Abstract
New evidence indicates that astrocytes of the central nervous system (CNS) are extensively infected with human immunodeficiency virus type 1 (HIV-1) in vivo. Although no new virus is produced, this nonproductive or restricted infection contributes to the pathogenesis of HIV-associated dementia (HAD) and compromises virus eradication strategies. The HIV-1 long terminal repeat (LTR) plays a critical role in regulating virus production from infected cells. Here, we determined whether LTRs derived from CNS and non-CNS compartments are genetically and functionally distinct and contribute to the restricted nature of astrocyte infection. CNS- and/or non-CNS-derived LTRs (n=82) were cloned from primary HIV-1 viruses isolated from autopsy tissues of seven patients who died with HAD. Phylogenetic analysis showed interpatient and intrapatient clustering of LTR nucleotide sequences. Functional analysis showed reduced basal transcriptional activity of CNS-derived LTRs in both astrocytes and T cells compared to that of non-CNS-derived LTRs. However, LTRs were heterogeneous in their responsiveness to activation by Tat. Therefore, using a relatively large, independent panel of primary HIV-1 LTRs derived from clinically well-characterized subjects, we show that LTRs segregate CNS- from non-CNS-derived tissues both genetically and functionally. The reduced basal transcriptional activity of LTRs derived from the CNS may contribute to the restricted HIV-1 infection of astrocytes and latent infection within the CNS. These findings have significance for understanding the molecular basis of HIV-1 persistence within cellular reservoirs of the CNS that need to be considered for strategies aimed at eradicating HIV-1.
The human immunodeficiency virus type 1 (HIV-1) long terminal repeat (LTR) regulates viral transcription and the expression of the HIV-1 genome.1 The LTR serves as a convergence point for transcription factors and other elements of the transcriptional machinery of the host cell, as well as the virally encoded Tat protein to enhance LTR activity and the subsequent expression of viral RNA and proteins.2,3 In addition, the LTR is involved in reverse transcription of the RNA genome, integration of the provirus into the host cell genome, and the generation of viral genomes for virus assembly.4,5
The HIV-1 provirus contains two identical LTRs, each comprising unique 3′ (U3), the flanking R, and unique 5′ (U5) regions at either end.5 The U3 region is further subdivided into modulatory, enhancer, and core segments according to transcription factor binding sites that populate the LTR and their impact on LTR activity and viral gene expression.6 The core region is principally defined as the TATAA box and the immediately 5′ three tandem GC-rich binding sites for Sp factors.7 The enhancer element is directly upstream of the core promoter and is primarily defined by the presence of two NF-κB binding sites.8 The modulatory region is upstream of the enhancer element and contains binding sites for a range of transcription factors that have been reviewed elsewhere.3
Several studies have previously described the compartmentalization of LTR sequences between different anatomical sites using phylogenetic analyses.9,10 This suggests that LTRs may evolve in a tissue-specific manner to reflect the different transcriptional factors present within their resident cells. Within T lymphocytes, inducible LTR regulation is predominantly linked to members of the NF-κB transcription factor family and is dependent on the Sp binding motif.8 Removal of NF-κB binding sites severely restricts basal as well as Tat trans-activated LTR activity in T cells.11 In contrast, LTR activity in monocyte/macrophages demonstrated a critical importance of C/EBP factors.12 Although C/EBP proteins are expressed in a variety of tissues including hepatocytes and adipocytes, their expression in hematologic cells is primarily limited to the myeloid lineage. Finally, the Sp transcription factor family (Sp1, Sp3, Sp4) has also been shown to modulate LTR activity via the three Sp binding sites in the core region.7 Studies have shown that Sp1 and Sp4 activate the LTR while Sp3 represses LTR activity.13 Sp family members are differentially expressed throughout the body, which could affect the activator:repressor ratio and overall LTR activity.14
While previous studies have analyzed LTR activity in the context of T cells and monocytes/macrophages, little is known about HIV-1 transcription within astrocytes. We recently showed that astrocyte infection is extensive in vivo, with proviral DNA detected in up to 20% of GFAP-positive cells of autopsy brain tissues.15 However, astrocytes undergo a restricted infection that leads to little or no virus production, and several blocks to viral replication have been described in the literature.16,17 Whether HIV-1 transcription of viruses that reside in the central nervous system (CNS) is regulated differently within astrocytes and contributes to their restricted infection remains unknown. Regardless, HIV-1 infection of astrocytes results in their dysfunction and subsequent loss of neuronal support.18 This in turn contributes to the onset of HIV-associated neurocognitive disorders including HIV-associated dementia (HAD), which can affect up to 50% of infected individuals.19 Astrocyte infection also has important implications for HIV eradication and cure strategies, with infected cells representing a significant latently infected viral reservoir within the CNS.20 To better understand viral transcription of CNS-derived LTRs within astrocytes, in this study we genetically and functionally characterized LTRs derived from matched CNS- and non-CNS-derived anatomical compartments of subjects with HAD.
