Abstract
Quiescent HIV-1 infection of resting CD4+ T cells is an obstacle to eradication of HIV-1 infection. These reservoirs are maintained, in part, by repressive complexes that bind to the HIV-1 long terminal repeat (LTR) and recruit histone deacetylases (HDACs). cMyc and YY1 are two transcription factors that are recruited as part of well-described, distinct complexes to the HIV-1 LTR and in turn recruit HDACs. In prior studies, depletion of single factors that recruit HDAC1 in various cell lines was sufficient to upregulate LTR activity. We used short hairpin RNAs (shRNAs) to test the effect of targeted disruption of a single transcription factor on quiescent proviruses in T cell lines. In this study, we found that depletion of YY1 significantly increases mRNA and protein expression from the HIV-1 promoter in some contexts, but does not affect HDAC1, HDAC2, HDAC3, or acetylated histone 3 occupancy of the HIV-1 LTR. Conversely, depletion of cMyc or cMyc and YY1 does not significantly alter the level of transcription from the LTR or affect recruitment of HDACs to the HIV-1 LTR. Furthermore, global inhibition of HDACs with the HDAC inhibitor suberoylanilide hydroxamic acid (SAHA) enhanced the increase in LTR transcription in cells that were depleted of YY1.These findings show that despite prior isolated findings, redundancy in repressors of HIV-1 LTR expression will require selective targeting of multiple restrictive mechanisms to comprehensively induce the escape of quiescent proviruses from latency.
Introduction
Substantial advances have been made in the treatment of human immunodeficiency virus (HIV-1) infection. However, due to the burdens of lifelong antiviral therapy, attention has recently focused on research directed at eradication of HIV-1 infection. To accomplish this challenging goal, techniques to target and eliminate a small population of quiescent proviral genomes that persist in infected individuals undergoing therapy are under study.
The transcription of quiescent proviral genomes is repressed by the presence of mature heterochromatic structures that are maintained through the recruitment of cellular transcription factors. The quiescent HIV-1 long terminal repeat (LTR) is occupied by the NURD and the NCoR histone deacetylase (HDAC) complexes in Jurkat and monocyte-derived macrophage cell lines, respectively.1,2 These HDAC complexes repress transcription by removing acetyl groups from histones, which results in a repressive chromatin structure at the HIV-1 LTR that is not permissive for transcription. HDAC inhibitors are consistently able to reactivate quiescent HIV-1 in cell line models and in patients' cells.3–6 However, HDACs regulate the expression of a broad range of cellular factors, and HDAC inhibitors affect the acetylation of lysines on many nonhistone proteins. Therefore, the aim of this study was to determine whether factors that specifically recruit HDACs to the HIV-1 promoter could be directly targeted to reverse HIV-1 quiescence. To date, several transcription factors have been described that recruit HDAC1 to the HIV-1 LTR, including cMyc, YY1, CBF1, NF-κB, and Sp1.3,7–9 Surprisingly, although each of these factors binds to the LTR at a distinct location and individually recruits HDACs, previous studies have found that single depletion of any one of these factors is sufficient to disrupt quiescent HIV-1 proviruses and occupancy of HDAC1 at the HIV-1 promoter. Furthermore, these findings indicate that in the absence of one HDAC recruiting complex, HDAC occupancy cannot be maintained at a level sufficient to prevent reactivation of HIV-1 transcription.
YY1 and cMyc are two transcription factors that are recruited to the HIV-1 promoter and are involved in the maintenance of transcriptional repression. YY1 can both activate and repress transcription depending on the promoter context.10 YY1 is recruited to the HIV-1 LTR by the transcription factor LSF and represses transcription by recruiting HDAC1 in HeLa cells.3,11 cMyc is a transcription factor that is most commonly known for its roles in regulating cellular proliferation and cell growth. Like YY1, it is able to act as both a transcriptional activator and repressor depending on the context of the promoter.12 The mbIII domain of cMyc is important for its ability to repress transcription and mediates the interaction between cMyc and HDAC3.13 cMyc is repressive when recruited to the HIV-1 LTR by the transcription factor Sp1.8 However, cMyc is required for Tat activated HIV-1 transcription.14 Because of their well-characterized role in the maintenance of quiescent HIV, cMyc and YY1 are promising targets for therapies to disrupt the silencing of integrated HIV-1 proviruses.
Many of the initial studies that were instrumental in elucidating the mechanisms of HIV-1 transcriptional repression have been performed in HeLa cell lines. HeLa cells are an epithelial cell line that was originally isolated from cervical cancer tissue.15 In vivo, HIV-1 infects CD4+ T cells of the immune system. The cell environment and transcriptional profiles of epithelial and T cells are very different. Therefore, mechanisms that maintain transcriptionally quiescent proviral genomes in T cells are of paramount relevance. Jurkat cells are a cell line derived from T cells with biochemical features similar to resting CD4+ T cells. Several cell lines that are derived from Jurkat cells and contain quiescent, but inducible HIV-1 proviruses have been created as models of HIV-1 latency, including 2D10 and J89 cells.16,17 We performed our studies in 2D10 and J89 cells because unique proviral insertion sites, and perhaps subtle differences in cellular gene programming, result in widely distinct sensitivity to viral induction strategies. These studies more accurately reflect the diversity of HIV latency in vivo than those performed in a single clonal model. The goal of this study was to determine whether the selective targeting of transcription factors, such as cMyc and YY1, that recruit HDACs to the LTR and maintain transcriptional repression of HIV-1, could disrupt latency in T cell lines. Furthermore, we wished to determine whether these factors play a unique or general role in the individual recruitment of HDAC1, HDAC2, and HDAC3 to the HIV-1 LTR. The mechanisms that maintain proviral quiescence are high-value therapeutic targets for the development of antilatency therapies.
Materials and Methods
Cell culture
J89 (a kind gift from Dr. David Levy at the University of Alabama at Birmingham) and 2D10 (a kind gift from Dr. Jonathan Karn at Case Western Reserve University) cells were maintained in RPMI supplemented with 10% fetal bovine serum (FBS) and 1% Pen/strep in a 37°C incubator containing CO2. Cells were passaged every 3-4 days, and all experiments were performed on cells that had been passaged fewer than 12 times. For the experiments in which suberoylanilide hydroxamic acid (SAHA, Merck Research Laboratories, West Point, PA) or phorbol 12-myristate 13-acetate (PMA, Sigma-Aldrich, St. Louis, MO) was used, 500 nM SAHA or 10 nM PMA was added to the cell media 54 h after transduction with short-hairpin RNAs (shRNAs) and 18 h prior to collection for flow cytometry.
