Abstract
The aim of this study was to determine the prevalence of transmitted drug resistance (TDR) in newly diagnosed and treatment-naive HIV-infected patients from Croatia and evaluate a possible contribution of transmission clusters to the spread of resistant virus. The study enrolled treatment-naive HIV-infected patients that entered clinical care at the Croatian Reference Center for HIV/AIDS between 2006 and 2008. The protease gene and a part of the reverse transcriptase gene of the HIV-1 genome were sequenced by using the Trugene HIV-1 Genotyping System. The prevalence of transmitted drug resistance was analyzed by using the surveillance drug resistance mutations (SDRM) list recommended by the WHO in 2009. We report findings for 118 of 180 eligible patients (65.6% coverage). SDRM were detected in 26 of 118 patients (22.0%) who were infected with subtype B and belonged mostly to the men having sex with men (MSM). The majority of patients with primary resistance carried SDRM associated with resistance to nucleoside analogues reverse transcriptase inhibitors (NRTIs, 23 of 118 patients, 19.5%). The most frequently found NRTI SDRM was T215S (17 of 118 patients, 14.4%). SDRM associated with resistance to nonnucleoside reverse transcriptase inhibitors were detected in three (2.5%) patients and primary resistance to protease inhibitors was not detected. Non-B subtypes were detected in 13/118 patients (11%). A total of 12 transmission pairs and eight distinct transmission clusters were identified with the largest cluster harboring sequences from 19 patients; among them all but two were carrying the T215S mutation. This study showed a high prevalence of TDR in newly diagnosed MSM from Croatia and is an important contribution concerning the relationship between local transmission clusters and the spread of resistant virus.
Introduction
Croatia is a small South European country with a population of 4.3 million people. 1 Despite numerous socioeconomic and political changes in the past decades, a transition toward a market-driven economy, as well as life loses and migrations during the war for independence (1991–1995), no increase in the prevalence of HIV infection has been observed in recent years.2 A total of 862 persons have been diagnosed with HIV infection in the period 1985–2010 in Croatia.3 However, the proportion of men who have sex with men (MSM) among newly diagnosed patients with HIV infection is increasing (up to 80% in recent years) and a concentrated epidemics among MSM might be emerging.4
Clinical care of HIV patients in Croatia is centralized and all patients are treated exclusively at the HIV/AIDS center of the University Hospital for Infectious Diseases (UHID) in Zagreb.2 The health care insurance system is universal and antiretroviral treatment is free of charge for all citizens. Noteworthy, there are fewer antiretroviral drugs available in Croatia compared to European Union (EU) countries. For example, in 2009, out of 26 antiretroviral drugs registered in the EU, only 14 were available in Croatia.5
The majority of HIV-1 infections in Croatia are associated with subtype B. Molecular analysis of HIV subtypes in 145 Croatian patients (2001–2003) from different risk groups showed that 26% of infections were due to non-B subtypes (predominantly CRF02_AG, subtype C, subtype A, and CRF10_CD).6 Non-B subtype infections were found only in Croatian patients with heterosexual exposure (predominantly seafarers and their steady female partners) whereas HIV epidemics in MSM were due to subtype B infections only.6 A more recent respondent-driven sampling (RDS) study on the prevalence of HIV, sexually transmitted infections, and risky sexual behaviors among MSM from the capital of Croatia (Zagreb) confirmed the predominance of subtype B infections within this risk group.7,8
Transmission of antiretroviral drug-resistant HIV strains from treated patients who have experienced a suboptimal response to treatment or treatment failure to treatment-naive patients has been reported in both developed countries with long-term access to antiretroviral drugs as well as in developing countries with limited project-driven access to treatment. However, the data on the prevalence of transmitted drug resistance (TDR) reported in various studies are often not directly comparable, mainly due to the different methodological approaches (sampling strategy, etc.) and criteria for interpretation of primary resistance mutation significance (surveillance drug resistance mutations list recommended by the World Health Organization in 2009 versus other algorithms for the analysis of drug resistance mutations).9,10
The reported prevalence of TDR in Europe ranges between 0% and 25%.11–33 The prevalence of transmitted drug resistance in Europe has been carefully monitored via the surveillance program SPREAD. A recent report on the SPREAD program by Vercauteren et al. (2009) that enrolled 2793 patients from 20 European countries and Israel showed an overall TDR prevalence of 8.4%.32 Although there was no time trend in the overall drug resistance or nucleoside analogues reverse transcriptase inhibitor (NRTI) resistance, a significant decrease in the prevalence of protease inhibitors (PIs) and nonnucleoside reverse transcriptase inhibitor (NNRTI) resistance was observed in the study leading to the conclusion that TDR appears to be stabilizing in Europe.32 Noteworthy, the SPREAD study reported TDR trends for the period from September 2002 through December 2005 and more recent data on TDR in Europe are available only via national studies.
We performed a retrospective study on a representative cohort of HIV-1 patients (according to the national distribution of transmission risk groups) providing a 65% national coverage of newly diagnosed HIV-1 patients in a 3-year period (2006–2008). The aim of the study was to determine the prevalence of TDR to NRTIs, NNRTIs, and PIs and patterns of primary resistance mutations in treatment-naive HIV-infected Croatian patients. To the best of our knowledge this is the study with the highest coverage of newly diagnosed HIV-1 patients on a national level in a 3-year period to date.