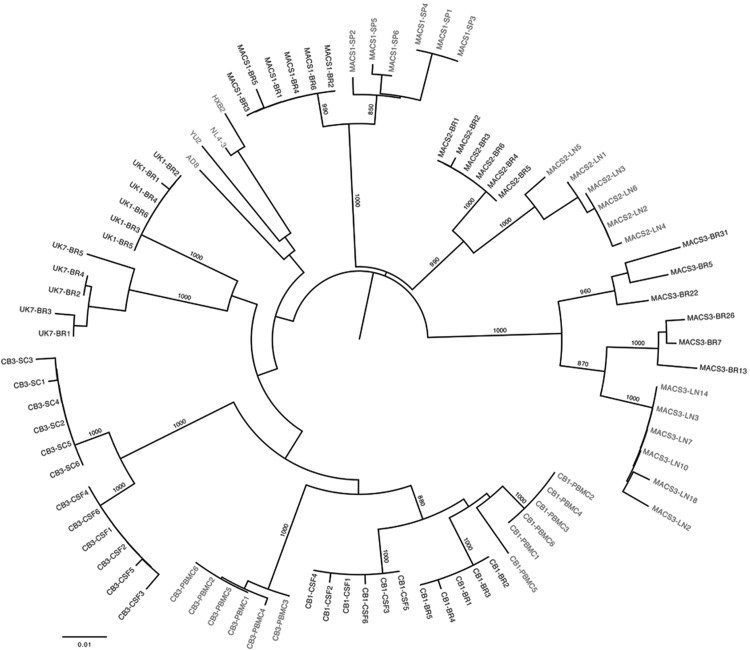
Primary HIV-1 viruses isolated from autopsy brain and/or cerebrospinal fluid, spinal cord, lymph node, spleen, or peripheral blood mononuclear cells (PBMCs) from subjects CB1, CB3, MACS1, MACS2, MACS3, UK1, and UK7 have been described in detail previously.21–23 Blast searching, detailed quasispecies analyses, and viral sequence comparisons to sequences derived directly from the autopsy tissues without culture indicate that the brain/CNS-derived primary isolates and cloned viral genes are not laboratory contaminants or derived from contaminating patient blood.21,22,24–26 The clinical characteristics of the subjects and the LTR clones generated are summarized in Table 1. A 0.6-kb fragment spanning the KpnI to HindIII restriction sites in the HIV-1 LTR (corresponding to nucleotides 9015 to 9621 in the HXB2 strain) was amplified from viral cDNA by polymerase chain reaction (PCR) and cloned into the pGL3-Basic luciferase reporter vector (Promega, USA). Between five and six LTR clones were generated for each virus, with a subset being unique clones (Table 1). The LTRs were sequenced and subjected to multiple sequence alignments (data not shown) and phylogenetic analysis (Fig. 1),27 which together showed that the LTRs were independent and formed distinct clusters according to their tissue of origin. Thus, we established and characterized a new panel of HIV-1 LTRs (n=82) derived from autopsy brain and other tissues of seven subjects who died from AIDS.
