Summary
C. albicans, an opportunistic human pathogen, grows as yeast, pseudohyphae and true hyphae. These cell types differ both in morphology and in aspects of cell cycle progression. In particular, polarized growth becomes uncoupled from other cell cycle events in hyphal cells. Yeast or pseudohyphae that undergo a cell cycle delay also exhibit polarized growth independent of cell cycle progression. The Spitzenkörper, an organelle also found in filamentous fungi, directs continuous hyphal elongation. A polarisome mediates cell cycle-dependent growth in yeast and pseudohyphae. Regulation of morphogenesis and cell cycle progression depends upon specific cyclins, all of which affect morphogenesis and some of which function specifically in yeast or hyphal cells.
Introduction
C. albicans, the most prevalent human fungal pathogen, causes life-threatening systemic infections in addition to superficial mucosal conditions such as thrush and vaginitis. A normal constituent of the gastrointestinal flora, it causes opportunistic infections, primarily in patients with compromised immunity.
Virulence is thought to require the ability to grow with the full repertoire of vegetative morphologic forms: yeast, pseudohyphae and true hyphae [1,2](Fig. 1). While it is difficult to distinguish the contributions of cell shape from those of gene expression, the observations that elongated hyphae evade or escape phagocytic cells, and that yeast cells disseminate in the bloodstream, suggest that morphology contributes to the survival of C. albicans in the broad range of host niches that it inhabits.
Figure 1.
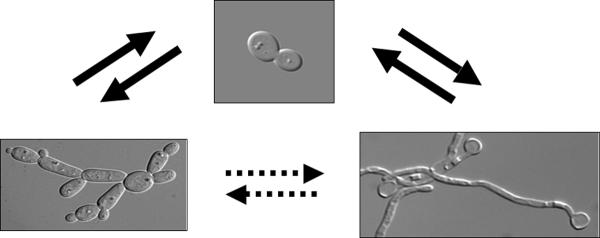
Vegetative morphology of C. albicans cells. Yeast cells (top center) can form both pseudohyphae (lower left) and true hyphae (lower right). Switching between the pseudohyphal and hyphal morphologies is less frequent.
The different morphologies are often treated as different developmental states. In the laboratory, cultures grown at low temperature and pH contain mostly ellipsoid yeast cells. Long, narrow hyphae develop from yeast cells at 37°C and neutral pH and in response to external stimuli such as serum. Elongated pseudohyphal cells develop at intermediate temperatures and pH. Pseudohyphae rarely form true hyphae [3] and hyphae rarely produce pseudohyphal buds (Fig. 1). Furthermore, pseudohyphal cultures always contain some yeast and/or some hyphal cells (Amornrattanapan et al., unpublished). Finally, C. albicans responds to cell cycle arrest by producing a filamentous cell type with properties of both pseudohyphae and true hyphae.
This review will focus on advances in our understanding of how cell cycle progression differs between yeast, pseudohyphae and true hyphae at the cellular and molecular level, highlighting the current view of how cyclins and other proteins regulate cell cycle progression and morphogenesis. It will also discuss changes to cell morphology that occur in response to cell cycle delays.
Cell biology of yeast, pseudohyphae and true hyphae
C. albicans yeast and pseudohyphal cells are very similar to S. cerevisiae yeast and pseudohyphae in shape, size and order of cell cycle events. As in S. cerevisiae [4], changes in actin patch distribution reflect a switch from polarized growth at the tip, to isotropic growth throughout the bud, to polarized deposition of cell wall material required for septation. This switch occurs early in the yeast cell cycle and later in pseudohyphae [5,6] (Finley et al., unpublished).
Yeast cells grow by asymmetric budding, forming smooth, round colonies (Fig. 2A). Septin rings appear prior to bud emergence [7], and nuclei divide across the mother-bud neck [8]. Bud site selection in C. albicans yeast cells is temperature dependent and cultures generally contain a mixture of cells that bud with an axial or bipolar budding pattern [9]. At START, the transition from G1 to S-phase of the cell cycle, bud emergence is coordinated with the onset of DNA replication and spindle pole body duplication [10]. Yeast cells separate after cytokinesis, when daughters have not yet reached the size of their mothers. Daughters enter the next cell cycle slightly later than their mothers, consistent with the idea that a cell size threshold affects the timing of START [6].
Figure 2.