Recently, we conducted a respondent-driven sampling (RDS) biobehavioral study on the prevalence of sexually transmitted infections including HIV as well as risky sexual behaviors among 360 MSM from Zagreb (the capital of Croatia).8 Phylogenetic analysis of sequences from HIV-infected RDS participants showed the presence of a transmission cluster suggesting an ongoing spread of HIV infection among MSM in Zagreb.8
The impact of transmission clusters on primary drug resistance in relatively closed populations has recently been evaluated by Yerly et al.24 A phylogenetic analysis of pol sequences from 637 newly diagnosed HIV patients from Geneva showed that transmission clusters were more frequent in patients with TDR.24 Yerly et al. suggested an important contribution of transmission clusters as a self-fuelling mechanism of TDR.24
In this study, by using phylogenetic analysis, we also evaluated the contribution of transmission clusters to the spread of resistant virus in newly diagnosed treatment-naive HIV-infected MSM from Croatia.
Materials and Methods
Study design and patients
The study enrolled treatment-naive patients who were diagnosed with HIV-1 infection at the Croatian Reference center for HIV/AIDS and UHID between January 2006 and December 2008, were ≥18 years old, provided a sample for genotyping resistance testing within 6 months of diagnosis, and had plasma viremia >1,000 HIV-1 RNA copies per ml. Selected demographic and epidemiological data on enrolled patients were obtained from patient's records. The study was approved by the Ethics committee of UHID and informed consent was signed by all patients.
Virological and immunological monitoring
Plasma viremia and absolute counts of CD4 T cells obtained on the date of the sample that was used for genotyping analysis were collected for all patients. Alternatively, the closest plasma viremia and CD4 T cell counts measured within 6 months of the date of the sample used for resistance testing were used for analysis.
HIV-1 RNA quantification was performed by using the COBAS AmpliPrep/COBAS Amplicor HIV-1 monitor assay (Roche Molecular Systems Inc., Branchburg, NJ) as recommended by the manufacturer. Quantification of CD4+ T cells in the peripheral blood was performed by using a Cytomics FC500 flow cytometer (Beckman Coulter, Fullerton, CA) and tetraONE system (CD45-FITC/CD4-PE/CD8-ECD/CD3-PC5 and CD45-FITC/CD56-PE/CD19-ECD/CD3-PC5 with Flow-Count Fluorospheres, Beckman Coulter).
Nucleic acid sequencing and analysis of primary resistance
HIV-1 RNA was extracted from plasma by using the QIAmp Viral RNA Kit (Qiagen, Hilden, Germany).
The HIV-1 protease gene (codons 1–99) and a part of the reverse transcriptase gene (codons 41–223) of the pol region were sequenced by using the TRUGENE HIV-1 Genotyping System (Visible Genetics, Toronto, Canada) as recommended by the manufacturer. Population-based nucleotide sequencing was performed by using an automated sequencing system (Long Read Tower, Visible Genetics). Sequences were aligned and compared with the reference strain HIV-1LAV-1 genome (GenBank number K02013) by using OpenGene DNA Sequencing System software (Visible Genetics).
Primary resistance to antiretroviral drugs was defined as the presence of ≥1 surveillance drug resistance mutations (SDRM) as recommended by Bennet et al. in 2009, which is applicable for all subtypes.10
HIV-1 subtypes were determined by the Rega HIV-1 subtyping tool (version 2.0, available at http://www.bioafrica.net/subtypetool/html/).34
The BioEdit package was used for aligning pol sequences (Clustal W) and manual editing of the alignment, from which a phylogeny was constructed by employing the maximum likelihood (ML) approach using PhyML 3.0 with 1,000 bootstrap replicates.35,36 The general time reversible (GTR) model with invariable sites and gamma distribution of rates among sites was chosen as the best fitting nucleotide substitution model, proposed by Akaike Information Criterion (AIC) of jModeltest 0.1.1.37,38 Posterior probability values of clades were obtained using MrBayes v3.1.2 with MCMC run for 1.5×106 generations and sampling frequency of 1,000 and trees were summarized after 25% burnin.39,40 Trees were viewed and edited by Dendroscope 3.41
Transmission clusters were defined as groups of three or more patients with at least two ancestors (to eliminate false clustering on account of rooting) having a bootstrap value of at least 980 and Bayesian posterior probability of 1. Pairs were not considered as transmission clusters, since they may not represent an on-going transmission.24,42
To eliminate false clustering due to selection of drug resistance mutations, major drug resistance positions according to the IAS table were removed from the alignment and phylogenetic trees were constructed as before.43
A total of 846-base pair-long sequences were used to construct ML phylogenies. Sequences from patients analyzed for primary resistance are available at the EMBL Nucleotide Sequence Database under accession numbers HE653276–HE653394 (sequences CRO529, CRO546, and CRO548 were excluded from phylogenetic analysis, due to bad quality). Additionally, 20 sequences from 16 epidemiologically unrelated Croatian HIV-1-infected patients obtained during routine clinical monitoring at UHID (resistance testing due to virological failure) were used as controls (designated 1-CRO, 2-CRO, 5-CRO, 7-CRO to 17-CRO, and 19-CRO to 24-CRO). These sequences are also available at the EMBL Nucleotide Sequence Database under accession numbers FN424270–FN424273, FN424275, FN424283–FN424284, FN424286–FN424292, and FN424294-FN424299. The sequence of subtype K (accession number AJ249239) was used as an outgroup.
Statistical analysis
We describe our data with frequencies, medians, and interquartile ranges. Frequencies were compared between groups by the Fisher exact test. Groups with continuous variables were compared with the Wilcoxon–Mann–Whitney U-test.
