Abstract
The thiopurine S-methyltransferase (TPMT) gene encoding thiopurine methyltransferase is a crucial enzyme in metabolism of thiopurine drugs: azathioprine and 6-mercoptopurine, which are used in the treatment of leukemia or inflammatory bowel diseases. Genetic polymorphism of the TPMT gene correlates with activity of this enzyme, individual reaction, and dosing of thiopurines. Thirty-one variants of the TPMT gene with low enzymatic activity have been described with three major alleles: TPMT*2 (c.238G>C), *3A (c.460 G>A, c.719A>G), and *3C (c.719A>G), accounting for 80% to 95% of inherited TPMT deficiency in different populations in the world. The aim of the study was to establish a rapid and highly sensitive method of analysis for the complete coding sequence of the TPMT gene and to determine the spectrum and prevalence of the TPMT gene sequence variations in the Polish population. Recently, high-resolution melting analysis (HRMA) has become a highly sensitive, automated, and economical technique for mutation screening or genotyping. We applied HRMA for the first time to TPMT gene scanning. In total, we analyzed 548 alleles of the Polish population. We found 11 different sequence variations, where two are novel changes: c.200T>C (p.P67S, TPMT*30) and c.595G>A (p.V199I, TPMT*31). Detection of these new rare alleles TPMT*30 and *31 in the Polish population suggests the need to analyze the whole TPMT gene and maybe also the extension of routinely used tests containing three major alleles, TPMT*2, *3A, and *3C. Identification of sequence variants using HRMA is highly sensitive and less time consuming compared to standard sequencing. We conclude that HRMA can be easy integrated into genetic testing of the TPMT gene in patients treated with thiopurines.
Introduction
The Thiopurine S-methyltransferase (TPMT) gene (MIM 187680) is located on chromosome 6 (6p22.3). It encompasses 35 kb of genomic DNA and contains 10 exons, where 9 provide coding information for protein synthesis, that is, the enzyme TPMT (Szumlanski et al., 1996). In 1980 Weinshilboum and Sladek (1980) reported that high, intermediate, and undetectable enzyme activity is correlated with the genetic variation and the autosomal codominant inheritance of the TPMT gene. TPMT (EC 2.1.1.67) catalyzes the S-methylation of thiopurines, including the immunosuppressant azathioprine and the anticancer agents 6-mercaptopurine (6MP) and 6-thioguanine (6TG) (Tai et al., 1996). These drugs are currently used in the treatment of inflammatory bowel disease (IBD), hematological malignancies, and in transplantation (Sahasranaman et al., 2008). However, intermediate and low TPMT enzyme activity leads to toxicity of thiopurine drugs (Yates et al., 1997; Pandya et al., 2002; Formea et al., 2004). Increased risk of adverse reactions of 6MP depends on the accumulation of thioguanine nucleotide (TGN) metabolites (6TGN), which concentration is inversely proportional to the activity of TPMT enzyme. Approximately 1 in 300 persons (0.3%) has low or undetectable TPMT enzyme activity, whereas about 11% are heterozygous for TPMT gene mutations resulting in intermediate TPMT activity (Weinshilboum, 2001). There are currently 31 known alleles responsible for TPMT deficiency (TPMT*2, *3A, *3B, *3C, *3D, *4, *5, *6, *7, *8, *9, *10, *11, *12, *13, *14, *15, *16, *17, *18, *19, *20, *21, *22, *23, *24, *25, *26, *27, *28, and *29) and 3 alleles associated with normal TPMT activity (TPMT*1 and two silent polymorphisms) (Derijks and Wong, 2010; Lee et al., 2012). The TPMT*2 (c.238G>C), *3A (c.460G>A, c.719A>G), and *3C (c.719A>G) alleles account for 80% to 95% of inherited TPMT deficiency in different populations all over the world (Yates et al., 1997). These results based on clinical and genetic study generate the need for efficient diagnostic tools for genotyping and screening of alleles in the TPMT gene. The guidelines developed by the Clinical Pharmacogenetics Implementation Consortium provide dosing recommendations (updates at www.pharmgkb.org) for azathioprine, MP, and TG based on the TPMT genotype (Relling et al., 2011). Standard methods (e.g., RFLP, denaturing high-performance liquid chromatography [DHPLC], and Sanger sequencing) for mutation detection in the TPMT gene are time consuming, laborious, and expensive. The aim of our study was to establish a highly sensitive, automated, and economical molecular approach for the determination of TPMT alleles and to check the prevalence of genetic changes in this gene in the Polish population. We present the application of fast and efficient high-resolution melting analysis (HRMA) for genotyping and screening of the TPMT gene in the Polish population.
