Abstract
Influenza A virus (IAV) infection leads to variable and imperfectly understood pathogenicity. We report that segment 3 of the virus contains a second open reading frame (‘X-ORF’), accessed via ribosomal frameshifting. The frameshift product, termed PA-X, comprises the endonuclease domain of the viral PA protein with a C-terminal domain encoded by the X-ORF and functions to repress cellular gene expression. PA-X also modulates IAV virulence in a mouse infection model, acting to decrease pathogenicity. Loss of PA-X expression leads to changes in the kinetics of the global host response, notably including increases in inflammatory, apoptotic and T-lymphocyte signaling pathways. Thus, we have identified a previously unknown IAV protein that modulates the host response to infection, a finding with important implications for understanding IAV pathogenesis.
Influenza A virus (IAV) is a single-stranded, segmented, negative-sense RNA virus of the Orthomyxoviridae family (1). Its ecology is complex, encompassing diverse host species, including both wild and domesticated birds, as well as several mammalian species, including humans, dogs, horses, and pigs. IAV virulence varies widely depending on virus and host, ranging from completely asymptomatic to nearly 100% lethality, and IAV pandemics over the past 100 years have shown case fatality rates varying from approximately 2% (1918 H1N1) to around 0.05% (2009 H1N1). While some aspects of this variability can be explained, IAV pathogenesis remains imperfectly understood and difficult to predict (2).
IAV genome segment 3 produces a single, unspliced mRNA that encodes a subunit of the viral RNA-dependent RNA polymerase complex (PA). PA provides an RNA-endonuclease activity, contained in an N-terminal domain, that cleaves capped RNA fragments from cellular pre-mRNAs to provide primers for viral transcription (3-5). Ectopic expression of PA from plasmid has been shown to inhibit the accumulation of other co-expressed proteins, a phenomenon proposed to result from proteolytic activity, either non-specific (6), or possibly resulting from degradation of RNA polymerase II (7, 8). A genome-wide survey of IAV synonymous codon usage aimed at identifying RNA packaging signals discovered a highly conserved internal region in segment 3 that did not correlate with any known or predicted RNA structural or functional motifs (9, 10). Here, we present evidence that the reduced synonymous variation in this region reflects a hitherto unrecognized overlapping open reading frame (termed here ‘X-ORF’) that is accessed by ribosomal frameshifting to produce a distinct PA-related polypeptide with a role in host cell shutoff, modulation of host gene expression, and consequently limitation of viral pathogenesis.
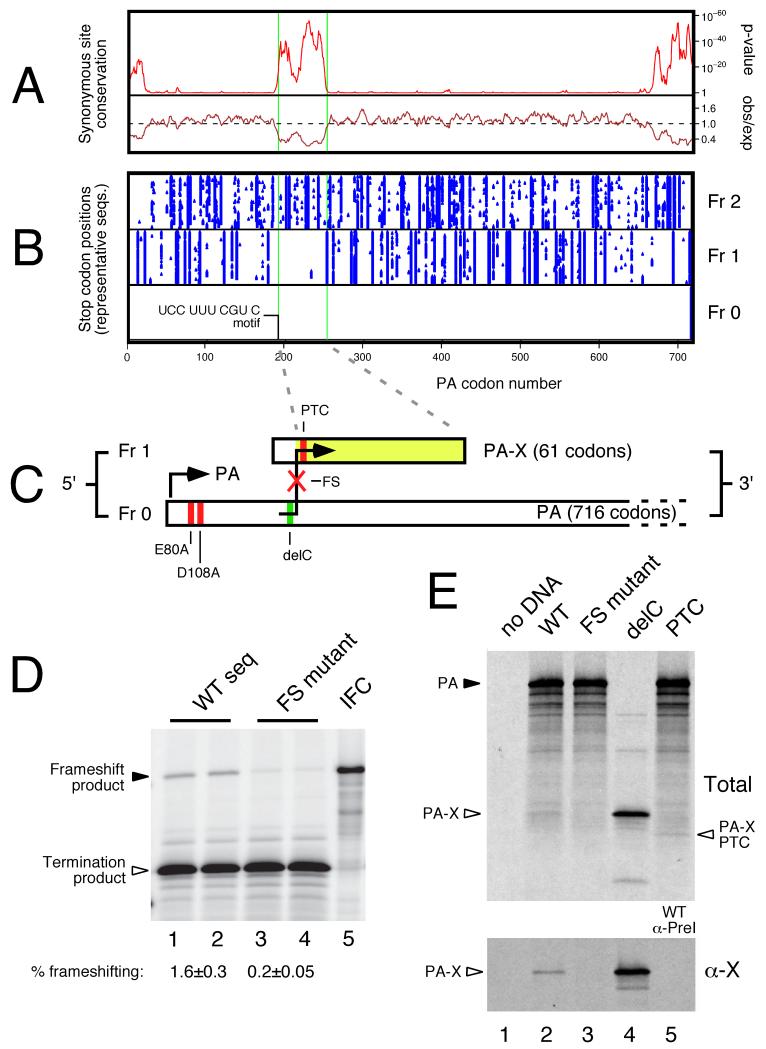
To further investigate the unexplained region of conservation in IAV segment 3 we measured synonymous site conservation in an alignment of >1,000 representative segment 3 sequences. The observed number of synonymous substitutions in a 9-codon sliding window was compared with the number expected under a null model of neutral evolution at synonymous sites (11). This analysis confirmed the presence of the 5′- and 3′-terminal conserved regions involved in genome packaging (10), as well as an additional region of prominent synonymous site conservation between