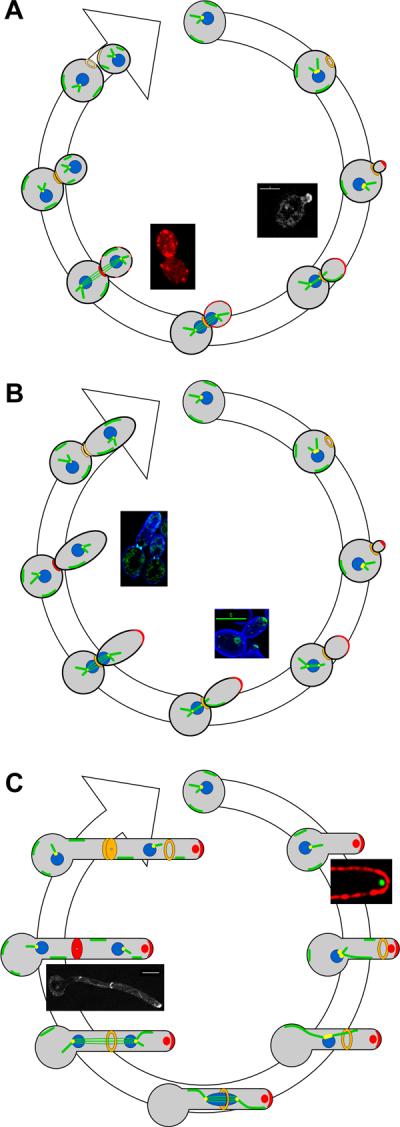
Models for cell cycle progression in yeast, pseudohyphal and hyphal cells.
A. Yeast cells traverse START by forming a septin ring (orange), initiating bud emergence directed by a polarisome (red crescent) and duplicating the spindle pole body (yellow). Growth becomes less polarized as sites of growth (red) become distributed around the bud. In G2 phase the nucleus (blue) moves to the neck assisted by astral microtubule (green) sliding along the cortex and, at anaphase, divides across the neck. At telophase, the spindle disassembles, growth is focused at the neck, the septin ring splits into two and then each ring disappears prior to appearance of the next ring in G1. Right inset, polarisome protein Mlc1p-YFP localizes to the tip during early bud growth. Left inset, delocalized actin (red) patches reflect isotropic growth.
B. Pseudohyphal cells have similar features to yeast cells with a few exceptions: the polarisome persists for longer and cells spend more time in G2 phase, becoming similar in size to mother cells; cells do not separate following cytokinesis. As in yeast cells, sites of growth are cell cycle dependent, leaving the tip and focusing at the bud neck prior to cytokinesis. Right inset, Mlc1p-GFP (green) appears at the tips of small and larger buds. Left inset, at cytokinesis, Mlc1p-GFP disappears from bud tip and localizes to the neck.
C. Upon induction of hyphal growth from a yeast cell, the Spitzenkörper (red circle) directs germ tube evagination, which persists throughout the cell cycle and initiates prior to START. A polarisome is also present at hyphal tips. Nuclei migrate to and divide across the presumptum, and the septin ring persists into the next cell cycle. Right inset, photomicrograph of Spitzenkörper protein Mlc1p-YFP (green); cell surface is labeled with Texas-red conjugated to Concanavalin A. Left inset, during cytokinesis Mlc1p-YFP remains at the growing tip and also appears at the septum.
C. albicans pseudohyphal cells bud in a unipolar pattern (Fig. 2B). Cells remain attached following cytokinesis, forming branched chains of elongated buds and colonies that are fuzzy or rough. Filaments invade the agar below the colony and extend across the agar from the colony edge. As in yeast cells, septin rings form prior to bud emergence and nuclei divide across the neck [8]. Like S. cerevisiae pseudohyphae [6], C. albicans pseudohyphal cells spend more time growing in a polarized manner and remain in G2 longer than do yeast cells (Finley et al., unpublished). Daughters and mothers reach START at a similar size and thus enter the next cell cycle with more synchrony than do yeast cells ([6], Finley et al., unpublished).
Hyphae are narrower than pseudohyphal cells (~2 μm) and have parallel walls with no obvious constriction at the site of septation [11] (Fig. 2C). Checkpoints that coordinate bud growth in S. cerevisiae do not appear to operate in C. albicans hyphae: evagination and elongation of the germ tube is continuous, beginning prior to other START events, continuing during cytokinesis and not responding to changes in Cdc28-K19 phosphorylation [10,12]. When hyphae are induced from yeast cells, a basal septin band, formed by a subset of septins not including Cdc3p and not requiring Gin4p, appears transiently at the mother-germ tube junction [3,9,13]. Septin ring formation, which occurs as the hyphal tip passes the site where the septum will form (the presumptum [14]), is coordinated with other events of START [10,14]. Nuclei migrate into and divide within the germ tube, usually across the presumptum [14].
Vacuole inheritance regulates hyphal branching frequency
Hyphae exhibit a linear growth rate because subapical cells remain quiescent in G1 for several cell cycles prior to branching [15]. This is due to the asymmetric inheritance of vacuoles such that the apical cell receives primarily cytoplasm and the subapical cell receives larger vacuoles [15]. The subapical compartments become competent to branch only when the ratio of vacuolar volume to cell volume decreases [15]. Consistent with the idea that a cytoplasmic volume threshold regulates the passage of START, perturbations of vacuolar inheritance alter branching frequencies [16,17]. It will be interesting to determine if factors such as Cln3p, which in S. cerevisiae regulate the size at which cells commit to START, will also regulate the frequency of hyphal branching.