Chromatin immunoprecipitation (ChIP) assays
Ten million cells that had been transduced with short hairpin RNAs (shRNAs) were collected, washed once with phosphate-buffered saline (PBS), and then fixed with 1% formaldehyde for 10 min. The cell nuclei were isolated according to the manufacture's instructions and sonicated for 24 min in a bioruptor (Diagenode, Denville, NJ) with intervals that consisted of 30 s of sonication and 15 s of rest. To bind the antibodies to the beads, protein A and protein G Dynal beads (Invitrogen, Grand Island, NY) were incubated with 5–10 μg of antibody. The following antibodies were used for the ChIP experiments: HDAC1 ChIP grade (Abcam, Cambridge, MA), HDAC2 ChIP grade (Abcam, Cambridge, MA), HDAC3 ChIP grade (Abcam, Cambridge, MA), histone 3 ChIP grade (Abcam, Cambridge, MA), and acetylated histone 3 (Millipore, Billerica, MA). The sonicated product was then added and incubated at 4°C overnight. The next day, the beads were washed once for 5 min with each of the following buffers: ChIP dilution buffer, low salt ChIP buffer, high salt ChIP buffer, LiCl ChIP buffer, and TE buffer. The samples were then eluted from the beads and incubated at 68°C for 2 h to decrosslink the proteins from the DNA and to degrade the protein. The DNA was purified using the Qiagen PCR purification kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. The enrichment of the HIV-1 LTR in the ChIP samples was assessed using quantitative PCR (qPCR). Independent transductions were performed for each ChIP replicate.
Quantitative PCR
One microliter of the DNA from the ChIP assay was used in a final volume of 20 μl containing 10 μl of SYBR green master mix (Applied Biosystems, Foster City, CA) and 0.5 μM each of the primers LTRrt8 (5′-TAGCCAGAGAGCTCCCAGGCTCAGA-3′) and LTRrt9 (5′-AGCCCTCAGATGCTACATATAA GCA-3′). The reaction was run on a 7900 quantitative PCR machine (Applied Biosystems, Foster City, CA) with the following parameters: 50°C for 2 min; 95.0°C for 10 min; and 40 replicates of 95.0°C for 15 s and 60°C for 1 min. A standard curve ranging from 50% to 0.0008% of the total ChIP input was run for each condition, and the data are displayed as the percent input with the background from the IgG condition subtracted. Experiments with less DNA than the IgG control sample were assigned zero quantity. Data are shown as the mean±standard error of the mean (SEM) from at least three independent ChIP experiments. The Student's t-test was used to assess significance, and a p-value of less than 0.05 was considered significant.
Quantification of gene expression
2D10 or J89 cells that had been transduced with shRNAs were collected 72 h after transduction, and RNA was extracted using the RNeasy mini kit (Qiagen, Valencia, CA). The RNA was treated with DNase, and then 380 ng of RNA was reverse transcribed using the SuperScript III First-Strand synthesis kit (Invitrogen, Grand Island, NY) according to the manufacturer's instructions. The reverse transcription quantitative polymerase chain reaction assay (RT-qPCR) was used to determine gene expression. The following primers and probes were used to amplify the cDNA: Gag-F: 5′ ACATCAAGCAGCCATGCAAAT, GAG-R: 5′ TCTGGCCTGGTGCAATAGG, GAG FAM PROBE: 5′ CTATCCCATTCTGCAGCTTCCTCATTGATG; EGFP-F: 5′ GGAGCGCACCATCTTCTTCA, EGFP-R: 5′ AGGGTGTCGCCCTCGAA, EGFP FAM Probe: CTACAAGACCCGCGCCGAGGTG; HDAC1-F: 5′ TGAGGACGAAGACGACCCT, HDAC1-R: 5′ CTCACAGGCAATTCGTTTGTC, and HDAC1 FAM probe: 5′ CAAGCGCATCTCGATCTGCTCCTC18; HDAC2-F: 5′ CTTTCCTGGCACAGGAGACTT, HDAC2-R: 5′ CTCATTGGAAAATTGACAGCATAGT, and HDAC2 FAM probe: 5′ AGGGATATTGGTGCTGGAAAAGGCAA; and HDAC3-F: 5′ GGTGGTTATACTGTCCGAAATGTT, HDAC3-R: 5′ GCTCCTCACTAATGGCCTCTTC, and HDAC3 FAM probe: 5′ AGCAGCGATGTCTCATATGTCCAGCA. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was amplified for each sample and used for normalization. The primers and probes used were as follows: GAPDH-F: 5′ GCACCACCAACTGCTTAGCACC, GAPDH-R: 5′ TCTTCTGGGTGGCAGTGATG, and GAPDH HEX probe: 5′ TCGTGGGAAGGACTCATGACCACAGTCC.19 Results are displayed as the fold increase over the control condition, which was calculated using the ΔΔct method. The values shown represent the mean of three independent experiments±the SEM.
Western blots
J89 or 2D10 cells were collected 72 h after being transduced with lentiviruses carrying shRNAs. Protein was extracted with radioimmunoprecipitation assay buffer containing 10 μl Protease Inhibitor Cocktail (Sigma-Aldrich, St. Louis, MO) and 10 mM NaF (Sigma-Aldrich, St. Louis, MO). The quantity of protein was determined using a Bradford protein assay according to the manufacture's instructions (Bio-Rad, Hercules, CA). Five to ten micrograms of protein was loaded onto a 4–12% bis-tris gel (Invitrogen, Grand Island, NY). The western blot was then run as previously described.20 The following primary antibodies were used: HDAC1, HDAC2, HDAC3, cMyc, and YY1 (all from Santa Cruz Biotechnology, Santa Cruz, CA) and alpha tubulin (Abcam, Cambridge, MA) was used as a loading control. The band intensity of the western blots was analyzed using Image J software (NIH, Bethesda, MD). Each western blot was performed using cells from independent transductions at least three times. The values shown represent the mean percent protein knockdown±SEM from three independent experiments.
Transduction of shRNAs
Three million cells were split into 12 ml of RMPI media containing 10% FBS and 1% pen/strep and incubated overnight. At 24 h the cells were transduced with lentiviral vectors carrying cMyc or YY1 shRNAs, and the cells were incubated at 37°C overnight. shRNA plasmids and lentiviruses were obtained from the UNC Lenti-shRNA core facility, which has the Open Biosystems shRNA library (Open Biosystems, Lafayette, CO). The plasmid TRCN0000039642 was used to deplete cMyc, and plasmid TRCN0000019898 was used to deplete YY1. A plasmid containing an shRNA to eGFP was used as the nonspecific (NS) control in the ChIP experiments (catalog number RHS4459, Open Biosystems Lafayette, CO). The pLKO.1 empty vector control (catalog number RHS4080, Open Biosystems, Lafayette, CO) was used as the negative control for the RT-qPCR and flow cytometry experiments. Twenty-four hours after the addition of the shRNA, new media and 2 μg/ml of puromycin were added to select for cells that had been transduced. The cells were then incubated at 37°C for 48 h before collection for downstream applications. New transductions were performed for each experiment, and cells were never frozen down and thawed for subsequent experiments. Knockdown was assessed by qPCR and western blot as described above.