Materials and Methods
DNA samples
For estimation of frequency of the TPMT alleles in the Polish population, 274 DNA samples (548 alleles) obtained from 91 patients with diagnosed IBD and treated with thiopurines, as well as 183 unrelated healthy persons (including 91 men and 92 women), were subjected for analysis using HRMA. In this study, all DNA samples were considered as part of a Polish population group without association to the clinical reaction to thiopurine drugs. DNA samples were provided from the DNA Bank of the Institute of Human Genetics Polish Academy of Sciences in Poznan and from the Department of Gastroenterology, Human Nutrition and Internal Diseases, University School of Medical Sciences in Poznan.
Informed consent of the subjects was given in written form. Ethics approval for this study was obtained from the Local Ethics Committee of the University of Medical Sciences in Poznan, Poland. DNA was isolated from peripheral blood according to standard procedures using the method with guanidine isothiocyanate and stored in a TE buffer at 4°C.
High-resolution melting analysis
We designed sequences of primers to cover the complete coding region of the TPMT gene using Primer3 software. Primer sequences and amplicon sizes are shown Table 1.
Table 1.
Primers Sequences and Amplicon Sizes Used in High-Resolution Melting Analysis
| Primer name | Product size (bp) | Sequence (5’→3’) |
|---|---|---|
| TPMT_exon2_For | 197 | CGGAAGACATATGCTTGTGAGA |
| TPMT_exon2_Rev | ACCAAATATTTCTTACTGATGTCCTTG | |
| TPMT_exon3_For | 213 | TGAATGAAAAGTGTTCACCTACC |
| TPMT_exon3_Rev | CATCCTGTTAAATCACCCAAAGA | |
| TPMT_exon4_For | 249 | GCAGGTTTGCAGACCGGGGA |
| TPMT_exon4_Rev | TGCGTGCTAAATAGGAACCATCGGA | |
| TPMT_exon5_For | 122 | TTCATTATTTCATTACAGAGTTCTTCG |
| TPMT_exon5_Rev | GCCTGGCAAGCATTCAAA | |
| TPMT_exon6_For | 173 | TGTTGAAGTACCAGCATGCAC |
| TPMT_exon6_Rev | CTTACCATTTGCGATCACCTG | |
| TPMT_exon7_For | 179 | CTTCCTTCCCTGCCTTTTGT |
| TPMT_exon7_Rev | CCCAACAACTTTACCTGGATG | |
| TPMT_exon8_For | 191 | GAGAAGAACATGCCACATCATCACCTA |
| TPMT_exon8_Rev | TTTGTTTAAAAAGTTACAGCATAAGT | |
| TPMT_exon9_For | 213 | GAATCCCTGATGTCATTCTTCA |
| TPMT_exon9_Rev | CCTCAAAAACATGTCAGTGTGA |
TPMT, thiopurine S-methyltransferase.
A commercially available kit for amplification was used: High-Resolution Master Mix (Roche Diagnostics GmbH, Mannheim, Germany). Polymerase chain reaction (PCR) was carried out in a volume of 10 μL containing 5 μL Master Mix (include FastStart Taq DNA Polymerase, reaction buffer, dNTP mix, and High-Resolution Melting Dye), 1.2 μL MgCl2 (25 mM), 0.5 μL of each primer (5 pmol), and 20 ng DNA. HRMA was performed using the Light Cycler® 480 (Roche Diagnostics Ltd., Rotkreuz, Switzerland) equipment with 96-well block.
Polymerase chain reaction (PCR) procedure started with preincubation at 95°C for 10 min, followed by 45 cycles of denaturation (95°C for 10 s), annealing (60°C for 15 s) and extension (72°C for 15 s), and a final extension step at 72°C for 5 min. After amplification, PCR products were denatured at 95°C for 1 min (transition rate, 4.4°C/s) and cooled to 40°C (transition rate, 2.2°C/s) to form heteroduplexes. Melting analysis of the amplicon was carried out at the temperature interval from 60°C to 95°C (ramping rate 1.0°C/s). The final stage of the experiment was to cool the PCR product to 4°C for storage. Melting curve analysis was performed on Light Cycler® 480 using Light Cycler® 480 Scanning software (Roche Diagnostics Ltd., Rotkreuz, Switzerland).
Sequencing of PCR products
The PCR products from the LightCycler PCR amplification were directly sequenced. Selected samples with different melting profiles were treated with the QIA quick™ PCR purification kit (Qiagen, Hilden, Germany) and sequenced bidirectionally on a MegaBACE™1000 DNA Analysis System (Amersham Biosciences, Buckinghamshire, United Kingdom) using the DYEnamic™ ET Dye Terminator technology.