Results
Study population
A total of 180 newly diagnosed patients with HIV-1 infection have enrolled into care at the Croatian Reference Center for HIV/AIDS and UHID in the period 2006–2008. Of those, 168 patients were eligible for the study (nine patients were excluded due to viremia <1,000 copies per ml, two patients were <18 years of age, and one patient did not provide a sample for virological monitoring). Of those, 125 patients were randomly selected for genotyping resistance testing (with respect to the risk factors for HIV transmission and sex). Nucleic acid sequencing was successfully performed in 118 of 125 patients (94.4%). Sequencing failures were associated with lack of amplification (n=4) as well as an incomplete sequence of the target region (only the protease sequence was available) in three patients.
We report findings for 118 of 180 eligible patients (65.6%, 111 men, 94.1% and seven women, 5.9%, median age 37.4 years, interquartile range IQR 28.4–43.6 years). Risk factors for HIV infection in the patients were MSM (n=80 of 118, 67.8%), heterosexual (n=24, 20.3%), intravenous drug users, IVDU (n=3, 2.5%), and unknown/declined to answer (n=11, 9.1%) (Table 1). A total of 37 patients (31.4%) included in the TDR analysis were diagnosed with HIV infection in 2006 followed by 42 patients (35.6%) diagnosed in 2007 and 39 patients (33.1%) in 2008.
Table 1.
Main Characteristics of 118 Newly Diagnosed HIV-Infected Patients in Croatia in the Period 2006–2008
| Variable | Patients with primary resistance N=26 | Patients without primary resistance N=92 | Difference between groups (p value) |
|---|---|---|---|
| Sex, n (%) | p=0.199 | ||
| Females | 0 | 7 (7.6%) | |
| Males | 26 (100%) | 85 (92.4%) | |
| Age (median, 25th and 75th percentile; years) | 30 (28–36) | 36 (29.0–46.0) | p=0.012 |
| CD4 T cell count (median, 25th and 75th percentile; cells/μl) | 318 (75–396) | 293 (100.0–511.0) | p=0.633 |
| Plasma viremia (median, 25th and 75th percentile; log copies of HIV-1 RNA per ml) | 5.2 (4.6–5.9) | 4.8 (4.2–5.5) | p=0.061 |
| Risk factor for infection | |||
| Men who have sex with men (MSM) | 20 (76.9%) | 60 (65.2%) | p=0.354a |
| Heterosexual group | 3 (11.5%) | 21 (22.8%) | |
| Intravenous drug users | 0 | 3 (3.3%) | |
| Unknown/declined to answer | 3 (11.5%) | 8 (8.7%) | |
| HIV subtypes | |||
| B | 26 (100%) | 79 (85.9%) | p=0.038b |
| A | 0 | 6 (6.5%) | |
| C | 0 | 3 (3.3%) | |
| CRF02_AG | 0 | 2 (2.2%) | |
| CRF01_AE | 0 | 1 (1.1%) | |
| D | 0 | 1 (1.1%) | |
Comparison of MSM versus non-MSM.
Comparison of subtype B versus non-B subtypes.
Percentages sums do not equal 100 due to rounding.
At the time of primary resistance analysis, the median CD4+ T cell count of patients was 311 cells/μl (IQR 100–506 cells/μl). The median plasma viremia was 78,450 copies/ml (IQR 19,600–377,000 copies/ml).
Distribution of HIV subtypes and TDR analysis
The majority of patients analyzed in this study were infected with subtype B of HIV-1 (105 of 118, 89%), while non-B subtypes were detected in 13 (11%) patients only: subtype A (n=6, 5%), subtype C (n=3, 2.5%), CRF02_AG (n=2 patients, 1.7%), CRF01_AE (n=1, 0.8%), and subtype D (n=1, 0.8%).
The majority of patients infected with subtype B (n=105) were males (n=102, 97.1%) and MSM (n=77, 73.3 %). Other risk factors for HIV transmission in patients infected with subtype B were heterosexual contacts (n=15, 14.3%) and intravenous drug use (IVDU) (n=2, 1.9%), with 11 patients (10.5%) classified as unknown/declined to answer.
Ten of 14 patients with non-B subtypes (nine males and five females) belonged to the heterosexual risk group, one patient was a heterosexual with a history of IVDU, and three patients were MSM.
SDRM were detected in 26 of 118 patients (22.0%). The majority of patients with primary resistance carried SDRM associated with resistance to NRTI (23/118 patients, 19.5%).
One SDRM was found in 17 of 23 patients with NRTI primary resistance. The most frequently found NRTI SDRM was 215S (17 of 118 patients, 14.4%) while four other SDRM to NRTI (F77L, M184V, T215C, and T215D) were found in one patient each (Table 2).
Table 2.
Patterns of Surveillance Drug Resistance Mutations in 26 Treatment-Naive Newly Diagnosed Patients from Croatia Infected with Genotype B of HIV-1 (2006–2008)
| Surveillance drug resistance mutations (SDRM) | Number of patients (n=26) |
|---|---|
| NRTI-associated SDRM | |
| T215S | 13 |
| F77L or M184V or T215C or T215D | 1 patient per each mutation |
| M41L+L210W+T215D | 2 |
| T215S+K219R | 1 |
| T215S+L210W | 2 |
| D67G+K219E | 1 |
| NNRTI-associated SDRM | |
| Y181DHNI | 1 |
| K101E | 1 |
| NRTI+NNRTI-associated SDRM | |
| T215S+K103N | 1 |
NRTI, nucleoside reverse transcriptase inhibitors; NNRTI, nonnucleoside reverse transcriptase inhibitors.