Table 1.
Study Subjects, HIV-1 Isolates, and Summary of Long Terminal Repeat Clones
| Subject | Risk factor | Last CD4 count (cells/μl) | Antiretroviral(s) | HIV-1 encephalitis | Tissues yielding HIV-1 isolates | Name of virus isolate | LTRs cloned from virus isolate (n) | Unique LTRs (n) |
|---|---|---|---|---|---|---|---|---|
| CB1 | MH | 10 | ddI (prior AZT) | Severe | Brain | CB1-BR | 5 | 2 |
| CSF | CB1-CSF | 6 | 3 | |||||
| PBMC | CB1-PBMC | 6 | 3 | |||||
| CB3 | MH | 5 | ddI (prior AZT and ddC) | Severe | S.Cord | CB3-SC | 6 | 4 |
| CSF | CB3-CSF | 6 | 3 | |||||
| PBMC | CB3-PBMC | 6 | 3 | |||||
| MACS1 | MH | 2 | None | Severe | Brain | MACS1-BR | 6 | 3 |
| Spleen | MACS1-SP | 6 | 4 | |||||
| MACS2 | MH | 52 | AZT | Moderate | Brain | MACS2-BR | 6 | 5 |
| L.Node | MACS2-LN | 6 | 4 | |||||
| MACS3 | MH | 95 | None | Moderate | Brain | MACS3-BR | 6 | 6 |
| L.Node | MACS3-LN | 6 | 4 | |||||
| UK1 | IVDU | 87 | ddC (1 mo) | Moderate | Brain | UK1-BR | 6 | 2 |
| UK7 | IVDU | 90 | AZT | Severe | Brain | UK7-BR | 5 | 4 |
The clinical and neuropathological details of the study subjects and the derivation and characterization of the primary tissue-derived HIV-1 isolates have been published previously21,23,26 and are summarized again here to assist in the interpretation of the data derived from the cloned long terminal repeats (LTRs). LTRs were amplified from primary virus isolates by PCR and cloned into the pGL3-Basic vector to act as a promoter for the luciferase gene. The LTRs described here have been assigned GenBank accession numbers JX289943 to JX290024. MH, male homosexual; IVDU, intravenous drug user; mo, month; ddI, didanosine; AZT, zidovudine; ddC, zalcitabine; CSF, cerebrospinal fluid; PBMC, peripheral blood mononuclear cells; S.Cord, spinal cord; L.Node, lymph node.
FIG. 1.
Phylogenetic analysis of long terminal repeat (LTR) nucleotide sequences. The phylogenetic tree, shown in radial representation, was constructed from an LTR nucleotide multiple sequence alignment, as described previously.27 Briefly, the phylogenetic analysis was performed using the neighbor-joining algorithm of MEGA 4 (MEGA Software, Tempe AZ), with a transition/transversion ratio of 2.0, empirical base frequencies, and a randomized input order of sequences. A consensus tree was generated from bootstrapping of 1,000 replicates, which was viewed in FigTree to produce the final figure (http://tree.bio.ed.ac.uk/software/figtree/). Clone names for central nervous system (CNS)-derived LTRs are shown in black and non-CNS-derived LTRs are shown in gray. The nucleotide sequences of HIV-1 AD8, YU2, NL4-3, and HXB2 LTRs are shown in gray and were included for comparison. Numbers associated with each branch are bootstrap values obtained from 1,000 replicates. Only values above 700 for the major branches are shown. Branch lengths are proportional to the amount of sequence divergence. The scale bar represents 1% genetic distance.