Results
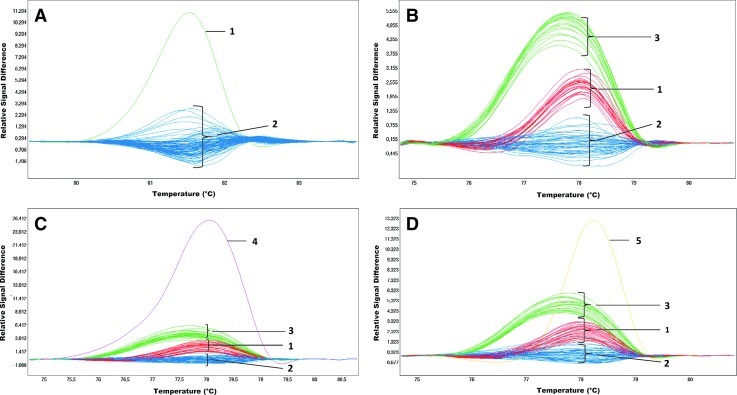
The Figure 1 presents an example of PCR product melting analysis for exons 2 and 3. A heterozygous substitution c.30T>A (p.Ile10Ile) was detected in exon 2 (Fig. 1A). Results for exon 3 show three repetitive, different melting profiles of the analyzed amplicons (Fig. 1B), which correspond to the alteration in the third intron in the position c.233+35C>T. Additionally, the melting analysis of exon 3 revealed the presence of 2 additional samples with a different profile compared to the remaining samples. Sequence analysis indicated two changes, c.219G>A (p.Ala73Ala) (Fig. 1C) and c.200T>C (p.Phe67Ser) (Fig. 1D), for individual samples (Table 2).
FIG. 1.
High-resolution melting analysis (HRMA) detection of thiopurine S-methyltransferase (TPMT) changes in exons 2 and 3. (A) exon 2: c.30T>A; line 1—genotype TA, and lines 2—wild type; (B) exon 3: c.233+35C>T; lines 1—genotype CC; lines 2—genotype CT; lines 3—genotype TT; (C) exon 3: c.200T>C; genotype TC—line 4; other lines like in (B); (D) exon 3: c.219G>A; genotype GC—line 5; other lines like in (B). Color images available online at www.liebertpub.com/gtmb
Table 2.
Observed Sequence Variations of the TPMT Gene
| Exon | Nucleotide change | Amino acid change | Ref SNP number | Number of alleles | Observed frequency (%) | Name of allele (reference) |
|---|---|---|---|---|---|---|
| 2 | 30T>A | Ile10Ile | rs200780293 | 1 | 0.18 | |
| 3 | 233+35C>T | Intron | rs4449636 | 292 | 53.28 | |
| 3 | 219G>A | Ala73Ala | — | 1 | 0.18 | |
| 3 | 200T>C | p.Phe67Ser | Novel | 1 | 0.18 | *30 (Present study) |
| 4 | 366+58T>C | Intron | rs2518463 | 201 | 36.68 | |
| 4 | 399C>T | Cys133Cys | — | 2 | 0.36 | |
| 6 | 460G>A | Ala154Thr | rs1800460 | 18 | 3.28 | *3A (Tai et al., 1996) |
| 9 | 719A>G | Tyr240Cys | rs1142345 | 18 | 3.28 | |
| 6 | 474C>T | Ile158Ile | rs2842934 | 431 | 78.65 | |
| 7 | 495-1G>A | Splice site | rs9333570 | 1 | 0.18 | *15 (Lindqvist et al., 2004) |
| 8 | 595G>A | Val199Ile | Novel | 1 | 0.18 | *31 (Present study) |
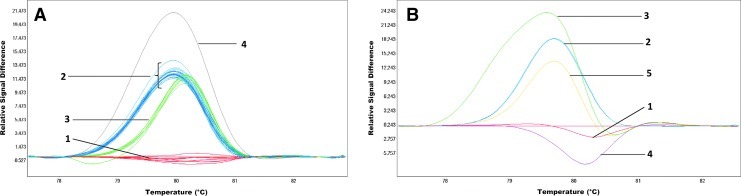
On the basis of the HRMA of exon 4, three repetitive melting profiles and additionally two different curves were observed within the entire group (Fig. 2A). Sequencing of the samples corresponding to each of the three profiles revealed the presence of a polymorphic site in intron c.366+58T>C (rs2518463) within the amplified fragment. There are three genotypes in the investigated group, which correspond to three melting curves, that is, T/T, T/C, and C/C. Two additional different melting curves were observed in the analysis of exon 4 resulting from a polymorphic change c.399C>T in this fragment (Fig. 2A and Table 2).
FIG. 2.