Flow cytometry
Cells were collected 72 h after transduction with lentiviruses, washed once with 1×PBS, and then fixed in 3.2% paraformaldehyde. The GFP expression of the cells in the fixed samples was measured using an Attune flow cytometer (Applied Biosystems, Foster City, CA) or a CyAn ADP analyzer (Beckman Coulter, Inc., Brea, CA, UNC flow cytometry core facility). At least 10,000 cells were collected for each condition. The analysis was performed using FlowJo flow cytometry analysis software (FlowJo, Ashland, OR).The values shown are the mean of three independent experiments±SEM from at least three independent experiments.
Results
Depletion of cMyc and YY1 in Jurkat cells does not affect cell viability
To determine whether cMyc and YY1 are required to maintain transcriptional repression of the HIV-1 provirus in Jurkat cells, the RNA interference pathway was used to deplete the mRNA transcripts of cMyc and YY1, and the downstream effects were monitored. Jurkat cells are difficult to transfect. Therefore, lentiviral vectors were used to transduce plasmids containing shRNAs into Jurkat cell lines to deplete the transcription factors cMyc and YY1. Transduction of cMyc-specific shRNAs reduced the amount of cMyc in 2D10 cells by 85% and in J89 cells by 72% (Fig. 1 and data not shown). Single depletion of YY1 with shRNAs reduced the amount of YY1 protein by 87% in 2D10 cells and 56% in J89 cells (Fig. 1 and data not shown). Furthermore, the transduction of both cMyc and YY1 shRNAs reduced protein levels by 57% and 83% in 2D10 cells and by 86% and 62% in J89 cells, respectively (Fig. 1 and data not shown). The transduced cells were then selected with puromycin to ensure that the study population of cells homogenously carried the shRNA-expressing vector. The CellTiter-Blue cell viability assay (Promega, Madison, WI) was used to determine the effect of YY1, cMyc, or YY1 and cMyc depletion on the viability of Jurkat cells. Transducing Jurkat cells with cMyc, YY1, or cMyc and YY1 shRNAs does not significantly affect the viability of 2D10 or J89 cells as compared to the cells that were transduced with the vector control plasmid (Fig. 1d and data not shown). Therefore, lentivirus transduction of shRNAs is an effective technique for the depletion of cMyc and YY1 in Jurkat cells and does not significantly affect cell viability.
FIG. 1.
Depletion of cMyc or YY1 does not affect Jurkat cell viability. Lentiviral shRNAs specific to cMyc, YY1, or cMyc and YY1 significantly deplete the protein levels of cMyc (a), YY1 (b), or cMyc and YY1 (c) in 2D10 cells as determined by western blot analysis. (d) Depletion of cMyc, YY1, or cMyc and YY1 does not affect the viability of 2D10 cells 72 h after transduction of short hairpin RNAs (shRNAs).
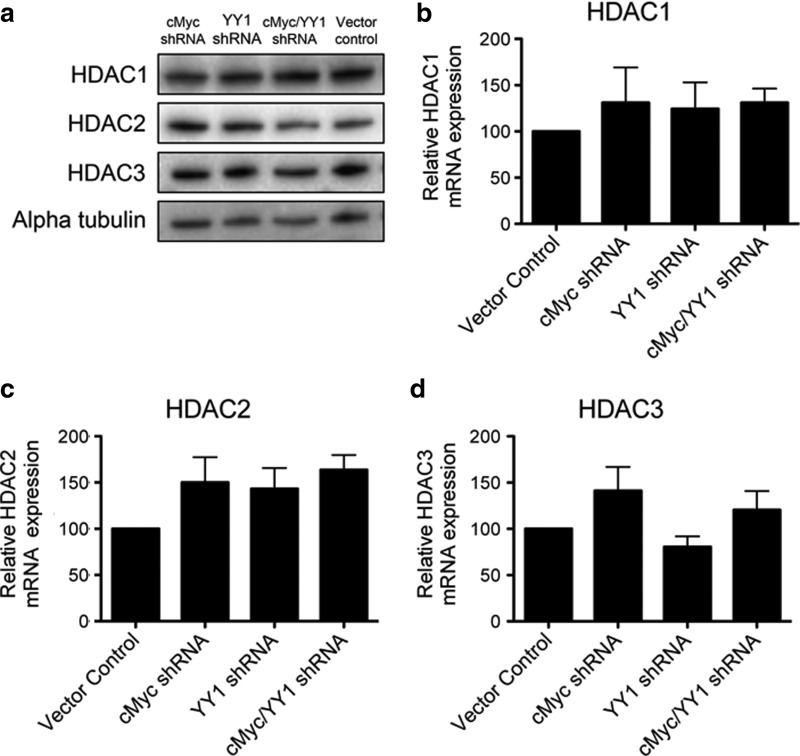
Knockdown of the transcription factors cMyc, YY1, or both does not affect protein or mRNA expression of HDAC1, HDAC2, or HDAC3
cMyc and YY1 both affect the expression and posttranscriptional regulation of a significant number of cellular genes.10,12 Because cMyc and YY1 are both broadly acting transcription factors, it is possible that they could affect HIV-1 transcription by regulating the expression of HDACs. Furthermore, cMyc has recently been shown to regulate the transcription of HDAC2 in mesenchymal stem cells.21 Therefore, to ensure that the effects of depleting cMyc and YY1 were not related to an effect on HDAC expression, we measured the levels of HDAC protein using western blot and mRNA using RT-qPCR after depletion of cMyc, YY1, or cMyc and YY1 (Fig. 2). No significant changes in the protein levels of HDAC1, HDAC2, or HDAC3 were observed by western blot after knockdown of cMyc, YY1, or cMyc and YY1 in 2D10 or in J89 cells (Fig. 2a and data not shown). Furthermore, depletion of cMyc, YY1, or cMyc and YY1 did not significantly affect HDAC1, HDAC2, or HDAC3 mRNA expression when compared to the cells transduced with the vector control (Fig. 2). Therefore, the effects of depleting cMyc, YY1, or cMyc and YY1 with shRNAs are not mediated by a secondary depletion of HDAC1, HDAC2, or HDAC3 in 2D10 or J89 cells (Fig. 2 and data not shown).