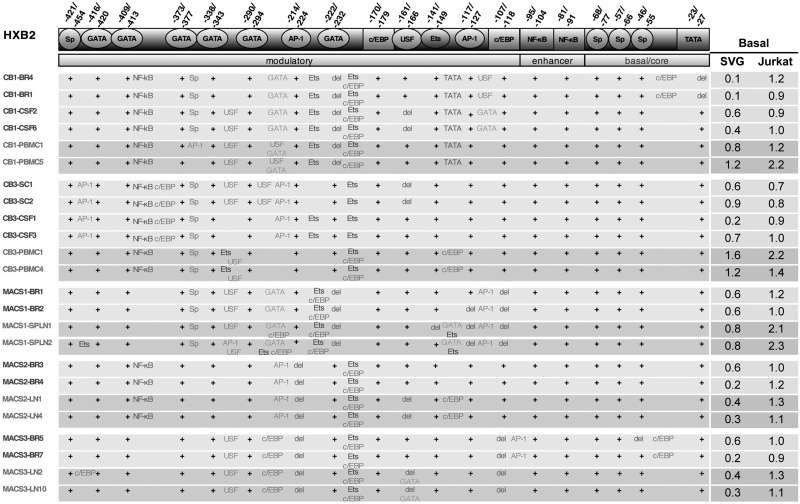
We next analyzed LTR clones for the presence or absence of transcription factor bindings sites using the web-based transcription factor binding prediction program, TF search (http://www.cbrc.jp/research/db/TFSEARCH.html). For these and subsequent experiments, a subset of unique LTR clones was chosen that represents the predominant sequences for the respective viral isolates and was limited to subjects with matched CNS and non-CNS isolates. In silico transcription factor motif predictions were performed with a minimum threshold of 75% homology to consensus sequences for a variety of transcription factors (AP-1, Ets, GATA, C/EBP, NFAT, NF-κB, Sp, TATAA, and USF) that have been shown to be important for HIV-1 transcriptional regulation.3 The output was analyzed against the HXB2 LTR and the numbering is relative to the start site of transcription in HXB2. Schematic diagrams of these alignments are presented in Fig. 2. The nucleotide sequence in the core/basal promoter and enhancer regions of patient LTR sequences showed considerable variation; however, the presence of predicted transcription factor binding sites within this region (TATAA box, three Sp-binding sites, and two NF-κB motif) was conserved across all isolates. Importantly, transcription factor binding site analyses did not identify a CNS-specific signature sequence that correlated to a unique promoter configuration associated with brain infection. However, despite the lack of a CNS-specific signature sequence, the observed changes and mutations may affect site affinities and result in a common CNS-specific transcriptional activity.
FIG. 2.
Transcription factor motif prediction of patient LTR sequences. LTR sequences were analyzed using “TF search” for the presence of a variety of transcription factor binding sites (TFBS) as compared to the HXB2 LTR. Sites with 75% homology to consensus or above are shown. A plus indicates that the respective site in HXB2 is retained in the patient LTR. Where deletion of a site occurs, this is represented by “del.” Text within the table represents additional TFBS. Clone names for CNS-derived LTRs are shown in black and non-CNS-derived LTRs are shown in gray. The corresponding relative basal transcriptional activities of each LTR in SVG and Jurkat cells are shown on the right. Basal values are shown relative to the wild-type NL4-3 LTR.