HRMA detection of TPMT changes in exons 4 and 6. (A) exon 4: c.366+58T>C; lines 1—genotype CC, lines 2—genotype TC, lines 3—genotype TT, c.399C>T; line 4—genotype TC. (B) exon 6: c.460G>A and c.474C>T; line 1—genotype CT(c.474) and GG(c.460), line 2—genotype TT, (c.474) and GA (c.460), line 3—genotype CT (c.474) and GA (c.460), line 4—genotype CC (c.474) and GG (c.460), line 5—genotype TT (c.474) and GG (c.460). Color images available online at www.liebertpub.com/gtmb
HRMA conducted for exon 5 has not revealed melting curves of different shape, suggesting polymorphic changes or mutation within the sequences of the examined fragments.
The HRM results for exon 6 revealed the presence of five different melting profiles of the analyzed fragment (Fig. 2B). Having conducted the sequencing of samples representing each of the melting profiles, it was concluded that the reason for such differentiation is the occurrence of two polymorphic sites within the analyzed sequence c.474C>T (p.Ile158Ile) and c.460G>A (p.Ala154Thr) (Fig. 2B). The frequency of c.460G>A was 3.28% (Table 2).
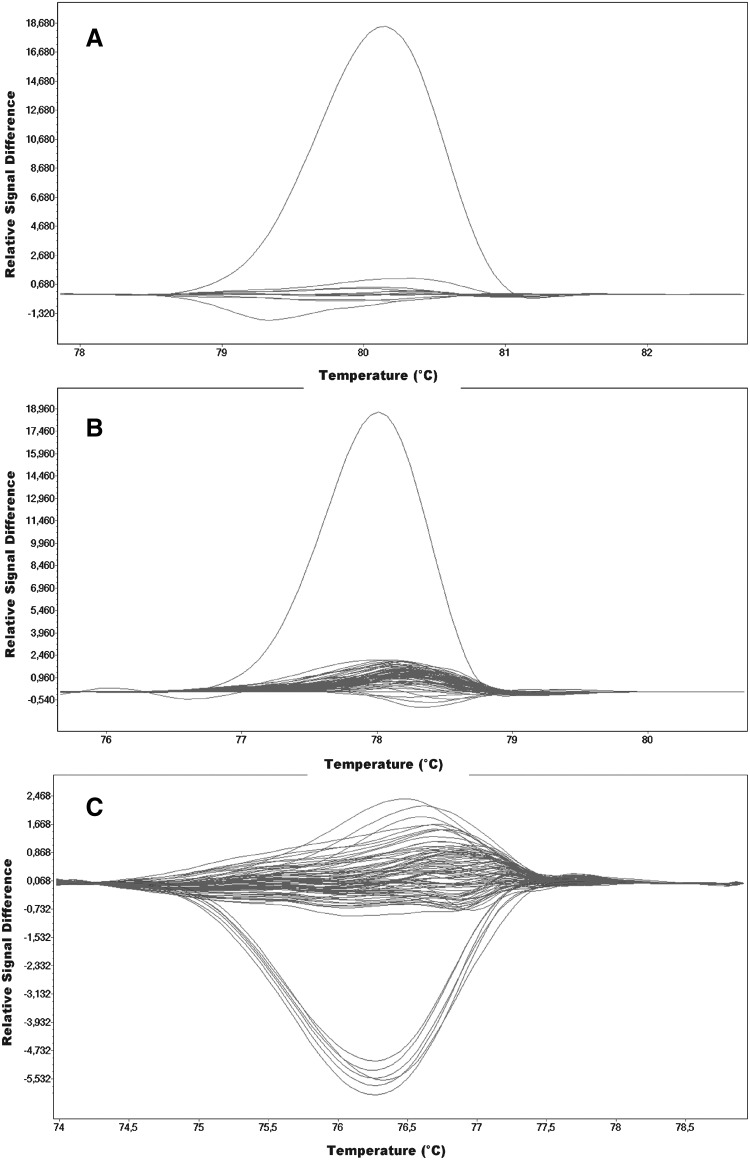
In the case of exon 7, a single case of a different melting profile was observed within the investigated group (Fig. 3A), while sequencing of this DNA fragment exposed an alteration within the splicing site c.495-1G>A.
FIG. 3.
HRMA detection of TPMT changes in exons 7, 8, and 9. (A) exon 7: c.495-1G>A; curved upper line—genotype GA and horizontal lines—wild type. (B) exon 8: c.595G>A; curved upper line—genotype GA and horizontal lines—wild type. (C) exon 9: c.719A>G; curved lower lines—genotype GA and horizontal lines—wild type.
