl, 100 μM β-mercaptoethanol, and 120 μM ZnCl2. The ligation reactions were quenched with 20 mM EDTA and 8 M urea, heated to 95°C for 4 min, and separated by denaturing PAGE gel. The gel was analyzed using a GE Healthcare (Amersham Bioscience) Phosphorimager and ImageQuant software (Amersham Bioscience).
Resonance assignment
All NMR spectra were acquired at 298 K on a Bruker spectrometer equipped with cryoprobe at 700MHz and Varian spectrometer at 600MHz. The samples were in buffer of 150 mM NaCl, 20 mM HEPES, 10 mM β-mercaptoethanol, and pH 7.5. Moreover, all protein samples were saturated with ZnCl2 by observing changes in HSQC spectra prior to other NMR experiments. Triple resonance spectra such as CBCA(CO)NH, HNCACB26–28 were used to assign peaks on 15N-HSQC. All resonances in these two 3D spectra and 15N-HSQC were picked and fed into the PISTACHIO program (National Magnetic Resonance Facility in Madison, WI, USA)29 to obtain preliminary assignments. Final complete assignments were done by manual checks and searches. Carbonyl groups and others side-chain carbons were assigned by HNCO and C(CO)NH-TOCSY30; side-chain protons were assigned by 15N-NOESY-HSQC, 15N-TOCSY-HSQC, and HC(CO)NH-TOCSY experiments30 with 150 ms mixing time, 60 ms mixing time, and 12 ms mixing time, respectively.
Distance restraints
All proton distance restraints were determined from the cross-peak intensities in the NOESY spectra by calibration with HN(i)Hα(i-1) distances located at the C-terminal region31, whose helix propensity was shown by chemical shift index and 3JHNHα coupling values31,32. The cross peaks from HN(i)Hα(i-1) distances in that region were categorized as medium NOEs, so the intensities of other cross peaks smaller than this intensity range were defined as weak NOEs and those larger than this range belonged to strong NOEs. The upper bounds of distance restraints of strong, medium, and weak NOEs were given as 2.9, 3.5, and 5 Å respectively, and lower bounds were set to 1.8 Å in all cases. Starting from unambiguously assigned NOEs at the beginning of calculation, mis-calibrated NOEs were adjusted and then ambiguously assigned NOEs were gradually added into the restraint table during iterative calculation.
Torsion angle restraints
Backbone Phi angle restraints were acquired from the HNHA experiment, and the quantitative 3JHNHα coupling values were calculated from the intensity ratios of cross peaks to diagonal peaks and corrected by 3.7% to account for relaxation33. The correction is proportional to the rotational correlation time of the protein (3 ns), which was measured from 1-dimensional TRACT experiment34. The Phi angle of residue i with J-coupling larger than 8.5 Hz was restrained from −160° to −80°, and that with J-coupling smaller than 6 Hz was restrained from −90° to −40°. Moreover, the Psi angle restraints were derived from the 15N-NOESY data. If the intensity ratio of the HN(i)Hα(i) cross peak to the HN(i)Hα(i-1) peak is smaller than one, the Psi(i-1) is restrained from 20° to 220°; otherwise, the Psi(i-1) is restrained from 80° to −140° 35,36.
RDC measurement
The stability of several alignment media for ligase 10C was tested. 5% neutral and negative-charged acrylamide gel was first attempted, but only weak residual dipolar couplings (absolute values < 5 Hz) were obtained. Additionally, the samples precipitated in both DMPC/D7PC and DMPC/D6PC bicelle preparations. We also tested the liquid crystalline medium formed by CPCl (cetylpyridinium chloride) and 1-hexanol, but had poor results in terms of sample stability. The sample was finally aligned in the other liquid crystalline medium made by the mixture of C12E5 (5% alkyl-poly(ethylene glycol)) and 1-hexanol (r=0.85)37. The residual dipolar couplings of amide groups were obtained by measuring the splitting difference between a decoupling HSQC peak and a TROSY peak in isotropic solution and anisotropic medium.