The Spitzenkorper: a hyphal-specific organelle
In filamentous fungi, the Spitzenkörper, a structure just behind the hyphal tip, mediates growth directionality and hyphal tip morphogenesis by concentrating the delivery of secretory vesicles [18,19]. C. albicans hyphae have a Spitzenkörper as well as a cap-shaped polarisome. In yeast and pseudohyphae a polarisome directs polarized growth in a cell cycle-dependent manner [5] (Fig. 2). Continuous polarized tip growth is associated with the presence of the Spitzenkörper, whereas cell cycle-dependent polarized growth is associated with the presence of the polarisome. Thus, hyphal growth has properties distinct from those of pseudohyphae and C. albicans hyphae resemble the hyphae of filamentous fungi.
Spindle dynamics and nuclear migration
Nuclear and spindle movement, including long distance migration of bipolar spindles in hyphae, occurs by repetitive sliding of astral microtubules along the cell cortex [14] that is mediated primarily by cytoplasmic dynein [20] (Finley et al., unpublished). In contrast, the mother nucleus returns to the mother cell primarily by spindle elongation forces. Furthermore, in hyphae, the timing of anaphase onset is coordinated with hyphal length and/or volume: hyphal length at anaphase onset remains constant in strains with decreased rates of hyphal elongation [14].
Induction of, and commitment to, hyphal growth
In the laboratory, stationary cells that have reached very high cell density (OD600>13 [21]) are most responsive to hyphal and pseudohyphal induction signals. This is due, in part, to release of the cells from exposure to farnesol, a quorum sensing inhibitor of hyphal growth [22]. Other factors, such as levels of available nitrogen likely affect the efficiency of induction as well [15].
A controversy remains regarding whether hyphae can be induced from all cell cycle stages. In classic experiments, Soll and co-workers found that, when released from starvation at 37°C, small budded cells formed hyphae, while large budded cells completed a cell cycle prior to forming hyphae [23]. The critical transition point occurred when buds reached a size at which they normally switch from polarized to isotropic growth [23], suggesting that buds that have switched to isotropic growth cannot form hyphae. In contrast, when Liu and coworkers [10] treated asynchronous yeast cultures with serum at 37°C, large-budded cells formed a tapered extension, which was interpreted as indicating hyphal elongation can be induced at any time in the cell cycle. These cells all had constrictions at the neck and may not have exhibited the hallmarks of true hyphae [11]. Importantly, exposure to serum stimulates cell elongation that is independent of hyphal growth: fkh2 mutants, which are constitutively pseudohyphal, form more polarized buds in the presence of serum than in other hyphal induction conditions [24]. Thus, serum may induce polarized growth, but not true hyphal growth, in large budded cells. This leaves open the attractive model that a cell cycle restriction point, corresponding to the switch to isotropic growth, limits hyphal formation to earlier stages of the cell cycle.
Cell cycle regulators: cyclins, cyclin-dependent kinases and CDC proteins
Although fundamental aspects of cyclin dependent kinase (CDK) activities and substrates are similar across yeast species, the global patterns of transcription for cell cycle genes are very different between S. cerevisiae and C. albicans [25]. Furthermore, several genes that are essential in S. cerevisiae are not required for viability in C. albicans (e.g., CDC4 [26], CDC14 [27], RAS1 [28]). Genes essential in C. albicans but not in S. cerevisiae (e.g, CLB4 [29], CLN3 [30,31]) can be explained by the genome duplication that resulted in many pairs of genes with redundant functions in S. cerevisiae [32].
The G1 cyclins have a very different division of labor in C. albicans than in S. cerevisiae. Ccn1p (formerly termed Cln1p [33]) has similarity to the ScCln3p cyclin box and was isolated by its dominant-negative effect on S. cerevisiae pheromone responses [34]. It is expressed in G1 and early S-phase [10,24] and is required for the maintenance of polarized growth but not for its initiation [35].
Hgc1p (formerly named Cln21p) is most similar to ScCln1p and ScCln2p. It associates with the Cdc28 cyclin-dependent kinase and weakly complements for START activity in S. cerevisiae. Importantly, it is expressed in hyphae and not in yeast cells and is co-regulated with other hyphal specific genes ([36], (Zirbes et al., unpublished). It is necessary, but not sufficient, for hyphal growth. It promotes the maintenance of actin and Spa2p, a polarisome component, at hyphal tips [36]. In addition, Hgc1p is required to inhibit the localization of Cdc14p at the septum [27]. In yeast and pseudohyphae, but not in hyphae, Cdc14p initiates a cascade of events leading to cell separation [27]. Thus, Cdc14p may be a (direct or indirect) target of the Hgc1/Cdc28 CDK [27].
CLN3 (formerly CLN2), the only essential G1 cyclin, is most similar to ScCLN3 and complements S. cerevisiae lacking G1 cyclins [33]. Loss of Cln3p also affects morphogenesis: depletion of Cln3p in yeast cells causes cells to first increase in diameter and then to form hyphae that continue to grow and divide [30,31]. Thus, Cln3p is essential for yeast growth and may be important for size control at G1. The timing of the transition to hyphal growth appears to depend upon the degree to which CLN3 is repressed and, thus, the rate of cell growth prior to the transition [30,31]. This also implies that a size or volume threshold must be crossed to induce this transition to hyphal growth. Interestingly, the levels of Cln3p are reduced in the presence of farnesol, which inhibits hyphal growth, suggesting that Cln3p may modulate cell cycle progression in both yeast and hyphal cells.