On the basis of HRMA of exon 8, one sample with a different melting profile was selected. The profile resulted from a change in the sequence c.595G>A (p.Val199Ile) (Fig. 3B).
Figure 3C presents the results of the melting analysis for exon 9, with two groups of samples representing different melting profiles. As a result of sequencing, a change in sequence was identified, c.719A>G (p.Tyr240Cys), with a frequency of 3.28% (Table 2).
Discussion
Routine diagnostics of TPMT, the main enzyme participating in thiopurine biotransformation, aim at optimal adjustment of drug dose, to reduce the number of undesired reactions to treatment with thiopurine drugs in patients with low TPMT activity, and at improving the treatment in patients with overproduction of this enzyme. However, the results of phenotyping through evaluating the enzyme's activity in erythrocytes can be false (Ford and Berg, 2010). It is known that the activity of thiopurine methylotransferase is determined by the genotype of the TPMT gene. The Clinical Pharmacogenetics Implementation Consortium offers recommendations regarding drug dosing based on the TPMT genotype (Relling et al., 2011). Until now, various methods have been used for that purpose, including restriction digest analysis (Yates et al., 1997), single-strand conformational polymorphism (Spire-Vayron de la Moureyre et al., 1998), amplification refractory mutation system (Roberts et al., 2004), DHPLC (Schaeffeler et al., 2001), pyrosequencing (Haglund et al., 2004; Okada et al., 2005), and fluorescence hybridization probe assays (Schütz et al., 2000). Many of these methods are very time consuming, and except for the real time PCR analyses, they require further processing of the amplification product. To improve and optimize the diagnostics of TPMT, we used HRMA for the first time to examine the entire coding area of the TPMT gene, including the splicing sites.
HRMA was developed in 2004 as a precise and fast alternative for standard screening methods for investigating polymorphisms and mutations (Willmore et al., 2004; Zhou et al., 2005). The researchers conducting the studies of large genes (e.g., BRCA1, BRCA2, and CFTR) also confirmed high sensitivity of HRMA, which amounted to over 96% (Chou et al., 2005; De Leeneer et al., 2008). These data combined with other advantages of the method: low labor input, fast results, omitting the gel technique stage, and make the method optimal for analyzing the TPMT gene, especially in the case of people who were not found to have the most common alleles and whose genetic analysis should be extended. In the HRMA, each fragment amplifies in real time and is analyzed for unknown sequence variants using a single, high-resolution melt dye. Allele-specific fluorescent probes are not necessary, which significantly reduces the costs of the analysis (Schütz et al., 2000). HRM amplification and analysis are performed on one device. It decreases the duration of the analysis as well as the risk of accidental swap of the samples. Additional advantage of the HRMA, as opposed to other automated DHPLC-screening methods, is detecting differences in homozygous sequences (Chou et al., 2005).
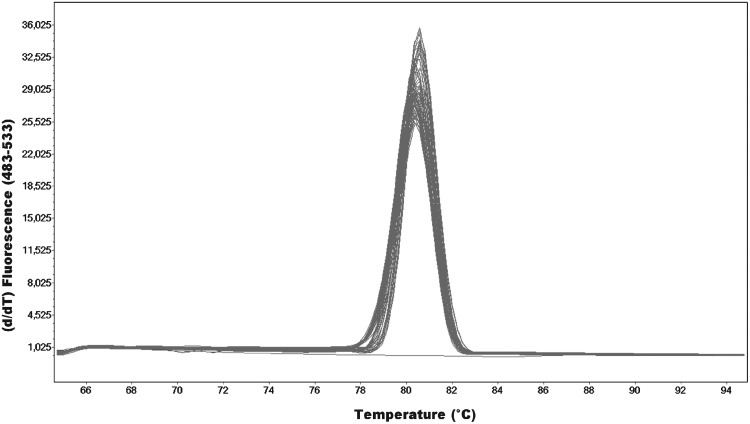
The optimization of the HRM method was conducted in accordance with the recommendations of the manufacturers and guidelines provided by other researchers (Vossen et al., 2009; Wittwer, 2009). The highest differentiation of changes in sequences is obtained for short fragments below 300 bp. We designed primers for all exons of the TPMT gene, which amplify fragments between 122 and 249 bp (Table 1). It is critical for the HRMA to obtain a strong, single amplification product without nonspecific reaction products, such as dimers of primers. This way, multiple complex melting domains difficult to interpret can be avoided. An example of optimal graph of the melting temperature (Tm)-calling analysis obtained from exon 4 of the TPMT gene has been presented below (Fig. 4). An additional advantage of the developed HRM method for the TPMT gene analysis is a simple PCR protocol that all fragments were amplified in the same conditions (annealing temperature and MgCl2 concentration). After PCR, the amplicons were analyzed in a wide spectrum of temperatures, between 60°C and 95°C, not to miss the melting point of the amplicon. Additionally, to verify the sensitivity of the optimized HRMA method, all samples were examined with the use of pyrosequencing for three most commonly observed changes in the Caucasian population TPMT*2 (c.238G>C), *3A (c.460G>A, c.719A>G), and *3C (c.719A>G) (data unpublished). Results of the analyses by both methods are consistent in 100%, which confirms high precision and sensitivity of the HRMA method optimized for the TPMT gene.