Two SDRM to NRTIs were found in four patients (T215S and K219R, n=1; T215S and L210W, n=2; D67G and K219E, n=1, Table 2).
Three SDRM to NRTIs were found in two patients only (M41L, L210W, and T215D in both patients, Table 2).
NNRTI-associated SDRM were detected in two patients (Y181I and K101E) and one patient had a combination of SDRM associated with resistance to both NRTIs and NNRTIs (T215S and K103N, Table 2).
SDRM to PI were not detected in Croatian patients analyzed for primary resistance.
All patients with SDRM were infected with subtype B (Table 1). Risk factors for HIV infection in patients with SDRM were MSM (n=20 of 26 patients), heterosexual (n=3), and unknown/declined to answer (n=3).
Phylogenetic analysis
Phylogenetic analysis was performed on 120 HIV-1 pol region sequences from newly diagnosed individuals as well as controls.
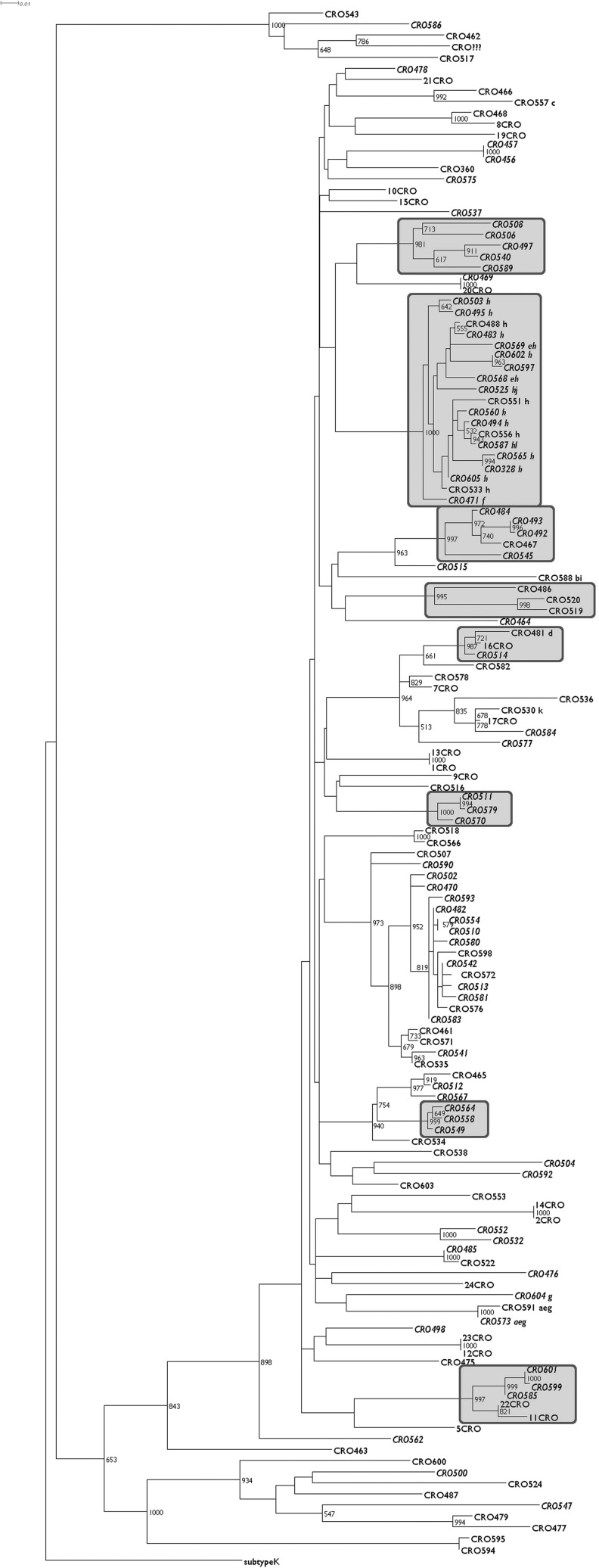
A total of 12 transmission pairs and eight distinct transmission clusters were identified (Fig. 1) with the largest cluster harboring sequences from 19 patients, among them all but two carrying the T215S mutation.
FIG. 1.
Maximum likelihood phylogenetic tree representing HIV-1 pol sequences from all included patients with bootstrap values >500 shown on corresponding nodes and significant clusters in boxes. Sequences from patients reporting men who have sex with men (MSM) status are in italic and the presence of surveillance drug resistance mutation (SDRM) in small letters on the right side of the taxon names representing the following nucleoside reverse transcriptase inhibitor (NRTI) mutations: M41L (a), D67G (b), F77L (c), M184V (d), L210W (e), T215C (f), T215D (g), T215S (h), K219E (i), K219R (j); and nonnucleoside reverse transcriptase inhibitor (NNRTI) mutations: K101E (k), K103N (l).
No differences were seen in the obtained topology when reviewing the phylogenetic tree constructed from the alignment with removed drug resistance mutations (data not shown). Furthermore, even the cluster mentioned with 17/19 included patients harboring T215S had a bootstrap support of 999.
The prevalence of TDR in patients who were not a part of the T215S transmission cluster was 7.6% (9/118 patients).
Discussion
The results of this study have shown a high prevalence TDR in newly diagnosed HIV-infected patients from Croatia. The majority of patients with documented TDR were MSM infected with genotype B and the most frequently found SDRM was T215S. The prevalence of TDR to NNRTI was low (three patients only) whereas no resistance to PIs was detected. Phylogenetic analysis of HIV strains representing 65% of newly diagnosed patients in the selected time period (2006–2008) showed the presence of a large transmission cluster composed of MSM carrying the T215S mutation that appears to play an important role in local transmission of resistant strains and affects the overall prevalence of TDR.