Pcl2p is a cyclin homolog that is expressed preferentially in yeast cells [22,25] and that is required for morphogenesis in S. cerevisiae [37]. Accordingly, its levels are increased in the presence of farnesol [22] and decreased in Cln3p-depleted cells that are forming hyphal-like extensions [30]. Given the opposite patterns of PCL2 and HGC1 expression, it is tempting to speculate that they have complementary roles in yeast and hyphal cells. Alternatively, they each may execute very different processes in the two cell types, given that Hgc1p associates with Cdc28 CDK [36] and Pcl2p is predicted to associate with the Pho85 CDK.
C. albicans has only two B-cyclins (homologs of ScCLB2 and ScCLB4), one of which (CLB2, formerly termed CYB1) is essential [29]. Both B-cyclins negatively regulate polarized growth, albeit to different degrees and with very different morphological phenotypes: cells lacking Clb4p (formerly termed Cyb99) grow slowly with a constitutively pseudohyphal morphology; Clb2p-depleted strains arrest in late anaphase with highly elongated cells and divided nuclei connected by long mitotic spindles. They elongate without completing a cell cycle and eventually die [29]. A similar phenotype is seen with cells depleted of Cdc28p, the CDK1 homolog [38]. This implies that, like S. pombe, C. albicans has one major mitotic cyclin, Clb2p, that associates with Cdc28p to mediate cell cycle progression.
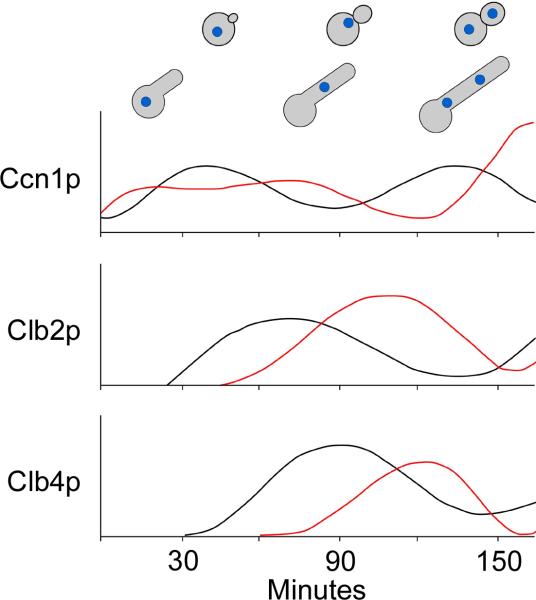
In yeast cells, Ccn1p levels are high in G1 and decline in early G2/M just as Clb2p levels peak [27,29] (Fig. 3). Clb4p levels peak ~15 minutes later, in mid G2/M, and levels of both B-cyclins decline at M-phase when nuclei divide. Interestingly, in hyphae, Ccn1p accumulates earlier and persists longer than Clb2p and Clb4p, which appear at later times that correspond with M-phase and disappear during the exit from mitosis. Thus, the cell cycle is significantly delayed in hyphal cells, especially when one considers that hyphae are growing at higher temperatures than yeast cells. This cell cycle delay also indicates that a G1 cyclin is present for a larger portion of the hyphal cell cycle than in the yeast cell cycle and suggests that the cyclins may have slightly modified roles in hyphae relative to yeast.
Figure 3.
Cell cycle progression and cyclin levels differ in yeast and hyphae. G1-phase yeast daughter cells were synchronized by elutriation and then released into yeast (30°C) or hyphal (37°C, 5% serum) growth conditions. Cell morphology and levels of G1 cyclin Ccn1p, and B-cyclins Clb2p and Clb4p were followed using epitope tagged proteins. Ccn1p levels persisted longer and B-cyclins appeared later in hyphae, relative to yeast. Adapted from Bensen et al. 2005.
There is no obvious difference in the phosphorylation state of Cdc28 Tyr19 phosphorylation between yeast and hyphal cells [10]. This implies that phosphorylation of Cdc28 Tyr19 may not be important for polarized growth in C. albicans and that Swe1p, the ortholog of ScSwe1p and S. pombe Wee1p, a checkpoint kinase that phosphorylates Tyr19 on Cdc28/cdc2, is not required for hyphal growth. Indeed, swe1△/△ cells form normal pseudohyphae and hyphae although swe1△/△ yeast cells are slightly rounder than wild-type cells [3].
Morphogenesis during cell cycle arrest or delay
Conditions that arrest cell cycle progression often result in a polarized growth phenotype (Fig. 4/Box 1) [10,39–41]. For example, treatment of cells with hydroxyurea (HU), which depletes ribonucleotides and thus impedes DNA replication elongation and S-phase, or with nocodazole (NZ), which depolymerizes microtubles and locks cells in mitosis, give rise to cells that continue to elongate despite their inability to divide [40,42]. These cells have some features that are pseudohyphal-like (they are constricted at the neck and >2 μm in width) and others that are hyphal-like (they elongate continuously, nuclei move into the elongating bud, and they eventually express some hyphal specific genes) [42]; however, unlike either cell type, they do not divide and they eventually die. Thus, they represent a terminal phenotype different from either pseudohyphae or true hyphae.