Structure calculations
Simulating annealing protocols were performed in the XPLOR package38. An extended structure was first generated and the initial temperature was set at 3,500 K, then the temperature was cooled down to 0 K with 15,000 steps. The structure with the lowest energy was used for refinement with the initial temperature of 5,000 K and 30,000 steps. The resulting structure was further refined with RDC data after optimization of the parameters Da and Rh. The angle restraints of the zinc coordination geometry were based on ideal geometries derived from X-ray data39,40, which are in quantitative agreement with the EXAFS experiments. Distances derived from EXAFS have previously been used as restraints in NMR refinement41,42. Here, we report a structural ensemble of 20 conformers. The PROCHECK statistics show that 76.4% of residues are in most favored regions and 21.1% of residues are in allowed regions.
Zn K-edge EXAFS
Ligase 10C protein was fully saturated with excess Zn2+ and then dialyzed to remove excess Zn2+. The final protein sample was 1.39 mM in 15 mM Tris, pH = 7.5 and 112.5 mM NaCl. 20% v/v) glycerol was added to the protein samples in order to form a glass required for the EXAFS experiments. The Zn K-edge X-ray absorption spectra of ligase 10C were measured at the Stanford Synchrotron Radiation Lightsource (SSRL) on the 16 pole, 2 T wiggler beamline 9–3 under standard ring conditions of 3 GeV and ~200 mA ring current. A Si(220) double-crystal monochromator was used for energy selection. Other optical components used for the experiments were a cylindrical Rh-coated bent focusing mirror. Spectra were collected in the fully tuned configuration of the monochromator. The solution samples were immediately frozen after preparation and stored under liquid N2 until measurement. During data collection, the samples were maintained at a constant temperature of ~6 K using an Oxford Instruments CF 1208 liquid helium cryostat. Data were measured to k= 16 Å−1 by using a Canberra Ge 100-element monolith detector. Internal energy calibration was accomplished by simultaneous measurement of the absorption of a Zn-foil placed between two ionization chambers situated after the sample. The first inflection point of the foil spectrum was fixed at 9,660.7 eV. Data presented here are a 15 scan average. The data were processed by fitting a second-order polynomial to the pre-edge region and subtracting this from the entire spectrum as background. A five-region spline of orders 2, 3, 3, 3 and 3 was used to model the smoothly decaying post-edge region. The data were normalized by subtracting the cubic spline and assigning the edge jump to 1.0 at 9,680 eV using the Pyspline program43. Theoretical EXAFS signals χ(k) were calculated by using FEFF (Macintosh version 8.4)44–46. Initial model was based on the Zn(Cys)2(His)2 active site in a zinc finger protein (1MEY)47. Based on the preliminary fits, the models were modified to accommodate a six-coordinate active site (4 Zn-N/O and 2 Zn-S(Cys)).
The theoretical models were fit to the data using EXAFSPAK48. The structural parameters varied during the fitting process were the bond distance (R) and the bond variance σ2, related to the Debye-Waller factor resulting from thermal motion, and static disorder of the absorbing and scattering atoms. The non-structural parameter E0 (the energy at which k=0) was also allowed to vary but was restricted to a common value for every component in a given fit. Coordination numbers were systematically varied in the course of the fit but were fixed within a given fit.
Supplementary Material
Acknowledgments
We thank M. Golynskiy and A. Pohorille for helpful discussions, Z. Sachs, F. P. Seebeck, J. W. Szostak and F. Hollfelder for comments on the manuscript and R. Majerle for ITC instrument use. This work was supported by NASA Agreement No. NNX09AH70A issued through NASA Astrobiology Institute/Ames Research Center (to F.-A.C., A.M., L.C. and B.S.); the Minnesota Medical Foundation, University of Minnesota Biocatalysis Initiative (to B.S); the NIH (T32 GM08347 to J.C.H., T32 DE007288 to L.R.M., and GM100310 to G.V.). NMR data were collected at the University of Minnesota NMR Center. SSRL operations are funded by DOE, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology program is supported by NIH-NCRR, Biomedical Technology Program, and DOE, Office of Biological and Environmental Research. This publication was made possible by NIH-NCRR Award P41 RR001209.