The prevalence of TDR in newly diagnosed HIV-infected persons on a European level is monitored via the SPREAD program.32 The prevalence of TDR in patients diagnosed with HIV infection in the years 2002–2005 in Europe analyzed as a part of SPREAD (20 European Union countries and Israel, excluding Croatia) was 8.4%.32 The prevalence of TDR in Croatia reported in this study is much higher (22%) compared with the overall prevalence reported by SPREAD. A high prevalence of TDR in Croatian patients is, in part, related to the contribution of the phylogenetically important cluster of MSM carrying the T215S mutation, which represented 77% of all patients with TDR in our study, the different time periods reported for Croatia and SPREAD, as well as the fact that Croatian data were not included in the 2002–2005 SPREAD analysis.
The prevalence of SDRM associated with NRTI resistance in the SPREAD study was 4.7%, whereas 2.3% patients showed SDRM associated with NNRTI resistance.32 Primary resistance to PIs was reported in only 2.9% of patients.32 The pattern of TDR in Croatia is different from that reported in the SPREAD study because no TDR associated with PIs was detected in our study. The prevalence of TDR to NNRTIs in Croatia (2.5%) is similar to that reported by the SPREAD study.
The majority of SDRM reported in the SPREAD study were at position 215 (RT215Y/F or revertant) with a prevalence of 2.51% followed by RT41L (1.5%) and RT103N/S (1.42%).32 These data are in accordance with our results demonstrating a high frequency of the T215S mutation in Croatian patients.
The prevalence of TDR in newly diagnosed patients from the neighboring countries (Slovenia, Hungary, Serbia, Bosnia and Herzegovina, Montenegro, Italy) has been reported in several national studies that are variable in design, national coverage, as well as interpretation algorithms (mostly IAS).
The highest national coverage for TDR analysis in the national studies was achieved by Babic et al. reporting results for 87% of newly diagnosed patients in Slovenia between 2000 and 2004.15 Only three of 77 treatment-naive HIV-infected Slovenian patients showed primary drug resistance mutations (scored according to the IAS algorithm). Despite the differences in the interpretation algorithm, it is clear that the prevalence of TDR reported for Slovenia was very low compared to Croatian data presented in this study. Similar to our results, no TDR associated with PIs was found in newly diagnosed patients from Slovenia.15
Riva et al. analyzed TDR in a cohort of 119 seroconverters (1992–2003) collected for the CASCADE study and 271 newly diagnosed individuals (2002–2005) collected for the SPREAD study in Italy.28 The prevalence of TDR in the CASCADE study was 15.1% and was slightly lower in the SPREAD study (12.2%).28 A higher prevalence of TDR in Italy (15.7%) was reported by Bonura et al. in a cohort of 108 treatment-naive HIV-infected patients from Sicily showing results more similar to the Croatian data.27 Interestingly, NNRTI-associated resistance was most frequently found in the Sicilian cohort (10.2%) with K103N being the most prevalent one (4.6%).27
Mezei et al. extensively analyzed the molecular diversity of the env and pol regions in 30 treatment-naive HIV-infected MSM from Hungary diagnosed between 2008 and 2010 showing a 16.6% prevalence of TDR.29 Similar to our results, treatment-naive Hungarian HIV-infected patients had TDR to NRTIs and NNRTIs whereas major mutations associated with primary resistance to PIs were absent.
Ciccozzi et al. reported no TDR in a small study (n=10 treatment-naive patients) from Montenegro.33
One of the prerequisites for the comparison of data from various studies on the prevalence of TDR is the use of identical resistance mutation scoring algorithms (the WHO algorithm was used in our study). Audelin et al. recently published one of the largest national studies on TDR in Europe (n=1,405 patients) by using the WHO algorithm and reported a 6.1% prevalence of primary resistance among newly diagnosed patients from Denmark.31 Similar to our study, Audelin et al. showed that the most frequent SDRM associated with NRTI resistance were 215 revertants (31.8% of individuals with transmitted resistance) followed by 103N/S for NNRTIs and 90M as well as 85V for PIs.31 Phylogenetic analysis of the sequences in that study showed 12 transmission chains involving 37 individuals with primary resistance mutations.31
The contribution of transmission clusters to the frequency of transmitted drug resistance in relatively closed populations has not been extensively evaluated. A recent study on 637 newly diagnosed HIV patients from Geneva showed that 34.9% of newly diagnosed patients and 52.7% of recent infections were parts of transmission clusters.24 Furthermore, 84% of newly diagnosed patients with TDR mutations were parts of clusters that were composed of only newly diagnosed individuals.24 The results of our study also showed clustering of the majority of patients carrying SDRM, suggesting an important contribution to the spread of resistant virus on a national level.
The majority of HIV-1 transmissions in Croatia occur among MSM who are primarily infected by subtype B (76% of reported cases for Croatia in 2008), similar to several other countries in South Eastern Europe.4,44 The SPREAD study as well as TDR in patients enrolled in the EUROSIDA cohort have shown that MSM infected with a subtype B virus were more likely than other patients to be infected with drug-resistant HIV-1.18,32 Our results showing that the majority of newly diagnosed HIV-infected patients with SDRM were from the MSM group are consistent with these reports.