Figure 4/Box 1.
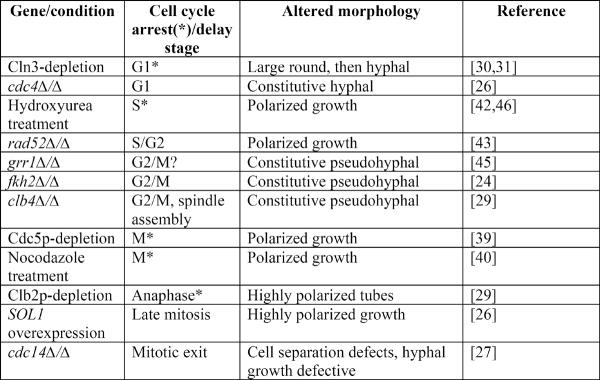
List of cell cycle conditions and mutants that cause changes in morphogenesis. Gene/conditions are ordered by approximate cell cycle stage at which arrest/delay occurs. Essential genes (asterisk) are terminally arrested. G1 arrested cells tend to be more hyphal-like, S/G2 and M arrests tend to be polarized pseudohyphal-like.
While the morphology of arrested cells is similar in cells treated with HU or depleted for Cdc5p [39], the gene expression patterns of the arrested cells have significant differences that reflect the cell cycle stage at which they are arrested [42]. They exhibit common expression of a few cell wall proteins and virulence factor genes (such as CSA2, PHR1 and DDR48) that are also expressed in elongating hyphal cells. Since pseudohyphae express low levels of hyphal specific genes (Amornrattanapan et al., unpublished), this expression pattern is not diagnostic of a specific cell type.
In general, arrest of the cell cycle triggers cell cycle checkpoints: in NZ the polarized growth response requires the Mad2p spindle assembly checkpoint [40] and in Cdc5-depleted cells the polarized growth response requires Bub2p, the mitotic spindle checkpoint [42]. The Swe1p morphogenesis checkpoint partially affects the elongation of HU-treated cells (Finley et al., unpublished) and rad52△/△ cells [43]. While S. cerevisiae Rad53p and Mec1p/Tel1p are required for the elongation of HU-arrested cells [44], the orthologous C. albicans genes have not been tested.
Interestingly, Ras1p is required for polarized growth in response to HU, possibly via a mechanism independent from its role in hyphal signaling [42]. An intriguing question is whether Ras1p has a role in the S-phase checkpoint. In summary, different cell cycle arrest conditions result in different gene expression patterns and trigger different checkpoints. Nonetheless, several arrest conditions result in similar morphologic outcomes. Perhaps the different checkpoints activate a common pathway (related to a pathway that operates in normal hyphal cells) that uncouples polarized growth from other cell cycle events.
Although several types of cell cycle arrest and/or checkpoint activation result in a similar polarized growth phenotype, this is not always the case. Most notably, depletion of Cln3 results in production of large round cells that later form hyphal-like tubes [30,31], suggesting that that arrest in late G1 has a different morphological outcome than does arrest in S, G2 or M phases of the cell cycle.
Polarized growth phenotypes are also seen in strains lacking genes that are not essential (CDC4, CLB4, CLB14, FKH2, GRR1, RAD52 and SOL1) [24,26,27,29,43,45] (Fig4 (Box1)). The shape of cells lacking these genes may be related to the length of the cell cycle delay, and thus to a delay in the switch to isotropic growth. In cases where it has been tested, this polarized growth does not require the Efg1p and Cph1p transcription factors necessary for normal hyphal growth, suggesting that it affects processes downstream of the signaling pathways that modulate Efg1p and Cph1p levels ([29]).
Importantly, the Mad2p spindle assembly checkpoint is required for virulence and polarized growth in the systemic mouse model of candidemia [40]. This suggests that C. albicans cells undergo cell cycle arrest during growth in the animal host and that the response to this arrest is required for survival and successful colonization and/or invasion of host niches. This is not true for all cell cycle checkpoint genes: deletion of SWE1 did not cause a significant decrease in virulence (Gale et al., unpublished). It will be important to determine if other cell cycle checkpoint proteins, such as Bub2p and, potentially, Rad53p, have an effect on virulence.
Conclusion
Hyphae, pseudohyphae and yeast differ from each other in the rate and order of cell cycle events. A major difference is the uncoupling of elongation from other cell cycle events both in hyphal cells as well as under conditions that arrest or delay cell cycle progression. Polarized growth in yeast and pseudohyphae appears to resemble that in S. cerevisiae, whereas polarization in hyphae requires a Spitzenkörper and is more analogous to hyphal growth in filamentous fungi. Regulation of morphogenesis involves cyclins, some of which function specifically in yeast or hyphal cells. The functions of, and relationships between, the different cyclins also appear to have diverged substantially from those of S. cerevisiae.