FIG. 2.
Depletion of cMyc, YY1, or cMyc and YY1 does not significantly affect expression of HDAC1, HDAC2, or HDAC3 in 2D10 cells. (a) HDAC1, HDAC2, and HDAC3 protein levels remain stable in 2D10 cells after depletion of cMyc, YY1, or cMyc and YY1. (b) HDAC1, (c) HDAC2, or (d) HDAC3 mRNA expression levels are not significantly affected by the depletion of cMyc, YY1, or cMyc and YY1 in 2D10 cells as measured by the reverse transcription quantitative polymerase chain reaction (PCR) assay (RT-qPCR).
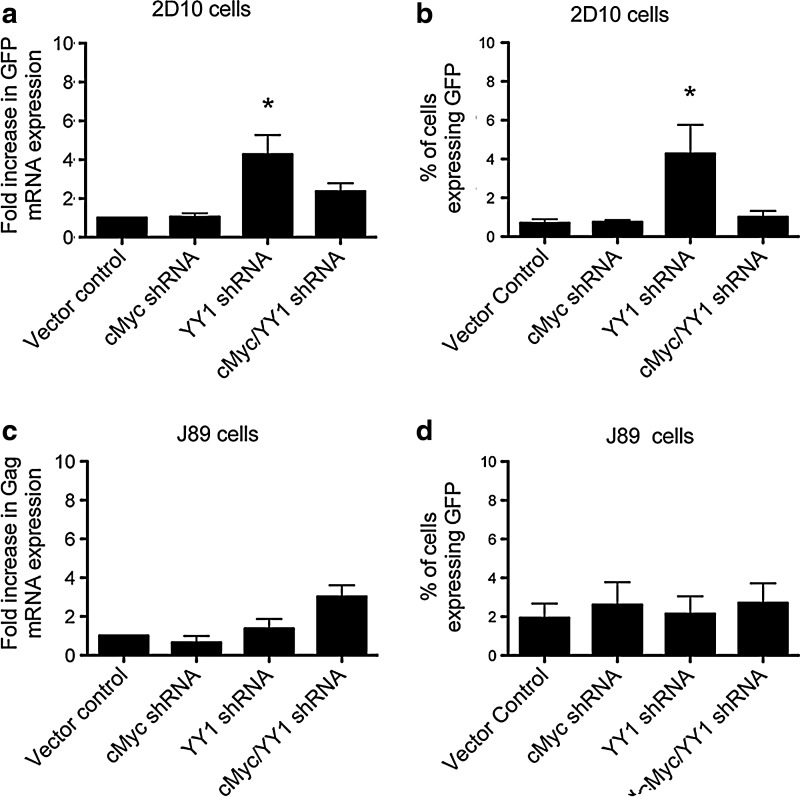
Individual depletion of the transcription factor YY1 significantly increases expression from the HIV-1 LTR
Targeting cMyc or YY1-containing complexes upregulates expression of HIV-1 mRNA in HeLa cell line models of HIV-1 latency.8,11,22 To determine whether these transcription factors are also important for maintaining HIV-1 transcriptional repression in a T cell line, cMyc, YY1, or cMyc and YY1 were depleted from J89 or 2D10 Jurkat T cells, and expression from the HIV-1 LTR was monitored using RT-qPCR and flow cytometry. The proviral genomes in both J89 and 2D10 cells contain GFP, which can be used to monitor expression from the HIV-1 LTR. In J89 cells, the GFP gene is inserted between the env and nef genes of the HIV-1 provirus.16 In 2D10 cells, the GFP gene is inserted between env and the 3′ LTR and nef is deleted.17 Additionally, 2D10 cells contain an attenuated tat and a truncated gag.17 Because the provirus in the 2D10 cells does not contain the complete gag sequence, we used RT-qPCR to monitor GFP mRNA expression in the 2D10 cells as a measure of HIV-1 LTR activity. Following depletion of YY1 from the 2D10 cells there was a significant 4.2-fold increase in GFP mRNA expression from the HIV-1 LTR (Fig. 3a, p-value <0.05). However, GFP mRNA expression did not increase significantly following depletion of cMyc or both cMyc and YY1 in the 2D10 cells (Fig. 3a). Furthermore, depletion of cMyc, YY1, or cMyc and YY1 did not result in a significant increase in expression of gag mRNA in the J89 cells (Fig. 3c).
FIG. 3.
Depletion of YY1 significantly increases HIV-1 expression in 2D10 cells. (a) GFP mRNA expression from the HIV-1 LTR significantly increased after depletion of YY1 from 2D10 cells. No changes were observed in GFP mRNA expression levels following depletion of cMyc or cMyc and YY1 from 2D10 cells. (b) The percentage of 2D10 cells expressing the GFP protein, as determined by flow cytometry, significantly increased after depletion of YY1, but not after depletion of cMyc or cMyc and YY1. (c) Expression of gag mRNA was not significantly affected by knockdown of cMyc, YY1, or both in J89 cells. (d) The percentage of J89 cells expressing the GFP protein, as determined using flow cytometry, did not significantly increase after knockdown of cMyc, YY1, or cMyc and YY1. *p-value of less than 0.05.
To further determine whether the effects seen at the mRNA level were reflected at the protein level, the percentage of cells expressing GFP was measured using flow cytometry. The percentage of 2D10 cells expressing GFP that were depleted of YY1 significantly increased over 6-fold when compared to the cells transduced with the vector control, which correlates with the observed increase in GFP mRNA that was observed (Fig. 3b, p-value <0.05). Following knockdown of cMyc or cMyc and YY1 there was not a significant increase in the percentage of 2D10 or J89 cells expressing GFP (Fig. 3b and d). Additionally, no increase in the percentage of J89 cells expressing GFP was observed following YY1 depletion (Fig. 3d).
Because cMyc and YY1 can both act as transcriptional activators as well as repressors, we next determined whether cMyc or YY1 was required for HIV-1 transcription by adding PMA to 2D10 or J89 cells that were depleted of YY1, cMyc, or both for 24 h and measuring the percentage of GFP-positive cells by flow cytometry. PMA induced a significant percentage of cells to express HIV-1-driven GFP over the cells that were treated with DMSO in the cells depleted of cMyc, YY1, or cMyc and YY1. Furthermore, there was no significant difference in the percentage of GFP-positive cells after exposure to PMA in the cells depleted of cMyc, YY1, or cMyc and YY1 as compared to the cells transduced with the control shRNA, which indicates that neither cMyc nor YY1 is required to induce transcription from the HIV-1 LTR (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/aid).