PA codons 190 – 253 (Fig 1A). Furthermore, there was a notable absence of stop codons in the +1 but not the +2 reading frame within this region (Fig 1B), suggestive of an overlapping ORF (X). As noted previously (10), there are no conserved AUG codons that could initiate independent translation of this ORF, nor could we identify any conserved splice donor-acceptor combinations that might allow access to it. However, a highly conserved UCC UUU CGU C motif was identified near the 5′ end of the X-ORF (Table S1), despite the fact that both Ser (UCC) and Arg (CGU) could each in principle be encoded by any of five other codons.
Fig. 1. Characterization of a ribosomal frameshift signal in IAV segment 3.
(A) PA-frame synonymous site conservation in an alignment of 1278 representative full-length PA coding sequences. Shown is the synonymous substitution rate relative to the PA segment average (observed/expected) and the corresponding statistical significance (p-value). (B) Positions of stop codons (blue triangles) in the 3 reading frames in 71 representative PA coding sequences. (C) PA-X ORF structure, showing full-length PA in frame 0 and the X-ORF in frame 1, with experimental mutations indicated. (D, E) Detection of frameshifting in rabbit reticulocyte lysate by SDS-PAGE and autoradiography of IVT reactions. (D) Dual reporter constructs separated by the putative ribosomal frameshifting signal from PR8 segment 3 along with 100 nt 5′- and 3′-flanking sequence. WT: wild-type sequence; FS mutant: shiftsite UUU CGU mutated to UUC AGA; IFC: in-frame control. The calculated frameshifting (FS) efficiencies (mean±SEM, n = 4) are shown. (E) IVT reactions were programmed with full-length segment 3 constructs from the 1918 virus and analyzed before (Total) or after (α-X) immunoprecipitation with rabbit anti-1918 X serum. Lane 5 of the bottom panel shows a WT IVT immunoprecipitated with a preimmune bleed. Polypeptides of interest are indicated.
We hypothesized that +1 ribosomal frameshifting on this motif could lead to expression of X as a fusion with the N-terminal domain of PA. +1 frameshifting can be stimulated by a slow-to-decode codon in the ribosomal A-site, which results in a pause in decoding that may allow a proportion of ribosomes to shift reading frame (12). Frameshifting efficiency depends on the relative speed with which the 0 frame and +1 frame A-site codons can be decoded, the potential for P-site codon:anticodon re-pairing in the +1 reading frame, and the relative stability of the P-site codon:anticodon duplex in the two alternative reading frames, as well as other poorly defined factors that may involve the E-site codon. Thus +1 slippage might occur when UUU is positioned in the ribosomal P-site with the rare codon CGU in the A-site. CGU is one of the most seldom-used codons in both mammalian and avian genomes. Both UUU and UUC are translated by a single tRNAPhe isoacceptor whose anticodon, GAA, has a higher affinity for UUC than for UUU, thus favoring P-site re-pairing to UUC in the +1 frame (13). Such a frameshift event would produce a 29kDa fusion protein, termed PA-X, comprising the N-terminal endonuclease domain of PA (4,5) and, in most isolates, a novel 61 amino acid C-terminus from the X-ORF (Fig 1C). In a minority of isolates, largely of the 2009 pandemic H1N1 lineage, a shorter 41 amino acid X domain is predicted.