However, the introduction of non-B subtypes into the MSM population in Europe has also been well documented. Giuliani et al. showed that 13.5% (15/111) of MSM from Rome analyzed between 2004 and 2008 harbored non-B subtypes (mostly subtypes C, A1, and F1).45 As expected, higher percentages of non-B subtypes were reported among non-Italian MSM (30.8%) compared with Italian MSM (8.2%).45 The results of the Rome study suggest that HIV molecular diversity increases among MSM patients as well.
Non-B subtypes were present in a small proportion of patients tested for TDR from Croatia (11%) and were associated with heterosexual transmission only. These results are in accordance with the results of our previous study showing that labor migrants, particularly seafarers from the Adriatic coast and their steady female partners, represent a gateway for the introduction of non-B subtypes into Croatia.6
A number of studies have demonstrated a lower prevalence of TDR (to all or some drug classes) in patients infected with non-B viruses compared with subtype B viruses.30,42,46–50 Chilton et al. analyzed results from 10,726 resistance tests conducted on treatment-naive individuals in the UK between 2001 and 2006 showing that 4.6% of samples with non-B subtypes carried TDR mutations compared with 11.6% of samples for subtype B.46 As the majority of patients carrying non-B subtypes likely acquired the infection in resource-limited countries in which treatment is available to the minority of infected persons, these results are to be expected.
Several studies employing the World Health Organization HIVDR threshold survey method to assess primary resistance in antennal clinics sites in several resource-limited countries (Swaziland, Malawi, Tanzania, and South Africa) confirmed the low (<5%) prevalence of TDR in women infected with non-B genotypes.51,52 Lack of TDR in Croatian newly diagnosed patients with non-B genotypes that were probably imported via labor migrants from similar resource-limited settings (seafarers on commercial ships on various international routes) is in accordance with the results from these studies.
In conclusion, the results of this study showed a high prevalence of TDR in newly diagnosed MSM from Croatia, mainly due to 215S SDRM in patients who were a part of a transmission cluster. The prevalence of TDR to NNRTIs in Croatia was low and no PI-associated resistance was detected. An important contribution of local transmission clusters to the spread of viruses carrying the 215S mutation found in Croatia emphasizes the need for continuous surveillance of TDR in the country as well as the need to develop programs to optimize the prevention and control the spread of resistant virus.
Sequence Data
Sequences analyzed in this study are available at the EMBL Nucleotide Sequence Database under accession numbers HE653276–HE653394, FN424270–FN424273, FN424275, FN424283–FN424284, FN424286–FN424292, and FN424294–FN424299.
Acknowledgments
This study was in part supported by grants from the Croatian Ministry of Science, Education and Sports to Dr. Zidovec Lepej and Prof. Begovac (grants 143-1080116-0097 and 108-1080116-0098) as well as a bilateral Slovenian-Croatian research project entitled “Molecular epidemiology of HIV-infection and resistance to antiretroviral drugs” to Prof. Poljak and Dr. Zidovec Lepej. The research leading to these results has received funding from the European Community's Seventh Framework Programme (FP7/2007-2013) under the project “Collaborative HIV and Anti-HIV Drug Resistance Network (CHAIN),” grant agreement no. 223131.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Croatian Bureau of Statistics: Census of Population, Households and Dwellings. First results by Settlements. 2011. http://www.dzs.hr. [Aug 29;2011 ]. http://www.dzs.hr
- 2.Begovac J. Zekan A. Skoko-Poljak D. Twenty years of human immunodeficiency virus infection in Croatia–an epidemic that is still in an early stage. Coll Antropol. 2006;30:17–23. [PubMed] [Google Scholar]
- 3.Croatian National Institute of Public Health Epidemiology of HIV infection AIDS in Croatia. http://www.hzjz.hr/epidemiologija/hiv.htm. [Aug 29;2011 ]. http://www.hzjz.hr/epidemiologija/hiv.htm
- 4.Bozicevic I. Begovac J. The emerging HIV epidemic among men who have sex with men in southeastern Europe. Expert Rev Anti Infect Ther. 2010;8:1351–1358. doi: 10.1586/eri.10.131. [DOI] [PubMed] [Google Scholar]
- 5.Romih V. Židovec Lepej S. Gedike K. Lukas D. Begovac J. Frequency of HIV-1 viral load monitoring of patients initially successfully treated with combination antiretroviral therapy. PLoS One. 2010;5:e15051. doi: 10.1371/journal.pone.0015051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramirez-Piedad MK. Lepej SZ. Yerly S. Begovac J. High prevalence of non-B HIV-1 subtypes in seamen and their sexual partners in Croatia. J Med Virol. 2009;81:573–577. doi: 10.1002/jmv.21433. [DOI] [PubMed] [Google Scholar]
- 7.Božičević I. Rode OD. Lepej SZ, et al. Prevalence of sexually transmitted infections among men who have sex with men in Zagreb, Croatia. AIDS Behav. 2009;13:303–309. doi: 10.1007/s10461-008-9436-7. [DOI] [PubMed] [Google Scholar]