Together, these results suggest that YY1 is involved in repression of the HIV-1 LTR, but that its effects predominate in select cellular contexts. The different response of the two cell lines to YY1 depletion may be due to differences between the locations of the proviral genomes in these two cell lines or the degree of repressive chromatin structures that have developed at the LTR. Furthermore, these results indicate that depletion of cMyc or cMyc and YY1 is not sufficient to induce expression from the HIV-1 LTR in these Jurkat cell lines. Furthermore, neither cMyc nor YY1 is required for expression from the HIV-1 LTR. Of relevance to the development of translational strategies to deplete persistent HIV-1 infection, these findings demonstrate some redundancy or overlap in epigenetic mechanisms that maintain proviral latency.
Depletion of cMyc or YY1 does not significantly affect HDAC occupancy of the HIV-1 LTR
Of the 11 human HDACs, only HDAC1, HDAC2, and HDAC3 have been shown to play a critical, direct role in the regulation of HIV-1 LTR expression.20 In HeLa cell lines containing quiescent proviral genomes, depletion of cMyc or YY1 leads to transcription from the HIV-1 LTR through depletion of HDAC1 occupancy.8,11 cMyc and YY1 are both associated with recruitment of HDACs and HDAC complexes to the LTR of HIV-1 and to the promoters of some cellular genes.13 Furthermore, HDAC inhibitors are consistently able to activate HIV-1 transcription in both cell line models and in patients' cells.4,5,23 Therefore, the next step of this study was to determine whether depleting factors that recruit HDACs to the HIV-1 promoter, cMyc and YY1, had an effect on the recruitment of HDACs to the HIV-1 LTR. Following depletion of cMyc or YY1, ChIP was used to measure the occupancy of HDAC1, HDAC2, and HDAC3 in 2D10 and J89 Jurkat cells (Fig. 4). Depletion of cMyc or YY1 from 2D10 and J89 cells did not significantly alter HDAC1, HDAC2, or HDAC3 occupancy of the HIV-1 LTR (Fig. 4). Although cMyc and YY1 are known to interact with HDACs and HDAC complexes, this result indicates that depletion of these factors is not sufficient to measurably block HDAC recruitment to the HIV-1 LTR in these Jurkat cell lines. This finding is consistent with earlier results that found that depletion of cMyc or cMyc and YY1 did not significantly affect HIV-1 expression from these cell lines. However, the lack of change in HDAC occupancy of the HIV-1 LTR following depletion of YY1 suggests that the induction of HIV-1 transcription after YY1 knockdown is induced by an effect that does not affect HDAC occupancy at the HIV-1 promoter.
FIG. 4.
Histone deacetylase (HDAC) occupancy of the HIV-1 promoter is not affected by depletion of cMyc or YY1. (a) HDAC1, (b) HDAC2, and (c) HDAC3 occupancy of the HIV-1 promoter was not significantly altered after depletion of cMyc or YY1 from 2D10 cells, as determined using a ChIP assay. (d) HDAC1, (e) HDAC2, and (f) HDAC3 occupancy of HIV-1 LTR did not change significantly after depletion of cMyc or YY1 from J89 cells as measured by ChIP assay.
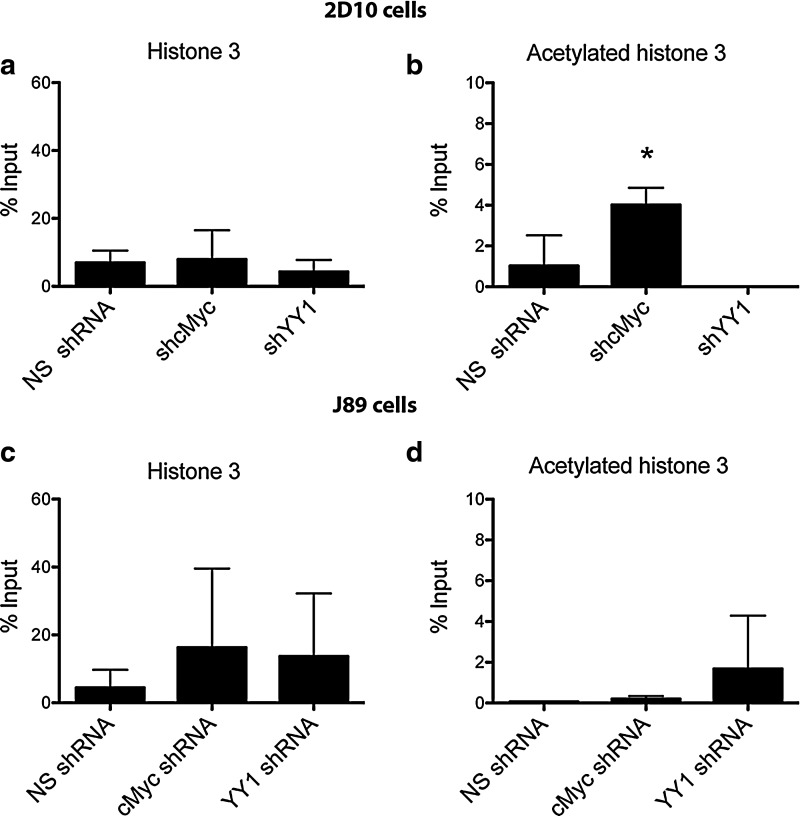
Acetylation of histone 3 increases at the promoters of activated genes, and given sufficient gene expression and chromatin remodeling, H3 histone occupancy may decrease.24,25 Therefore, to further examine the landscape of the HIV-1 LTR following depletion of cMyc, YY1, or cMyc and YY1 the occupancy of histone 3 and acetylated histone 3 was assessed.24 A significant increase in histone acetylation was observed following depletion of cMyc in 2D10 cells (Fig. 5b, p-value <0.05). This upregulation in histone acetylation was not accompanied by a significant increase in HIV-1 expression or a significant change in HDAC occupancy of the HIV-1 LTR. No significant changes in histone acetylation were observed in J89 cells following cMyc depletion or in either cell line following YY1 depletion (Fig. 5). The lack of change in histone 3 acetylation at the HIV-1 LTR following YY1 depletion is surprising given the increase in mRNA expression that was observed and indicates that depletion of YY1 alone is sufficient to disrupt repression of the HIV-1 LTR even in the presence of HDACs. The lack of change in HDAC occupancy following selective disruption of the YY1/LSF complex or the cMyc/Sp1 complex indicates that other factors are present at the HIV-1 LTR that are able to recruit HDACs in the absence of these complexes.8,11
FIG. 5.