Footnotes
Accession codes
The Protein Data Bank accession code for ligase 10C is 2LZE, and the BMRB accession number is 18749.
Author contributions
G.V. and B.S. designed the project; A.M., J.C.H., L.C. and L.N.H. expressed and purified proteins and carried out functional assays; F.-A.C. carried out all NMR experiments and the ITC; F.-A.C. and L.S. calculated the structure; R.S. performed the EXAFS measurements, all authors analyzed the data; F.-A.C., L.R.M., G.V. and B.S. wrote the paper.
Competing financial interests
The authors declare no competing financial interests.
Additional information
Supplementary results are available in the online version of the paper. Reprints and permissions information is available online at http://www.nature.com/reprints/index.html. Correspondence and requests for materials should be addressed to B.S.
References
- 1.Chothia C. Nature. 1992;357:543–544. doi: 10.1038/357543a0. [DOI] [PubMed] [Google Scholar]
- 2.Murzin AG, Brenner SE, Hubbard T, Chothia C. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 3.Ohno S. Evolution by gene duplication. Springer-Verlag; New York, USA: 1971. [Google Scholar]
- 4.Chothia C, Gough J, Vogel C, Teichmann SA. Science. 2003;300:1701–1703. doi: 10.1126/science.1085371. [DOI] [PubMed] [Google Scholar]
- 5.James LC, Tawfik DS. Trends Biochem Sci. 2003;28:361–368. doi: 10.1016/S0968-0004(03)00135-X. [DOI] [PubMed] [Google Scholar]
- 6.Tokuriki N, Tawfik DS. Science. 2009;324:203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- 7.Cordes MHJ, Walsh NP, McKnight CJ, Sauer RT. Science. 1999;284:325–327. doi: 10.1126/science.284.5412.325. [DOI] [PubMed] [Google Scholar]
- 8.Kaplan J, DeGrado WF. Proc Natl Acad Sci USA. 2004;101:11566–11570. doi: 10.1073/pnas.0404387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuinstra RL, et al. Proc Natl Acad Sci USA. 2008;105:5057–5062. doi: 10.1073/pnas.0709518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryan PN, Orban J. Curr Opin Struct Biol. 2010;20:482–488. doi: 10.1016/j.sbi.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith BA, Hecht MH. Curr Opin Chem Biol. 2011;15:421–426. doi: 10.1016/j.cbpa.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Keefe AD, Szostak JW. Nature. 2001;410:715–718. doi: 10.1038/35070613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mansy SS, et al. J Mol Biol. 2007;371:501–513. doi: 10.1016/j.jmb.2007.05.062. [DOI] [PubMed] [Google Scholar]
- 14.Seelig B, Szostak JW. Nature. 2007;448:828–831. doi: 10.1038/nature06032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seelig B. Nat Protoc. 2011;6:540–552. doi: 10.1038/nprot.2011.312. [DOI] [PubMed] [Google Scholar]
- 16.Holmbeck SMA, et al. J Mol Biol. 1998;281:271–284. doi: 10.1006/jmbi.1998.1908. [DOI] [PubMed] [Google Scholar]
- 17.Cho GS, Szostak JW. Chem Biol. 2006;13:139–147. doi: 10.1016/j.chembiol.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Q, et al. J Mol Biol. 2000;296:509–520. doi: 10.1006/jmbi.1999.3457. [DOI] [PubMed] [Google Scholar]
- 19.Maret W, Li Y. Chem Rev. 2009;109:4682–4707. doi: 10.1021/cr800556u. [DOI] [PubMed] [Google Scholar]