FIG. 4.
Melting peaks for exon 4 amplicons of the TPMT gene.
The second aim of the study was to determine the spectrum and the frequency of the TPMT alleles for the Polish population. Usually, the genetic examination of the TPMT gene involved defining of five most common alleles TPMT*1, *2, *3A, 3B,and *3C. Based on the HRMA and sequencing in the investigated group, two most frequent alleles for the Caucasian population were found: TPMT*1 with the frequency of 96.72% and TPMT*3A with the frequency of 3.28%. Slightly lower frequency of the *3A allele was observed in the Polish population in earlier studies on this gene by Kurzawski et al. (2004) with the use of RFLP and ASO PCR, where in the group of 350 subjects (716 alleles), the wild-type allele TPMT*1 was observed with the frequency of 96.8%, TPMT*3A in 2.7%, TPMT*2 in 0.4%, and TPMT*3C in 0.1% of the subjects. The research team of Peregud-Pogorzelski et al. (2011), based on the study of 203 Polish children suffering from acute lymphoblastic leukemia and subjected to thiopurine drug treatment, managed to determine the wild-allele frequency of 95.81%, TPMT*3A of 4.2%, and TPMT*3C of 0.25%. Similarly to our study, the rare TPMT*2 allele was not observed. Generally, the distribution of the most common alleles *3A, *2, and *3C of the TPMT gene corresponds with the general pattern for other White populations: the French population (5.7%, 0.5%, and 0.8% respectively), the British population (4.5%, 0.4%, and .1%), the Italian population (3.9%, 0.5%, and 1.0%), the Russian population (2.6%, 0.14%, and 0.36%), and White Americans (3.2%, 0.17%, and 0.17%) (Spire-Vayron De la Moureyre et al., 1998; Hon et al., 1999; McLeod et al., 1999; Rossi et al., 2001; Nasedkina et al., 2006). The HRMA method allowed us to determine the most frequent, routinely examined alleles TPMT*2, *3A, and *3C, and extend the analysis to find sequence variations in the entire coding section of the TPMT gene along with the splicing sites. Therefore, we discovered 11 substitutions (Table 2), including 6, which did not cause alterations of the amino acid sequence, leaving the TPMT enzyme activity unchanged. Apart from two known most popular variants in Caucasians (c.460G>A, c.719A>G, and TPMT*3A), one rare known variant in the acceptor splice site c.495-1G>A was found, which lead to TPMT deficiency. This change was first described in Sweden and named the TPMT*15 allele (Lindqvist et al., 2004). Moreover, the other two substitution, which so far remained unknown, were identified: in exon 3 c.200T>C (p.Phe67Ser) and in exon 8 c.595G>A (p.Val199Ile). These 2 novel TPMT variants were named TPMT*30 and *31, following to the last-described TPMT*29 allele (Lee et al., 2012). Both changes are located in the conservative region of the TPMT gene (Salavaggione et al., 2005) and cause the amino acid exchange, which indicates possible influence on lowered enzyme activity and is a subject for further investigation. In the current study, 2 new rare alleles TPMT*30 and *31 were also detected, but the rare TPMT*2 allele, which is included in the routine pharmacogenetic tests, was not observed among 274 subjects. To develop a more specific and sensitive TPMT test dedicated for the Polish population, it would be reasonable to screen larger cohort of Polish samples in the future to determine the rare alleles, if that are recurrent.
Finally, the HRM technique proved to be highly effective and sensitive in the analysis of the entire coding part of the TPMT gene. It can be successfully used for TPMT genetic testing in patients for whom treatment with thiopurine drugs is planned.
Acknowledgments
For support we thank the Polish Ministry of Science and Higher Education (grant no. N N402 209835).
Author Disclosure Statement
No competing financial interests exist.