Histone 3 and acetylated histone 3 levels on the HIV-1 promoter did not significantly change following depletion of cMyc or YY1 in Jurkat cells. (a, c) Histone 3 occupancy of the HIV-1 promoter in 2D10 (a) and J89 (c) cells did not significantly change following depletion of cMyc or YY1. (b) The levels of histone 3 acetylation significantly increased after depletion of cMyc in 2D10 cells. However, the level of acetylated histone 3 did not change following knockdown of cMyc or YY1 in J89 cells (d) or after depletion of YY1 in 2D10 cells (b). *p-value of less than 0.05.
Depletion of cMyc and YY1 does not affect quiescent HIV-1 expression in Jurkat cells
Given the possibility that LTR quiescence is maintained by redundant systems capable of recruiting HDACs to the HIV-1 LTR, both cMyc and YY1 were depleted from J89 and 2D10 cells. However, after knockdown of both cMyc and YY1, there was not a significant increase in HIV-1 mRNA expression in J89 or 2D10 cells (Fig. 3a and b). Correspondingly, there was not a significant change in the percentage of cells expressing GFP following depletion of both factors from either cell line (Fig. 3c and d). Therefore, depletion of these two HDAC recruiting factors is not sufficient to disrupt the maintenance of quiescent HIV.
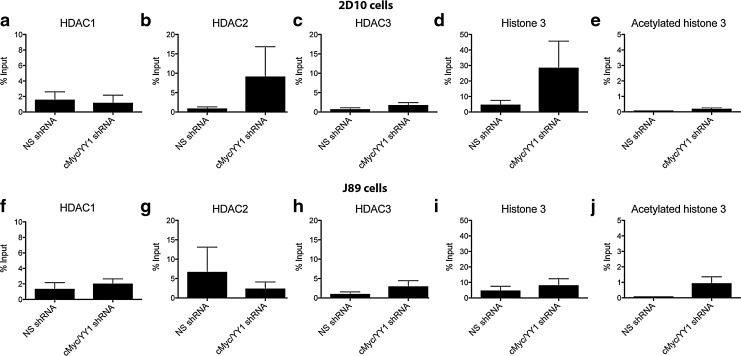
Furthermore, after depletion of both cMyc and YY1 there was not a significant change in the occupancy of HDAC1, HDAC2, or HDAC3 at the HIV-1 LTR as measured by ChIP assay (Fig. 6). This suggests that derepression of the HIV-1 LTR in T cell lines, unlike in HeLa cells, may require targeting of additional or different transcription factors. Furthermore, there were no significant changes in histone 3 occupancy or acetylation at the HIV-1 LTR in either 2D10 or J89 cells following depletion of cMyc and YY1 (Fig. 6). Altogether, these data indicate that a complex network of factors is involved in the maintenance of HIV-1 quiescence in Jurkat cells and that in the absence of cMyc and YY1, other mechanisms maintain recruitment of HDACs to the HIV-1 LTR.
FIG. 6.
Concurrent knockdown of cMyc and YY1 did not significantly affect HDAC1, 2, or 3 recruitment to the HIV-1 promoter or the levels of histone 3. Depletion of cMyc and YY1 from 2D10 cells did not affect the levels of HDAC1 (a), HDAC2 (b), HDAC3 (c), histone 3 (d), or acetylated histone 3 (e) at the HIV-1 LTR. Similarly, no changes in HDAC occupancy or histone 3 occupancy or acetylation was observed at the HIV-1 LTR following depletion of cMyc and YY1 in J89 cells (f–j) as measured by ChIP.
The effect of YY1 depletion is enhanced by HDAC inhibition
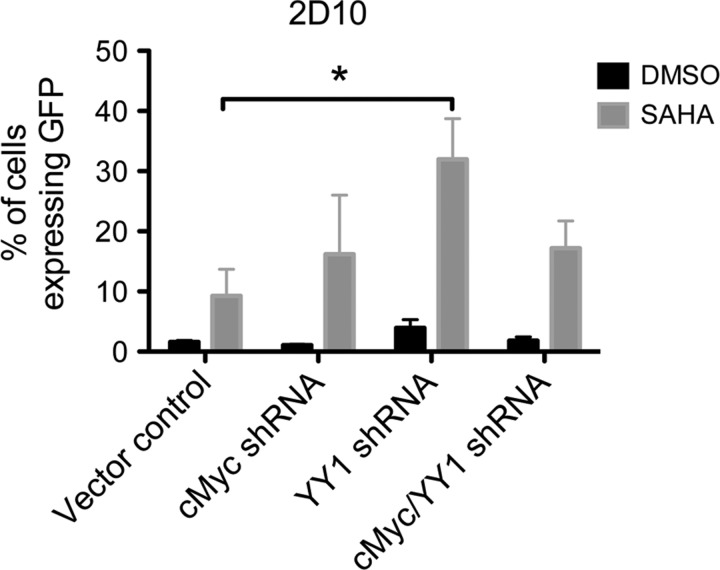
The modest effects of cMyc and YY1 depletion on HDAC recruitment to the HIV-1 LTR were surprising because both have previously been shown to repress transcription of the HIV-1 LTR through HDAC recruitment. However, as ChIP assays are only semiquantitative, an alteration of HDAC occupancy that is functionally important might not be detected. Therefore, because RNAi depletion did not achieve complete inhibition of protein expression and to further determine whether cMyc and YY1 were involved in repression of the HIV-1 promoter, 2D10 and J89 cells were treated with the HDAC inhibitor SAHA and the effects of depleting cMyc, YY1, or cMyc and YY1 from 2D10 and J89 cells were assayed. Eighteen hours after the addition of a maximal concentration of SAHA (500 nM) to cells that had been transduced with the vector control, approximately 9.23% of the 2D10 cells were found to be expressing GFP. However, the addition of SAHA in combination with the depletion of YY1 significantly increased the effect of YY1 knockdown and induced GFP expression in approximately 32% of the 2D10 cells (Fig. 7, p-value <0.05). Because the concentration of SAHA used is able to completely inhibit HDACs, the significant increase in expression when SAHA is added to YY1 depleted cells indicates that YY1 mediates repression of HIV-1 transcription through a non-HDAC mechanism.
FIG. 7.
Targeting YY1 and HDAC activity significantly increases expression from the HIV-1 LTR. Depletion of YY1 in conjunction with the addition of 500 nM SAHA for 18 h resulted in a significant increase in the percentage of 2D10 cells expressing GFP as compared to cells that were transduced with the vector control and treated with SAHA as measured using flow cytometry. Depletion of cMyc or cMyc and YY1 in combination with SAHA did not significantly increase the percentage of 2D10 cells expressing GFP. *p-value of less than 0.05.