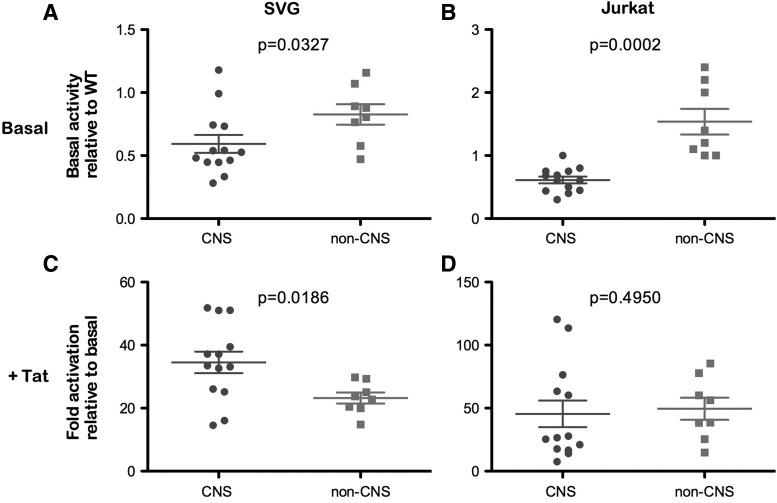
Therefore, to determine whether alterations in the LTR sequence influence promoter activity, we next performed transcriptional assays in the human fetal astrocyte cell line SVG, and in Jurkat T cells under basal and Tat-activated conditions, using the same panel of LTR clones shown in Fig. 2. In these assays, we observed lower basal and Tat-activated transcription in astrocytes compared to T cells, irrespective of the origin of the LTR (data not shown). These results likely reflect differences in the activation states between astrocytes and T cells and the different transcription factors that are available within these cells. The basal transcriptional activity of the individual LTRs in SVG and Jurkat cells is shown in Fig. 2. When we compared the transcriptional activity of CNS- and non-CNS-derived LTRs under basal conditions, the CNS-derived LTRs showed significantly lower basal transcription compared to non-CNS-derived LTRs in both astrocytes and T cells (Fig. 3). Under Tat-activated conditions, CNS-derived LTRs showed greater activation than non-CNS-derived LTRs in astrocytes, but this difference was not observed in T cells where CNS- and non-CNS-derived LTRs showed equivalent activation. It is important to recognize that these results are in the context of activation by Tat derived from the HXB2 strain of HIV-1, and may not reflect the activation by their cognate Tat proteins. Further studies analyzing the activation of these LTRs with titrating amounts of their matching Tat proteins are required to provide a more complete understanding of the in vivo function of the Tat/TAR axis within the CNS. However, we recently showed that Tat proteins derived from CNS- and non-CNS-derived viruses have similar activity in SVG cells,24 providing evidence that alterations in the Tat/TAR axis described here are likely to be at the level of LTR activity. Together, these results suggest that LTRs derived from the CNS have altered transcriptional activity that may contribute to the restricted HIV-1 infection of astrocytes. Furthermore, these data provide a greater understanding of the nature of the astrocyte viral reservoir and highlight its consideration in virus eradication/cure strategies.
FIG. 3.
Transcriptional activity of patient LTRs under basal and Tat-activated conditions in astrocytes and T cells. Matched CNS- and non-CNS-derived LTRs were analyzed for their transcriptional activity under basal (A, B) and Tat-activated conditions (C, D). Basal conditions were assessed in both a human fetal astrocyte cell line (SVG, A) and a T cell line (Jurkat, B), and results are shown relative to the wild-type NL4-3 LTR. Similarly, Tat-activated conditions were also assessed in SVG (C) and Jurkat (D) cell lines using saturating amounts of HXB2 Tat, and results are shown as fold activation over basal. CNS-derived LTRs are shown in black and non-CNS LTRs are shown in gray. Error bars represent mean±standard error (SE). The data shown are a compilation of the means of four independent experiments performed in triplicate. Statistical analyses were performed using the Mann–Whitney U-test with p values<0.05 considered statistically significant.
In conclusion, using a relatively large, independent panel of LTRs, we showed that LTR sequences exhibit distinct clustering between CNS and non-CNS tissues. We further show, for the first time, that these distinct LTRs correlate with functional differences in their transcriptional activity, with CNS-derived LTRs having lower basal transcriptional activity. This was evident in both an astrocyte and a T cell model, suggesting that CNS-derived LTRs have alterations in their core promoter that influence basal transcriptional activity. Analyses of the LTR sequences for the presence or absence of transcription factor binding sites failed to identify a common CNS-specific sequence motif responsible for the lower basal transcriptional activity, suggesting adaption to a common activity rather than a specific promoter configuration. Thus, our study provides new insights into the functional activity of CNS-derived LTRs, and suggests a molecular mechanism contributing to the restricted infection of astrocytes and their role as a significant viral reservoir in the CNS. Finally, we describe and characterize here a new and relatively large panel of functional LTRs from brain and other tissues, which will enhance the capacity of investigators to undertake neuroAIDS research.