Acknowledgements
I thank Ken Finley for producing Fig. 2 and Pete Sudbery for providing photomicrographs for Fig. 2. I am grateful to Kelly Bouchonville, Ken Finley, Cheryl Gale, Neil Gow and Pete Sudbery for helpful discussions and for comments on the manuscript. I apologize to the many authors whose work could not be cited because of space limitations. This work was supported by an award from the National Institutes of Health (R01 AI/DE14666).
References
- 1.Gow N, Brown A, Odds F. Fungal morphogenesis and host invasion. Curr Opin Microbiol. 2002;5:366. doi: 10.1016/s1369-5274(02)00338-7. [DOI] [PubMed] [Google Scholar]
- 2.Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous frms of Candida albicans during infection. Eukaryot Cell. 2003;2:1053–1060. doi: 10.1128/EC.2.5.1053-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wightman R, Bates S, Amornrrattanapan P, Sudbery P. In Candida albicans, the Nim1 kinases Gin4 and Hsl1 negatively regulate pseudohypha formation and Gin4 also controls septin organization. J Cell Biol. 2004;164:581–591. doi: 10.1083/jcb.200307176. [DOI] [PMC free article] [PubMed] [Google Scholar]; •This paper describes kinases that act at the septin ring and affect morphogenesis. It also shows that gin4Δ/Δ cells, which are pseudohyphal, cannot form hyphae, while Gin4p-depleted cells, which start in the yeast form, can form hyphae.
- 4.Lew DJ, Reed SI. Cell cycle control of morphogenesis in budding yeast. Curr Opin Genet Dev. 1995;5:17–23. doi: 10.1016/s0959-437x(95)90048-9. [DOI] [PubMed] [Google Scholar]
- 5.Crampin H, Finley KR, Gerami-Nejad M, Court H, Gale CA, Berman J, Sudbery PE. Candida albicans hyphae have a Spitzenkörper that is distinct from the polarisome found in yeast and pseudohyphae. J. Cell Sci. 2005;118:2935–2947. doi: 10.1242/jcs.02414. [DOI] [PubMed] [Google Scholar]; ••This manuscript demonstrates that C. albicans hyphae have a Spitzenörper in addition to a polarisome and that it persists throughout hyphal growth. It shows that components of polarisomes are found primarily in the Spitzenkörper (Mlc1p and Bni1p), while others (Spa2p, Bud6p and Cdc43p) are primarily found in the polarisome.
- 6.Kron SJ, Styles CA, Fink GR. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1994;5:1003–1022. doi: 10.1091/mbc.5.9.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warenda AJ, Konopka JB. Septin function in Candida albicans morphogenesis. Mol. Biol. Cell. 2002;13:2732–2746. doi: 10.1091/mbc.E02-01-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sudbery PE. The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localization. Mol Microbiol. 2001;41:19–31. doi: 10.1046/j.1365-2958.2001.02459.x. [DOI] [PubMed] [Google Scholar]
- 9.Gale CA, Gerami-Nejad M, McClellan M, Vandoninck S, Longtine MS, Berman J. Candida albicans Int1p interacts with the septin ring in yeast and hyphal cells. Mol. Biol. Cell. 2001;12:3538–3549. doi: 10.1091/mbc.12.11.3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazan I, Sepulveda-Becerra M, Liu H. Hyphal elongation is regulated independently of cell cycle in Candida albicans. Mol. Biol. Cell. 2002;13:134–145. doi: 10.1091/mbc.01-03-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]; ••This review describes the characteristics that can be used to distinguish yeast, pseudohyphal and hyphal cells.
- 12.Staebell M, Soll DR. Temporal and spatial differences in cell wall expansion during bud and mycelium formation in Candida albicans. J Gen Microbiol. 1985;131:1467–1480. doi: 10.1099/00221287-131-6-1467. [DOI] [PubMed] [Google Scholar]
- 13.Hausauer DL, Gerami-Nejad M, Kistler-Anderson C, Gale CA. Hyphal guidance and invasive growth in Candida albicans require the Ras-like GTPase Rsr1p and its GTPase-activating protein Bud2p. Eukaryot Cell. 2005;4:1273–1286. doi: 10.1128/EC.4.7.1273-1286.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finley KR, Berman J. Microtubules in Candida albicans hyphae drive nuclear dynamics and connect cell cycle progression to morphogenesis. Eukaryot Cell. 2005;4:1697–1711. doi: 10.1128/EC.4.10.1697-1711.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barelle CJ, Bohula EA, Kron SJ, Wessels D, Soll DR, Schafer A, Brown AJ, Gow NA. Asynchronous cell cycle and asymmetric vacuolar inheritance in true hyphae of Candida albicans. Eukaryot Cell. 2003;2:398–410. doi: 10.1128/EC.2.3.398-410.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]; • •
- 16.Barelle CJ, Richard ML, Gaillardin C, Gow NA, Brown AJ. Candida albicans VAC8 is required for vacuolar inheritance and normal hyphal branching. Eukaryot Cell. 2006;5:359–367. doi: 10.1128/EC.5.2.359-367.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]; •Together with ref. 17, provides evidence that vacuolar volume determines the time of hyphal branching
- 17.Veses V, Casanova M, Murgui A, Dominguez A, Gow NA, Martinez JP. ABG1, a novel and essential Candida albicans gene encoding a vacuolar protein involved in cytokinesis and hyphal branching. Eukaryot Cell. 2005;4:1088–1101. doi: 10.1128/EC.4.6.1088-1101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; •See ref. 16.