Inhibition of HDACs with 500 nM SAHA did not augment the effects of depleting cMyc or cMyc and YY1 in 2D10 cells (Fig. 7). Furthermore, no significant changes in LTR expression were observed in J89 cells that were treated with SAHA in combination with depletion of cMyc, YY1, or cMyc and YY1 (data not shown). This finding indicates that there are indeed multiple mechanisms involved in the repression of HIV-1 transcription and that targeting multiple factors can result in increased HIV-1 expression.
Discussion
cMyc and YY1 bind to the HIV-1 LTR and regulate transcription through recruitment of HDACs.8,11 We sought to determine whether selective antilatency therapy could be specifically targeted to transcription factors that recruit HDACs to the HIV-1 LTR. Depletion of cMyc, YY1, or cMyc and YY1 did not significantly affect transcription or protein levels of the HDAC proteins, which indicates that the effects on HIV-1 transcription are not a secondary effect of changes in HDAC expression. Depletion of YY1 resulted in a significant increase in mRNA expression and the percentage of cells expressing GFP protein from the HIV-1 LTR in the 2D10 cells but not in J89 cells (Fig. 3). However, in contrast to studies in HeLa cells, single knockdown of cMyc did not have a significant effect on transcription of mRNA from the HIV-1 LTR or on the induction of GFP protein expression from the HIV-1 LTR (Fig. 3). Furthermore, depletion of cMyc and YY1 together did not induce HIV-1 transcription in either cell line. As cMyc has been show to be required for Tat-mediated elongation of the HIV-1 promoter,14 it is possible that cMyc's role in elongation may account for the lack of HIV-1 transcription that was observed after depletion of YY1and cMyc and may explain why the significant increase in histone 3 acetylation did not correlate with an increase in expression. Depletion of cMyc, YY1, or cMyc and YY1, followed by activation with PMA, induced a significant amount of expression from the HIV-1 LTR in 2D10 and J89 cells (Supplementary Fig. S1). Therefore, we can conclude that cMyc is not absolutely required for activation of the HIV-1 promoter by PMA. However, because depletion of cMyc and YY1 resulted in less activation of the HIV-1 promoter than depletion of YY1 alone, the mechanism of repression that is mediated by YY1 may act through a pathway that requires cMyc. These results are particularly surprising in light of previous studies that found disruption of cMyc or YY1 was sufficient to disrupt quiescent HIV-1 proviruses. Because YY1 depletion did not activate HIV-1 transcription in both cell lines, targeting YY1 as part of a future antilatency therapy may not broadly disrupt latency in all proviral integrants.
Knockdown of cMyc, YY1, or cMyc and YY1 did not significantly affect histone 3 occupancy of the HIV-1 LTR. Although cMyc and YY1 are known to recruit HDACs to the HIV-1 LTR in HeLa cells, the results from this study indicate that they are not absolutely required to recruit HDAC1, HDAC2, or HDAC3 to the HIV-1 LTR in Jurkat cells. Contrary to previous findings, these results indicate that in T cells other transcription factors that bind to the HIV-1 LTR may be able to compensate for the loss of cMyc, YY1, or both and maintain HDAC occupancy. Specifically, NF-κB and CBF1 have been implicated in recruitment of HDAC1 to the HIV-1 LTR and may be able to compensate for the loss of cMyc and YY1.7,9 Furthermore, these or other proteins may contribute to the repression of HIV-1 transcription in T cells. Additionally, these finding highlight an important difference between HeLa cell line models of HIV-1 latency and T cell line models of HIV-1 latency and indicate that the epigenetic environment surrounding the HIV-1 LTR in HeLa cells may be less stable than in T cells. Although Jurkat cells more closely resemble primary CD4+ T cells, it is still important to develop more advanced tools and model systems for the future study of the epigenetics of quiescent HIV-1 proviruses.
Interestingly, targeting HDAC recruitment through depletion of YY1 in conjunction with HDAC inhibition resulted in a significant increase in GFP protein expression from the HIV-1 promoter. This finding indicates that targeting multiple restrictive mechanisms at the HIV-1 LTR may be an innovative method for disrupting HIV-1 latency. YY1 is involved in several malignancies and some new classes of chemotherapeutics have been demonstrated to decrease expression of YY1.26 Such approaches might be used for the treatment of latent HIV-1 infection, and may be able to augment the effects of SAHA (N. Archin, unpublished results). However, additional development of drugs that directly target the protein interaction domains of YY1 could be pursued.
Previous studies have found that the chromatin environment surrounding the HIV-1 insertion site may affect transcription. Furthermore, in vivo infected CD4+ T cells have a range of insertion sites. To account for the effect of insertion site differences, we performed our studies in two Jurkat cells lines with distinct proviral insertion sites. The differences seen between the two cells lines may be attributable to the difference in proviral locations. However, apart from the effects of YY1 depletion, most of the results were seen in both cell lines, indicating that the proviral location may have had only modest effects. The differences in results found in this study as compared to previous studies in HeLa cells highlight the importance of conducting studies in multiple lineages of cell lines. Although Jurkat cells are derived from CD4+ T cells, there are still some important differences between these cells and in vivo HIV-1-infected cells. Therefore, our laboratory and others are working to develop models of HIV-1 quiescence that more accurately mimic the in vivo environment of HIV-1 quiescence.
In conclusion, we find that depletion of the transcription factor YY1 using transduction of shRNAs in Jurkat cells is sufficient to disrupt the repression of the HIV-1 promoter in select cellular contexts. However, depletion of the transcription factor cMyc does not induce HIV-1 expression in Jurkat cells. Importantly, we found that depletion of cMyc, YY1, or cMyc and YY1 is not sufficient to disrupt the binding of HDAC complexes to the HIV-1 LTR in Jurkat cells. Therefore, we can conclude that the mechanism of maintenance of HIV-1 transcriptional repression is complex and may require a combination of therapies that target multiple levels or several factors to reverse transcriptional repression of the HIV-1 LTR.
Supplementary Material
Acknowledgments
We thank Dr. Kara Keedy and Dr. Nancie Archin for their assistance with experiments and review of the manuscript, and members of the Karn laboratory for additional review. We also would like to thank Dr. Jonathan Karn for the gift of the 2D10 cells, Dr. David Levy for the gift of the J89 cells, and Merck Research Laboratories for providing the SAHA used in the study. This study was supported by the National Institutes of Health's Grants DA030156 to D.M.M., RR024383 to the UNC TRaCS Institute, AI50410 to the UNC Center for AIDS Research, and an equipment grant from the James B. Pendleton Charitable Trust. The UNC Flow Cytometry Core Facility is supported in part by an NCI Center Core Support Grant (P30CA06086) to the UNC Lineberger Comprehensive Cancer Center.
Author Disclosure Statement
D.M.M. is a consultant for and has received grants from Merck Research Laboratories.