Acknowledgments
We thank Dana Gabuzda for provision of primary HIV-1 isolates. This study was supported by a grant from the Australian National Health and Medical Research Council (NHMRC) to M.J.C., S.L.W., and P.R.G. (603708). L.R.G. is the recipient of an Australian NHMRC Postdoctoral Training Fellowship. D.C. was the recipient of an NHMRC Dora Lush Postgraduate Scholarship. P.R.G. is the recipient of an Australian NHMRC Level 2 Biomedical Career Development Award. The authors gratefully acknowledge the contribution to this work of the Victorian Operational Infrastructure Support Program received by the Burnet Institute. Lachlan R. Gray and Daniel Cowley contributed equally to this work.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Sodroski J. Rosen C. Wong-Staal F, et al. Trans-acting transcriptional regulation of human T-cell leukemia virus type III long terminal repeat. Science. 1985;227(4683):171–173. doi: 10.1126/science.2981427. [DOI] [PubMed] [Google Scholar]
- 2.Cullen BR. Regulation of human immunodeficiency virus replication. Annu Rev Microbiol. 1991;45:219–250. doi: 10.1146/annurev.mi.45.100191.001251. [DOI] [PubMed] [Google Scholar]
- 3.Pereira LA. Bentley K. Peeters A. Churchill MJ. Deacon NJ. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 2000;28(3):663–668. doi: 10.1093/nar/28.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilhelm M. Wilhelm FX. Reverse transcription of retroviruses and LTR retrotransposons. Cellular and molecular life sciences. CMLS. 2001;58(9):1246–1262. doi: 10.1007/PL00000937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coffin JM. Molecular mechanisms of nucleic acid integration. J Med Virol. 1990;31(1):43–49. doi: 10.1002/jmv.1890310109. [DOI] [PubMed] [Google Scholar]
- 6.Gaynor R. Cellular transcription factors involved in the regulation of HIV-1 gene expression. AIDS. 1992;6(4):347–363. doi: 10.1097/00002030-199204000-00001. [DOI] [PubMed] [Google Scholar]
- 7.Jones KA. Kadonaga JT. Luciw PA. Tjian R. Activation of the AIDS retrovirus promoter by the cellular transcription factor, Sp1. Science. 1986;232(4751):755–759. doi: 10.1126/science.3008338. [DOI] [PubMed] [Google Scholar]
- 8.Nabel G. Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326(6114):711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 9.Ait-Khaled M. McLaughlin JE. Johnson MA. Emery VC. Distinct HIV-1 long terminal repeat quasispecies present in nervous tissues compared to that in lung, blood and lymphoid tissues of an AIDS patient. AIDS. 1995;9(7):675–683. doi: 10.1097/00002030-199507000-00002. [DOI] [PubMed] [Google Scholar]
- 10.Churchill M. Sterjovski J. Gray L, et al. Longitudinal analysis of nef/long terminal repeat-deleted HIV-1 in blood and cerebrospinal fluid of a long-term survivor who developed HIV-associated dementia. J Infect Dis. 2004;190(12):2181–2186. doi: 10.1086/425585. [DOI] [PubMed] [Google Scholar]
- 11.Zeichner SL. Kim JY. Alwine JC. Linker-scanning mutational analysis of the transcriptional activity of the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1991;65(5):2436–2444. doi: 10.1128/jvi.65.5.2436-2444.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henderson AJ. Connor RI. Calame KL. C/EBP activators are required for HIV-1 replication and proviral induction in monocytic cell lines. Immunity. 1996;5(1):91–101. doi: 10.1016/s1074-7613(00)80313-1. [DOI] [PubMed] [Google Scholar]