References
- Chou LS. Lyon E. Wittwer CT. A comparison of high-resolution melting analysis with denaturing high-performance liquid chromatography for mutation scanning. Am J Clin Pathol. 2005;124:330–338. doi: 10.1309/BF3M-LJN8-J527-MWQY. [DOI] [PubMed] [Google Scholar]
- De Leeneer K. Coene I. Poppe B, et al. Rapid and sensitive detection of BRCA1/2 mutations in a diagnostic setting: comparison of two high-resolution melting platforms. Clin Chem. 2008;54:982–989. doi: 10.1373/clinchem.2007.098764. [DOI] [PubMed] [Google Scholar]
- Derijks LJJ. Wong DR. Pharmacogenetics of thiopurines in inflammatory bowel disease. Curr Pharmaceut Des. 2010;16:145–154. doi: 10.2174/138161210790112773. [DOI] [PubMed] [Google Scholar]
- Ford LT. Berg JD. Thiopurine S-methyltransferase (TPMT) assessment prior to starting thiopurine drug treatment; a pharmacogenomic test whose time has come. J Clin Pathol. 2010;63:288–295. doi: 10.1136/jcp.2009.069252. [DOI] [PubMed] [Google Scholar]
- Formea CM. Myers-Huentelman H. Wu R, et al. Thiopurine S-methyltransferase genotype predicts azathioprine-induced myelotoxicity in kidney transplant recipients. Am J Transplant. 2004;4:1810–1817. doi: 10.1111/j.1600-6143.2004.00575.x. [DOI] [PubMed] [Google Scholar]
- Haglund S. Lindqvist M. Almer S, et al. Pyrosequencing of TPMT alleles in a general Swedish population and in patients with inflammatory bowel disease. Clin Chem. 2004;50:288–295. doi: 10.1373/clinchem.2003.023846. [DOI] [PubMed] [Google Scholar]
- Hon YY. Fessing M. Pui CH, et al. Polymorphism of the thiopurine S-methyltransferase gene in African-Americans. Hum Mol Genet. 1999;8:371–376. doi: 10.1093/hmg/8.2.371. [DOI] [PubMed] [Google Scholar]
- Kurzawski M. Gawronska-Szklarz B. Drozdzik M. Frequency distribution of thiopurine S- methyltransferase alleles in a polish population. Ther Drug Monit. 2004;26:541–545. doi: 10.1097/00007691-200410000-00013. [DOI] [PubMed] [Google Scholar]
- Lee CK. Loh TP. Wong ST, et al. Detection of a novel single nucleotide polymorphism of the human thiopurine S-methyltransferase gene in a Chinese individual. Drug Metab Pharmacokinet. 2012;27:559–561. doi: 10.2133/dmpk.dmpk-12-sc-008. [DOI] [PubMed] [Google Scholar]
- Lindqvist M. Haglund S. Almer S, et al. Identification of two novel sequence variants affecting thiopurine methyltransferase enzyme activity. Pharmacogenetics. 2004;14:261–265. doi: 10.1097/00008571-200404000-00006. [DOI] [PubMed] [Google Scholar]
- McLeod HL. Pritchard SC. Githang'a J, et al. Ethnic differences in thiopurine methyltransferase pharmacogenetics: evidence for allele specificity in Caucasian and Kenyan individuals. Pharmacogenetics. 1999;9:773–776. doi: 10.1097/00008571-199912000-00012. [DOI] [PubMed] [Google Scholar]
- Nasedkina TV. Fedorova OE. Glotov AS, et al. Rapid genotyping of common deficient thiopurine S-methyltransferase alleles using the DNA-microchip technique. Eur J Hum Genet. 2006;14:991–998. doi: 10.1038/sj.ejhg.5201647. [DOI] [PubMed] [Google Scholar]
- Okada Y. Nakamura K. Wada M, et al. Genotyping of thiopurine methyltransferase using pyrosequencing. Biol Pharm Bull. 2005;28:677–681. doi: 10.1248/bpb.28.677. [DOI] [PubMed] [Google Scholar]
- Pandya B. Thomson W. Poulton K, et al. Azathioprine toxicity and thiopurine methyltransferase genotype in renal transplant patients. Transplantation Proceedings. 2002;34:1642–1645. doi: 10.1016/s0041-1345(02)02963-9. [DOI] [PubMed] [Google Scholar]
- Peregud-Pogorzelski J. Tetera-Rudnicka E. Kurzawski M, et al. Thiopurine S-methyltransferase (TPMT) polymorphisms in children with acute lymphoblastic leukemia, and the need for reduction or cessation of 6-mercaptopurine doses during maintenance therapy: the Polish multicenter analysis. Pediatr Blood Cancer. 2011;57:578–582. doi: 10.1002/pbc.23013. [DOI] [PubMed] [Google Scholar]