- 20.van Tilborg PJ, et al. Biochemistry. 2000;39:8747–8757. doi: 10.1021/bi991550g. [DOI] [PubMed] [Google Scholar]
- 21.Yang W, Lee JY, Nowotny M. Mol Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Bhabha G, et al. Science. 2011;332:234–238. doi: 10.1126/science.1198542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baldwin AJ, Kay LE. Nat Chem Biol. 2009;5:808–814. doi: 10.1038/nchembio.238. [DOI] [PubMed] [Google Scholar]
- 24.Henzler-Wildman K, Kern D. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 25.Golynskiy MV, Seelig B. Trends Biotechnol. 2010;28:340–345. doi: 10.1016/j.tibtech.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 26.Grzesiek S, Bax A. J Magn Reson. 1992;96:432–440. [Google Scholar]
- 27.Muhandiram DR, Kay LE. J Magn Reson, Ser B. 1994;103:203–216. [Google Scholar]
- 28.Wittekind M, Mueller L. J Magn Reson, Ser B. 1993;101:201–205. [Google Scholar]
- 29.Eghbalnia HR, Bahrami A, Tonelli M, Hallenga K, Markley JL. J Am Chem Soc. 2005;127:12528–12536. doi: 10.1021/ja052120i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grzesiek S, Anglister J, Bax A. J Magn Reson, Ser B. 1993;101:114–119. [Google Scholar]
- 31.Wuthrich K. NMR of proteins and nucleic acids. John Wiley and Sons; New York, USA: 1986. [Google Scholar]
- 32.Wishart DS, Sykes BD, Richards FM. J Mol Biol. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- 33.Vuister GW, Bax A. J Am Chem Soc. 1993;115:7772–7777. [Google Scholar]
- 34.Lee D, Hilty C, Wider G, Wuthrich K. J Magn Reson. 2006;178:72–76. doi: 10.1016/j.jmr.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 35.Gagne SM, et al. Protein Sci. 1994;3:1961–1974. doi: 10.1002/pro.5560031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Zhao S, Somerville RL, Jardetzky O. Protein Sci. 2001;10:592–598. doi: 10.1110/ps.45301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruckert M, Otting G. J Am Chem Soc. 2000;122:7793–7797. [Google Scholar]
- 38.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 39.Alberts IL, Nadassy K, Wodak SJ. Protein Sci. 1998;7:1700–1716. doi: 10.1002/pro.5560070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viles JH, et al. J Mol Biol. 1998;279:973–986. doi: 10.1006/jmbi.1998.1764. [DOI] [PubMed] [Google Scholar]
- 41.Ohlenschlager O, et al. Oncogene. 2006;25:5953–5959. doi: 10.1038/sj.onc.1209584. [DOI] [PubMed] [Google Scholar]
- 42.Banci L, Bertini I, Del Conte R, Mangani S, Meyer-Klaucke W. Biochemistry. 2003;42:2467–2474. doi: 10.1021/bi0205810. [DOI] [PubMed] [Google Scholar]
- 43.Tenderholt A. Pyspline. Stanford University; Stanford, USA: 2007. [Google Scholar]
- 44.Deleon JM, Rehr JJ, Zabinsky SI, Albers RC. Phys Rev B. 1991;44:4146–4156. doi: 10.1103/physrevb.44.4146. [DOI] [PubMed] [Google Scholar]
- 45.Rehr JJ, Albers RC. Rev Mod Phys. 2000;72:621–654. [Google Scholar]
- 46.Rehr JJ, Deleon JM, Zabinsky SI, Albers RC. J Am Chem Soc. 1991;113:5135–5140. [Google Scholar]
- 47.Kim CA, Berg JM. Nat Struct Biol. 1996;3:940–945. doi: 10.1038/nsb1196-940. [DOI] [PubMed] [Google Scholar]
- 48.George GN. EXAFSSPAK and EDG-FIT. Stanford Synchrotron Radiation Lightsource; Menlo Park, USA: 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.