- 13.Majello B. De Luca P. Hagen G. Suske G. Lania L. Different members of the Sp1 multigene family exert opposite transcriptional regulation of the long terminal repeat of HIV-1. Nucleic Acids Res. 1994;22(23):4914–4921. doi: 10.1093/nar/22.23.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suske G. The Sp-family of transcription factors. Gene. 1999;238(2):291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- 15.Churchill MJ. Wesselingh SL. Cowley D, et al. Extensive astrocyte infection is prominent in human immunodeficiency virus-associated dementia. Ann Neurol. 2009;66(2):253–258. doi: 10.1002/ana.21697. [DOI] [PubMed] [Google Scholar]
- 16.Gorry PR. Ong C. Thorpe J, et al. Astrocyte infection by HIV-1: Mechanisms of restricted virus replication, and role in the pathogenesis of HIV-1-associated dementia. Current HIV Res. 2003;1(4):463–473. doi: 10.2174/1570162033485122. [DOI] [PubMed] [Google Scholar]
- 17.Ong CL. Thorpe JC. Gorry PR, et al. Low TRBP levels support an innate human immunodeficiency virus type 1 resistance in astrocytes by enhancing the PKR antiviral response. J Virol. 2005;79(20):12763–12772. doi: 10.1128/JVI.79.20.12763-12772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez-Scarano F. Martin-Garcia J. The neuropathogenesis of AIDS. Nature Rev Immunol. 2005;5(1):69–81. doi: 10.1038/nri1527. [DOI] [PubMed] [Google Scholar]
- 19.Heaton RK. Franklin DR. Ellis RJ, et al. HIV-associated neurocognitive disorders before and during the era of combination antiretroviral therapy: Differences in rates, nature, and predictors. J Neurovirol. 2011;17(1):3–16. doi: 10.1007/s13365-010-0006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chun TW. Fauci AS. Latent reservoirs of HIV: Obstacles to the eradication of virus. Proc Natl Acad Sci USA. 1999;96(20):10958–10961. doi: 10.1073/pnas.96.20.10958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorry PR. Bristol G. Zack JA, et al. Macrophage tropism of human immunodeficiency virus type 1 isolates from brain and lymphoid tissues predicts neurotropism independent of coreceptor specificity. J Virol. 2001;75(21):10073–10089. doi: 10.1128/JVI.75.21.10073-10089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorry PR. Taylor J. Holm GH, et al. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J Virol. 2002;76(12):6277–6292. doi: 10.1128/JVI.76.12.6277-6292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas ER. Dunfee RL. Stanton J, et al. Macrophage entry mediated by HIV Envs from brain and lymphoid tissues is determined by the capacity to use low CD4 levels and overall efficiency of fusion. Virology. 2007;360(1):105–119. doi: 10.1016/j.virol.2006.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cowley D. Gray LR. Wesselingh SL. Gorry PR. Churchill MJ. Genetic and functional heterogeneity of CNS-derived tat alleles from patients with HIV-associated dementia. J Neurovirol. 2011;17(1):70–81. doi: 10.1007/s13365-010-0002-5. [DOI] [PubMed] [Google Scholar]
- 25.Gray L. Sterjovski J. Ramsland PA. Churchill MJ. Gorry PR. Conformational alterations in the CD4 binding cavity of HIV-1 gp120 influencing gp120-CD4 interactions and fusogenicity of HIV-1 envelopes derived from brain and other tissues. Retrovirology. 2011;8(1):42. doi: 10.1186/1742-4690-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray LR. Gabuzda D. Cowley D, et al. CD4 and MHC class 1 down-modulation activities of nef alleles from brain- and lymphoid tissue-derived primary HIV-1 isolates. J Neurovirol. 2011;17(1):82–91. doi: 10.1007/s13365-010-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gray L. Churchill MJ. Sterjovski J, et al. Phenotype and envelope gene diversity of nef-deleted HIV-1 isolated from long term survivors infected from a single source. Virology J. 2007;4:75. doi: 10.1186/1743-422X-4-75. [DOI] [PMC free article] [PubMed] [Google Scholar]