- Relling MV. Gardner EE. Sandborn WJ, et al. Clinical pharmacogenetics implementation consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing. Clin Pharmacol Ther. 2011;89:387–391. doi: 10.1038/clpt.2010.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RL. Barclay ML. Gearry RB, et al. A multiplexed allele-specific polymerase chain reaction assay for the detection of common thiopurine s-methyltransferase (TPMT) mutations. Clin Chim Acta. 2004;341:49–53. doi: 10.1016/j.cccn.2003.10.029. [DOI] [PubMed] [Google Scholar]
- Rossi AM. Bianchi M. Guarnieri C, et al. Genotype-phenotype correlation for thiopurine S-methyltransferase in healthy Italian subjects. Eur J Clin Pharmacol. 2001;57:51–54. doi: 10.1007/s002280000246. [DOI] [PubMed] [Google Scholar]
- Sahasranaman S. Howard D. Roy S. Clinical pharmacology and pharmacogenetics of thiopurines. Eur J Clin Pharmacol. 2008;64:753–767. doi: 10.1007/s00228-008-0478-6. [DOI] [PubMed] [Google Scholar]
- Salavaggione OE. Wanga L. Wiepertb M, et al. Thiopurine S-methyltransferase pharmacogenetics: variant allele functional and comparative genomics. Pharmacogenet Genomics. 2005;15:801–815. doi: 10.1097/01.fpc.0000174788.69991.6b. [DOI] [PubMed] [Google Scholar]
- Schaeffeler E. Lang T. Zanger UM, et al. High-throughput genotyping of thiopurine S-methyltransferase by denaturing HPLC. Clin Chem. 2001;47:548–555. [PubMed] [Google Scholar]
- Schütz E. von Ahsen N. Oellerich M. Genotyping of eight thiopurine methyltransferase mutations: three-color multiplexing, “two-color/shared” anchor, and fluorescence-quenching hybridization probe assays based on thermodynamic nearest-neighbor probe design. Clin Chem. 2000;46:1728–1737. [PubMed] [Google Scholar]
- Spire-Vayron de la Moureyre C. Debuysere H. Mastain B, et al. Genotypic and phenotypic analysis of the polymorphic thiopurine S-methyltransferase gene (TPMT) in a European population. Br J Pharmacol. 1998;125:879–887. doi: 10.1038/sj.bjp.0702152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szumlanski C. Otterness D. Her C, et al. Thiopurine methyltransferase pharmacogenetics: human gene cloning and characterization of a common polymorphism. DNA Cell Biol. 1996;15:17–30. doi: 10.1089/dna.1996.15.17. [DOI] [PubMed] [Google Scholar]
- Tai H-L. Krynetski EY. Yates CR, et al. Thiopurine S-methyltransferase deficiency: two nucleotide transitions define the most prevalent mutant allele associated with loss of catalytic activity in Caucasians. Am J Hum Genet. 1996;58:694–702. [PMC free article] [PubMed] [Google Scholar]
- Vossen RHAM. Emmelien A. Roos A, et al. High resolution melting analysis (HRMA)—more than just sequence variant screening. Hum Mutat. 2009;30:860–866. doi: 10.1002/humu.21019. [DOI] [PubMed] [Google Scholar]
- Weinshilboum R. Thiopurine pharmacogenetics: clinical and molecular studies of thiopurine methyltransferase. Drug Metab Dispos. 2001;29:601–605. [PubMed] [Google Scholar]
- Weinshilboum RM. Sladek SL. Mercaptopurine pharmacogenetics: monogenic inheritance of erythrocyte thiopurine methyltransferase activity. Am J Hum Genet. 1980;32:651–662. [PMC free article] [PubMed] [Google Scholar]
- Willmore C. Holden JA. Zhou L, et al. Detection of c-kit–activating mutations in gastrointestinal stromal tumors by high-resolution amplicon melting analysis. Am J Clin Pathol. 2004;122:206–216. doi: 10.1309/4E6U-YBY6-2N2F-CA6N. [DOI] [PubMed] [Google Scholar]
- Wittwer CT. High-resolution DNA melting analysis: advancements and limitations. Hum Mutat. 2009;30:857–859. doi: 10.1002/humu.20951. [DOI] [PubMed] [Google Scholar]
- Yates CR. Krynetski EY. Loennechen T, et al. Molecular diagnosis of thiopurine S-methyltransferase deficiency: genetic basis for azathioprine and mercaptopurine intolerance. Ann Intern Med. 1997;126:608–614. doi: 10.7326/0003-4819-126-8-199704150-00003. [DOI] [PubMed] [Google Scholar]
- Zhou L. Wang L. Palais R, et al. High-resolution DNA melting analysis for simultaneous mutation scanning and genotyping in solution. Clin Chem. 2005;51:1770–1777. doi: 10.1373/clinchem.2005.054924. [DOI] [PubMed] [Google Scholar]