- 18.Harris SD, Read ND, Roberson RW, Shaw B, Seiler S, Plamann M, Momany M. Polarisome meets spitzenkorper: microscopy, genetics, and genomics converge. Eukaryot Cell. 2005;4:225–229. doi: 10.1128/EC.4.2.225-229.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; •A nice review of Spitzenkörper studies including classic and modern analyses.
- 19.Virag A, Harris SD. The Spitzenkorper: a molecular perspective. Mycol Res. 2006;110:4–13. doi: 10.1016/j.mycres.2005.09.005. [DOI] [PubMed] [Google Scholar]; •A review of molecular models for Spitzenkörper function in filamentous fungi.
- 20.Martin R, Walther A, Wendland J. Deletion of the dynein heavy-chain gene DYN1 leads to aberrant nuclear positioning and defective hyphal development in Candida albicans. Eukaryot Cell. 2004;3:1574–1588. doi: 10.1128/EC.3.6.1574-1588.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kadosh D, Johnson AD. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol Biol Cell. 2005;16:2903–2912. doi: 10.1091/mbc.E05-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enjalbert B, Whiteway M. Release from quorum-sensing molecules triggers hyphal formation during Candida albicans resumption of growth. Eukaryot Cell. 2005;4:1203–1210. doi: 10.1128/EC.4.7.1203-1210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; •This work documents the effect of farnesol, a quorum sensing factor, on the induction of hyphal growth.
- 23.Soll DR, Herman MA, Staebell MA. The involvement of cell wall expansion in the two modes of mycelium formation of Candida albicans. J Gen Microbiol. 1985;131:2367–2375. doi: 10.1099/00221287-131-9-2367. [DOI] [PubMed] [Google Scholar]
- 24.Bensen ES, Filler SG, Berman J. A forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans. Eukaryot Cell. 2002;1:77–98. doi: 10.1128/EC.1.5.787-798.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ihmels J, Bergmann S, Berman J, Barkai N. Comparative Gene Expression Analysis by a Differential Clustering Approach: Application to the Candida albicans Transcription Program. PLoS Genetics. 2005;1:0380–0393. doi: 10.1371/journal.pgen.0010039. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••An analysis of the global transcription patterns from >200 microarray experiments for C. albicans genes and compares the overall patterns to those in S. cerevisiae. Specific examples of cell cycle and amino acid gene expression patterns are presented.
- 26.Atir-Lande A, Gildor T, Kornitzer D. Role for the SCFCDC4 Ubiquitin Ligase in Candida albicans Morphogenesis. Mol Biol Cell. 2005 doi: 10.1091/mbc.E05-01-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clemente-Blanco A, Gonzalez-Novo A, Machin F, Caballero-Lima D, Aragon L, Sanchez M, de Aldana CR, Jimenez J, Correa-Bordes J. The Cdc14p phosphatase affects late cell-cycle events and morphogenesis in Candida albicans. J Cell Sci. 2006;119:1130–1143. doi: 10.1242/jcs.02820. [DOI] [PubMed] [Google Scholar]; •Demonstrates that Cdc14p in C. albicans differs from S. cerevisiae Cdc14p, that it is required for cell separation and that its presence at the septin ring is inhibited by Hgc1p.
- 28.Feng Q, Summers E, Guo B, Fink G. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J Bacteriol. 1999;181:6339–6346. doi: 10.1128/jb.181.20.6339-6346.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bensen ES, Clemente-Blanco A, Finley KR, Correa-Bordes J, Berman J. The Mitotic Cyclins Clb2p and Clb4p Affect Morphogenesis in Candida albicans. Mol Biol Cell. 2005;16:387–400. doi: 10.1091/mbc.E04-12-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]; •This paper describes the phenotypes and protein levels of the two B-cyclins in C. albicans.
- 30.Bachewich C, Whiteway M. Cyclin Cln3p links G1 progression to hyphal and pseudohyphal development in Candida albicans. Eukaryot Cell. 2005;4:95–102. doi: 10.1128/EC.4.1.95-102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; •Together with ref. 31, this paper describes the phenotypes of cells depleted for C. albicans Cln3p.
- 31.Chapa y Lazo B, Bates S, Sudbery P. The G1 cyclin Cln3 regulates morphogenesis in Candida albicans. Eukaryot Cell. 2005;4:90–94. doi: 10.1128/EC.4.1.90-94.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; •See ref. 30.