- 8.Lepej SZ. Vrakela IB. Poljak M. Bozicevic I. Begovac J. Phylogenetic analysis of HIV sequences obtained in a respondent-driven sampling study of men who have sex with men. AIDS Res Hum Retroviruses. 2009;25:1335–1338. doi: 10.1089/aid.2009.0130. [DOI] [PubMed] [Google Scholar]
- 9.Johnson VA. Brun-Vézinet F. Clotet B, et al. Update of the drug resistance mutations in HIV-1: December 2010. Top HIV Med. 2010;18:156–163. [PubMed] [Google Scholar]
- 10.Bennett DE. Camacho RJ. Otelea D, et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One. 2009;4:e4724. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jørgensen LB. Christensen MB. Gerstoft J, et al. Prevalence of drug resistance mutations and non-B subtypes in newly diagnosed HIV-1 patients in Denmark. Scand J Infect Dis. 2003;35:800–807. doi: 10.1080/00365540310016916. [DOI] [PubMed] [Google Scholar]
- 12.Paraskevis D. Magiorkinis E. Katsoulidou A, et al. Prevalence of resistance-associated mutations in newly diagnosed HIV-1 patients in Greece. Virus Res. 2005;112:115–122. doi: 10.1016/j.virusres.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Cane P. Chrystie I. Dunn D, et al. Time trends in primary resistance to HIV drugs in the United Kingdom: Multicentre observational study. BMJ. 2005;331:1368. doi: 10.1136/bmj.38665.534595.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciccozzi M. Gori C. Boros S, et al. Molecular diversity of HIV in Albania. J Infect Dis. 2005;192:475–479. doi: 10.1086/431599. [DOI] [PubMed] [Google Scholar]
- 15.Babic DZ. Zelnikar M. Seme K, et al. Prevalence of antiretroviral drug resistance mutations and HIV-1 non-B subtypes in newly diagnosed drug-naïve patients in Slovenia, 2000–2004. Virus Res. 2006;118:156–163. doi: 10.1016/j.virusres.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Sagir A. Oette M. Kaiser R, et al. Trends of prevalence of primary HIV drug resistance in Germany. J Antimicrob Chemother. 2007;60:843–848. doi: 10.1093/jac/dkm274. [DOI] [PubMed] [Google Scholar]
- 17.Paraschiv S. Otelea D. Dinu M. Maxim D. Tinischi M. Polymorphisms and resistance mutations in the protease and reverse transcriptase genes of HIV-1 F subtype Romanian strains. Int J Infect Dis. 2007;11:123–128. doi: 10.1016/j.ijid.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Bannister WP. Cozzi-Lepri A. Clotet B, et al. Transmitted drug resistant HIV-1 and association with virologic and CD4 cell count response to combination antiretroviral therapy in the EuroSIDA Study. J Acquir Immune Defic Syndr. 2008;48:324–333. doi: 10.1097/QAI.0b013e31817ae5c0. [DOI] [PubMed] [Google Scholar]
- 19.Vercauteren J. Derdelinckx I. Sasse A, et al. Prevalence and epidemiology of HIV type 1 drug resistance among newly diagnosed therapy-naive patients in Belgium from 2003 to 2006. AIDS Res Hum Retroviruses. 2008;24:355–362. doi: 10.1089/aid.2007.0212. [DOI] [PubMed] [Google Scholar]
- 20.Vercauteren J. Deforche K. Theys K, et al. The incidence of multidrug and full class resistance in HIV-1 infected patients is decreasing over time (2001–2006) in Portugal. Retrovirology. 2008;5:12. doi: 10.1186/1742-4690-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santoro MM. Ciccozzi M. Alteri C, et al. Characterization of drug-resistance mutations in HIV type 1 isolates from drug-naive and ARV-treated patients in Bulgaria. AIDS Res Hum Retroviruses. 2008;24:1133–1138. doi: 10.1089/aid.2008.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madsen TV. Lohse N. Jensen ES, et al. High prevalence of drug-resistant human immunodeficiency virus type 1 in treatment-naïve patients in Greenland. AIDS Res Hum Retroviruses. 2008;24:1073–1077. doi: 10.1089/aid.2008.0049. [DOI] [PubMed] [Google Scholar]
- 23.Chaix ML. Descamps D. Wirden M, et al. Stable frequency of HIV-1 transmitted drug resistance in patients at the time of primary infection over 1996–2006 in France. AIDS. 2009;23:717–724. doi: 10.1097/QAD.0b013e328326ca77. [DOI] [PubMed] [Google Scholar]
- 24.Yerly S. Junier T. Gayet-Ageron A, et al. The impact of transmission clusters on primary drug resistance in newly diagnosed HIV-1 infection. AIDS. 2009;23:1415–1423. doi: 10.1097/QAD.0b013e32832d40ad. [DOI] [PubMed] [Google Scholar]
- 25.Avi R. Huik K. Sadam M, et al. Absence of genotypic drug resistance and presence of several naturally occurring polymorphisms of human immunodeficiency virus-1 CRF06_cpx in treatment-naive patients in Estonia. J Med Virol. 2009;81:953–958. doi: 10.1002/jmv.21482. [DOI] [PubMed] [Google Scholar]
- 26.Stańczak GP. Stańczak JJ. Marczyńska M, et al. Evolving patterns of HIV-1 transmitted drug resistance in Poland in the years 2000–2008. J Med Virol. 2010;82:1291–1294. doi: 10.1002/jmv.21782. [DOI] [PubMed] [Google Scholar]
- 27.Bonura F. Tramuto F. Vitale F. Perna AM. Viviano E. Romano N. Group for HIV-1 Antiretroviral Studies in Sicily: Transmission of drug-resistant HIV type 1 strains in HAART-naive patients: A 5-year retrospective study in Sicily, Italy. AIDS Res Hum Retroviruses. 2010;26:961–965. doi: 10.1089/aid.2009.0250. [DOI] [PubMed] [Google Scholar]