References
- 1.Hanley TM. Viglianti GA. Nuclear receptor signaling inhibits HIV-1 replication in macrophages through multiple trans-repression mechanisms. J Virol. 2011;85(20):10834–10850. doi: 10.1128/JVI.00789-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cismasiu VB. Paskaleva E. Suman Daya S. Canki M. Duus K. Avram D. BCL11B is a general transcriptional repressor of the HIV-1 long terminal repeat in T lymphocytes through recruitment of the NuRD complex. Virology. 2008;380(2):173–181. doi: 10.1016/j.virol.2008.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He G. Margolis DM. Counterregulation of chromatin deacetylation and histone deacetylase occupancy at the integrated promoter of human immunodeficiency virus type 1 (HIV-1) by the HIV-1 repressor YY1 and HIV-1 activator Tat. Mol Cell Biol. 2002;22(9):2965–2973. doi: 10.1128/MCB.22.9.2965-2973.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Archin NM. Keedy KS. Espeseth A. Dang H. Hazuda DJ. Margolis DM. Expression of latent human immunodeficiency type 1 is induced by novel and selective histone deacetylase inhibitors. AIDS. 2009;23(14):1799–1806. doi: 10.1097/QAD.0b013e32832ec1dc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Archin NM. Espeseth A. Parker D. Cheema M. Hazuda D. Margolis DM. Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res Hum Retroviruses. 2009;25(2):207–212. doi: 10.1089/aid.2008.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Archin N. Liberty A. Kashuba A, et al. Administration of vorinostat disrupts HIV-1 latency in patients on ART. Paper presented at the 18th Annual Conference on Retroviruses and Opportunistic Infections; Seattle, WA. Mar, 2012. [Google Scholar]
- 7.Williams SA. Chen LF. Kwon H. Ruiz-Jarabo CM. Verdin E. Greene WC. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 2006;25(1):139–149. doi: 10.1038/sj.emboj.7600900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang G. Espeseth A. Hazuda DJ. Margolis DM. c-Myc and Sp1 contribute to proviral latency by recruiting histone deacetylase 1 to the human immunodeficiency virus type 1 promoter. J Virol. 2007;81(20):10914–10923. doi: 10.1128/JVI.01208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tyagi M. Karn J. CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. EMBO J. 2007;26(24):4985–4995. doi: 10.1038/sj.emboj.7601928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castellano G. Torrisi E. Ligresti G, et al. The involvement of the transcription factor Yin Yang 1 in cancer development and progression. Cell Cycle. 2009;8(9):1367–1372. doi: 10.4161/cc.8.9.8314. [DOI] [PubMed] [Google Scholar]
- 11.Coull JJ. Romerio F. Sun JM, et al. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J Virol. 2000;74(15):6790–6799. doi: 10.1128/jvi.74.15.6790-6799.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang CV. O'Donnell KA. Zeller KI. Nguyen T. Osthus RC. Li F. The c-Myc target gene network. Sem Cancer Biol. 2006;16(4):253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 13.Kurland JF. Tansey WP. Myc-mediated transcriptional repression by recruitment of histone deacetylase. Cancer Res. 2008;68(10):3624–3629. doi: 10.1158/0008-5472.CAN-07-6552. [DOI] [PubMed] [Google Scholar]
- 14.Bres V. Yoshida T. Pickle L. Jones KA. SKIP interacts with c-Myc and Menin to promote HIV-1 Tat transactivation. Mol Cell. 2009;36(1):75–87. doi: 10.1016/j.molcel.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lucey BP. Nelson-Rees WA. Hutchins GM. Henrietta Lacks, HeLa cells, and cell culture contamination. Arch Pathol Lab Med. 2009;133(9):1463–1467. doi: 10.5858/133.9.1463. [DOI] [PubMed] [Google Scholar]
- 16.Kutsch O. Benveniste EN. Shaw GM. Levy DN. Direct and quantitative single-cell analysis of human immunodeficiency virus type 1 reactivation from latency. J Virol. 2002;76(17):8776–8786. doi: 10.1128/JVI.76.17.8776-8786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim YK. Bourgeois CF. Pearson R, et al. Recruitment of TFIIH to the HIV LTR is a rate-limiting step in the emergence of HIV from latency. EMBO J. 2006;25(15):3596–3604. doi: 10.1038/sj.emboj.7601248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poleshko A. Palagin I. Zhang R, et al. Identification of cellular proteins that maintain retroviral epigenetic silencing: Evidence for an antiviral response. J Virol. 2008;82(5):2313–2323. doi: 10.1128/JVI.01882-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verhoeven D. Sankaran S. Silvey M. Dandekar S. Antiviral therapy during primary simian immunodeficiency virus infection fails to prevent acute loss of CD4+ T cells in gut mucosa but enhances their rapid restoration through central memory T cells. J Virol. 2008;82(8):4016–4027. doi: 10.1128/JVI.02164-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keedy KS. Archin NM. Gates AT. Espeseth A. Hazuda DJ. Margolis DM. A limited group of class I histone deacetylases acts to repress human immunodeficiency virus type 1 expression. J Virol. 2009;83(10):4749–4756. doi: 10.1128/JVI.02585-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhandari DR. Seo KW. Jung JW. Kim HS. Yang SR. Kang KS. The regulatory role of c-MYC on HDAC2 and PcG expression in human multipotent stem cells. J Cell Mol Med. 2011;15(7):1603–1614. doi: 10.1111/j.1582-4934.2010.01144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stojanova A. Caro C. Jarjour RJ. Oster SK. Penn LZ. Germinario RJ. Repression of the human immunodeficiency virus type-1 long terminal repeat by the c-Myc oncoprotein. J Cell Biochem. 2004;92(2):400–413. doi: 10.1002/jcb.20065. [DOI] [PubMed] [Google Scholar]
- 23.Contreras X. Schweneker M. Chen CS, et al. Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J Biol Chem. 2009;284(11):6782–6789. doi: 10.1074/jbc.M807898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacDonald VE. Howe LJ. Histone acetylation: Where to go and how to get there. Epigenetics. 2009;4(3):139–143. doi: 10.4161/epi.4.3.8484. [DOI] [PubMed] [Google Scholar]
- 25.Govind CK. Qiu H. Ginsburg DS, et al. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell. 2010;39(2):234–246. doi: 10.1016/j.molcel.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baritaki S. Huerta-Yepez S. Sakai T. Spandidos DA. Bonavida B. Chemotherapeutic drugs sensitize cancer cells to TRAIL-mediated apoptosis: Up-regulation of DR5 and inhibition of Yin Yang 1. Mol Cancer Ther. 2007;6(4):1387–1399. doi: 10.1158/1535-7163.MCT-06-0521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.