- 32.Wolfe K. Evolutionary genomics: yeasts accelerate beyond BLAST. Curr Biol. 2004;14:R392–394. doi: 10.1016/j.cub.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 33.Sherlock G, Bahman AM, Mahal A, Shieh JC, Ferreira M, Rosamond J. Molecular cloning and analysis of CDC28 and cyclin homologues from the human fungal pathogen Candida albicans. Mol Gen Genet. 1994;245:716–723. doi: 10.1007/BF00297278. [DOI] [PubMed] [Google Scholar]
- 34.Whiteway M, Dignard D, Thomas DY. Dominant negative selection of heterologous genes: isolation of Candida albicans genes that interfere with Saccharomyces cerevisiae mating factor-induced cell cycle arrest. Proc Natl Acad Sci U S A. 1992;89:9410–9414. doi: 10.1073/pnas.89.20.9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loeb JD, Sepulveda-Becerra M, Hazan I, Liu H. A G1 cyclin is necessary for maintenance of filamentous growth in Candida albicans. Mol Cell Biol. 1999;19:4019–4027. doi: 10.1128/mcb.19.6.4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng X, Wang Y. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. Embo J. 2004;23:1845–1856. doi: 10.1038/sj.emboj.7600195. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••This paper shows that Hgc1p, a protein most similar to S. cerevisiae G1 cyclins Cln1p and Cln2p, is expressed only in hyphae and is required for hyphal growth.
- 37.Moffat J, Andrews B. Late-G1 cyclin-CDK activity is essential for control of cell morphogenesis in budding yeast. Nat Cell Biol. 2004;6:59–66. doi: 10.1038/ncb1078. [DOI] [PubMed] [Google Scholar]; •Demonstrates that a burst of G1 cyclin activity from Cln1p, Cln2p, Pcl1p and Pcl2p is required for morphogenesis but not for other events in cell cycle progression in S. cerevisiae.
- 38.Umeyama T, Kaneko A, Niimi M, Uehara Y. Repression of CDC28 reduces the expression of the morphology-related transcription factors, Efg1p, Nrg1p, Rbf1p, Rim101p, Fkh2p and Tec1p and induces cell elongation in Candida albicans. Yeast. 2006;23:537–552. doi: 10.1002/yea.1373. [DOI] [PubMed] [Google Scholar]
- 39.Bachewich C, Thomas DY, Whiteway M. Depletion of a Polo-like Kinase in Candida albicans Activates Cyclase-dependent Hyphal-like Growth. Mol Biol Cell. 2003;14:2163–2180. doi: 10.1091/mbc.02-05-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bai C, Ramanan N, Wang YM, Wang Y. Spindle assembly checkpoint component CaMad2p is indispensable for Candida albicans survival and virulence in mice. Mol Microbiol. 2002;45:31–44. doi: 10.1046/j.1365-2958.2002.02995.x. [DOI] [PubMed] [Google Scholar]
- 41.Bedell GW, Werth A, Soll DR. The regulation of nuclear migration and division during synchronous bud formation in released stationary phase cultures of the yeast Candida albicans. Exp Cell Res. 1980;127:103–113. doi: 10.1016/0014-4827(80)90418-8. [DOI] [PubMed] [Google Scholar]
- 42.Bachewich C, Nantel A, Whiteway M. Cell cycle arrest during S or M phase generates polarized growth via distinct signals in Candida albicans. Mol Microbiol. 2005;57:942–959. doi: 10.1111/j.1365-2958.2005.04727.x. [DOI] [PubMed] [Google Scholar]; ••This work shows that cell cycle arrest by HU or by depletion of Cdc5p leads to similar morphologies but different expression patterns that reflect the different cell cycle arrest stages. Expression of a few hyphal specific genes is seen in both conditions, which may reflect genes necessary for cell elongation, a feature shared by arrested and hyphal cells.
- 43.Andaluz E, Ciudad T, Gomez-Raja J, Calderone R, Larriba G. Rad52 depletion in Candida albicans triggers both the DNA-damage checkpoint and filamentation accompanied by but independent of expression of hypha-specific genes. Mol Microbiol. 2006;59:1452–1472. doi: 10.1111/j.1365-2958.2005.05038.x. [DOI] [PubMed] [Google Scholar]; •Demonstrates that cell cycle delays in response to DNA damage result in polarized growth that involves, but does not require, the expression of hyphal-specific genes or the Efg1p and Cph1p transcription factors and that is partially dependent upon Swe1p.
- 44.Jiang YW, Kang CM. Induction of S. cerevisiae filamentous differentiation by slowed DNA synthesis involves Mec1, Rad53 and Swe1 checkpoint proteins. Mol Biol Cell. 2003 doi: 10.1091/mbc.E03-06-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Butler DK, All O, Goffena J, Loveless T, Wilson T, Toenjes KA. The GRR1 gene of Candida albicans is involved in the negative control of pseudohyphal morphogenesis. Fungal Genet Biol. 2006 doi: 10.1016/j.fgb.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 46.Soll DR, Stasi M, Bedell G. The regulation of nuclear migration and division during pseudo- mycelium outgrowth in the dimorphic yeast Candida albicans. Exp Cell Res. 1978;116:207–215. doi: 10.1016/0014-4827(78)90077-0. [DOI] [PubMed] [Google Scholar]