- 28.Riva C. Lai A. Caramma I, et al. Transmitted HIV Type 1 drug resistance and Non-B subtypes prevalence among seroconverters and newly diagnosed patients from 1992 to 2005 in Italy. AIDS Res Hum Retroviruses. 2010;26:41–49. doi: 10.1089/aid.2009.0057. [DOI] [PubMed] [Google Scholar]
- 29.Mezei M. Ay E. Koroknai A, et al. Molecular epidemiological analysis of env and pol sequences in newly diagnosed HIV Type 1-infected, untreated patients in Hungary. AIDS Res Hum Retroviruses. 2011;27(11):1243–1247. doi: 10.1089/aid.2011.0077. [DOI] [PubMed] [Google Scholar]
- 30.Yebra G. de Mulder M. del Romero J. Rodríguez C. Holguín A. HIV-1 non-B subtypes: High transmitted NNRTI-resistance in Spain and impaired genotypic resistance interpretation due to variability. Antiviral Res. 2010;85:409–417. doi: 10.1016/j.antiviral.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 31.Audelin AM. Gerstoft J. Obel N, et al. Molecular phylogenetics of transmitted drug resistance in newly diagnosed HIV type 1 individuals in Denmark, a nation-wide study. AIDS Res Hum Retroviruses. 2011;27:1–8. doi: 10.1089/aid.2010.0368. [DOI] [PubMed] [Google Scholar]
- 32.Vercauteren J. Wensing AM. van de Vijver DA, et al. Transmission of drug-resistant HIV-1 is stabilizing in Europe. J Infect Dis. 2009;200:1503–1508. doi: 10.1086/644505. [DOI] [PubMed] [Google Scholar]
- 33.Ciccozzi M. Vujošević D. Lo Presti A, et al. Genetic diversity of HIV type 1 in Montenegro. AIDS Res Hum Retroviruses. 2011;27:921–924. doi: 10.1089/aid.2010.0323. [DOI] [PubMed] [Google Scholar]
- 34.De Oliveira T. Deforche K. Cassol S, et al. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics. 2009;4:e4724. doi: 10.1093/bioinformatics/bti607. [DOI] [PubMed] [Google Scholar]
- 35.Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 36.Guindon S. Dufayard J-F. Lefort V, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 37.Posada D. jModelTest: Phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 38.Guindon S. Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 39.Huelsenbeck JP. Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 40.Ronquist F. Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 41.Huson DH. Scornavacca C. Dendroscope 3: An interactive viewer for rooted phylogenetic trees, networks. Syst Biol. 2012 doi: 10.1093/sysbio/sys062. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 42.Chalmet K. Staelens D. Blot S, et al. Epidemiological study of phylogenetic transmission clusters in a local HIV-1 epidemic reveals distinct differences between subtype B and non-B infections. BMC Infect Dis. 2010;10:262. doi: 10.1186/1471-2334-10-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson VA. Brun-Vézinet F. Clotet B, et al. Update of the drug resistance mutations in HIV-1: December 2010. Top HIV Med. 2010;18:156–163. [PubMed] [Google Scholar]
- 44.Stanojevic M. Alexiev I. Beshkov D, et al. HIV-1 molecular epidemiology in the Balkans–a melting pot for high genetic diversity. AIDS Rev. 2012;14:28–36. [PubMed] [Google Scholar]
- 45.Giuliani M. Montieri S. Palamara G, et al. Non-B HIV type 1 subtypes among men who have sex with men in Rome, Italy. AIDS Res Hum Retroviruses. 2009;25:157–164. doi: 10.1089/aid.2008.0175. [DOI] [PubMed] [Google Scholar]
- 46.Chilton DN. Castro H. Lattimore S, et al. HIV type-1 drug resistance in antiretroviral treatment-naive adults infected with non-B subtype virus in the United Kingdom. Antivir Ther. 2010;15:985–991. doi: 10.3851/IMP1658. [DOI] [PubMed] [Google Scholar]
- 47.Fox J. Hill S. Kaye S, et al. Prevalence of primary genotypic resistance in a UK centre: Comparison of primary HIV-1 and newly diagnosed treatment-naive individuals. AIDS. 2007;21:237–239. doi: 10.1097/01.aids.0000247577.26375.ef. [DOI] [PubMed] [Google Scholar]
- 48.Booth CL. Garcia-Diaz AM. Youle MS. Johnson MA. Phillips A. Geretti AM. Prevalence and predictors of antiretroviral drug resistance in newly diagnosed HIV-1 infection. J Antimicrob Chemother. 2007;59:517–524. doi: 10.1093/jac/dkl501. [DOI] [PubMed] [Google Scholar]
- 49.Maphalala G. Okello V. Mndzebele S, et al. Surveillance of transmitted HIV drug resistance in the Manzini-Mbabane corridor, Swaziland, in 2006. Antivir Ther. 2008;13(Suppl 2):95–100. [PubMed] [Google Scholar]
- 50.Kamoto K. Aberle-Grasse J. Malawi HIV Drug Resistance Task Force: Surveillance of transmitted HIV drug resistance with the World Health Organization threshold survey method in Lilongwe, Malawi. Antivir Ther. 2008;13(Suppl 2):83–87. [PubMed] [Google Scholar]
- 51.Somi GR. Kibuka T. Diallo K, et al. Surveillance of transmitted HIV drug resistance among women attending antenatal clinics in Dar es Salaam, Tanzania. Antivir Ther. 2008;13(Suppl 2):77–82. [PubMed] [Google Scholar]
- 52.Pillay V. Ledwaba J. Hunt G, et al. Antiretroviral drug resistance surveillance among drug-naive HIV-1-infected individuals in Gauteng Province, South Africa in 2002 and 2004. Antivir Ther. 2008;13(Suppl 2):101–107. [PubMed] [Google Scholar]