The frameshifting hypothesis was tested by cloning the putative frameshift site and flanking sequences from influenza A/Puerto Rico/8/1934 (H1N1) (PR8) segment 3 between Renilla and firefly luciferase ORFs such that a +1 frameshift event would give rise to a fusion product of the two reporter polypeptides (14). These constructs were in vitro translated in rabbit reticulocyte lysates, which demonstrated frameshifting at an efficiency of ~ 1.3% in a wildtype (WT) construct, while mutation of the frameshift motif from UUU CGU to UUC AGA reduced frameshifting to ~ 0.2% (Fig. 1D). Further mutagenesis of the region supported the proposed mechanism of frameshifting stimulated by a combination of the rare (A-site) codon CGU, the (E-site) codon UCC, and possibly more distal elements in the 5′ flanking sequence (Fig S1).
To test if frameshifting occurred in the context of full-length IAV segment 3, we in vitro translated plasmids containing WT and mutant A/Brevig Mission/1/1918 (1918) segment 3. The WT segment produced full-length PA and several smaller polypeptide species, including a ~29 kDa product (Fig 1E, lane 2). This latter species co-migrated with the polypeptide produced from a plasmid in which cytosine 598 (the base predicted to be ‘skipped’ during the frameshift event; see Figs 1C and S2 for details on the 1918 mutations employed) had been deleted to put the X-ORF in frame with the PAN coding region (delC; Fig. 1E, lane 4). This band was much reduced when the frameshift site was mutated (lane 3) while a faster migrating species was seen when a premature termination codon was introduced into the X-ORF (Fig. 1E, lane 5). Next, we raised an antiserum against an X-derived peptide and subjected selected in vitro translation products to immunoprecipitation. Both the delC product and the corresponding species from WT 1918 translation reactions were precipitated by anti-X, but not preimmune antisera (Fig 1E; Fig S3A-C). However, no anti-X-specific product was precipitated from the FS mutant samples. We also detected X-ORF expression in plasmid transfected cells that again was dependent on an intact frameshift site and was sensitive to the introduction of a premature X-ORF stop codon (Fig S3D, E). Thus the X-ORF is accessed by ribosomal frameshifting.
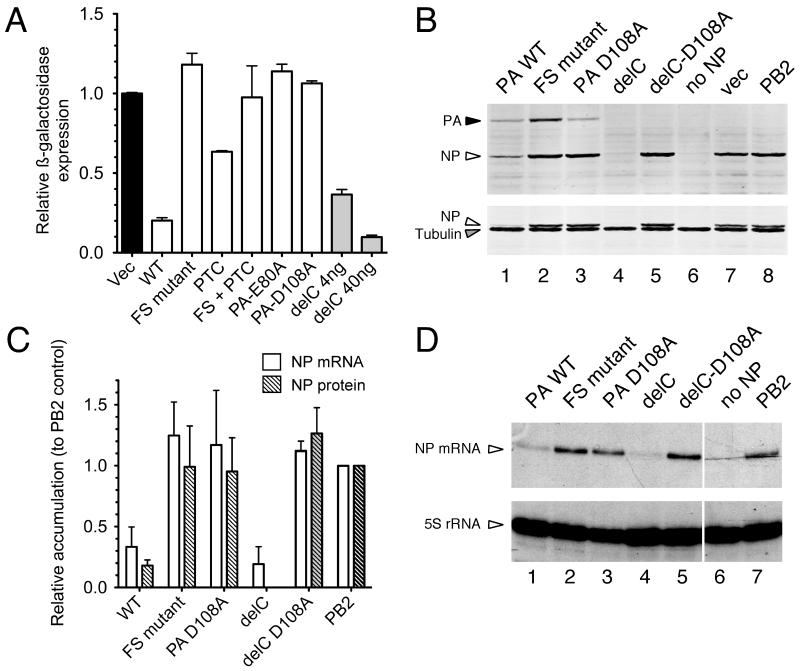
Next, we considered the potential function of the PA-X polypeptide. Since it incorporates the PA endonuclease domain but not the PA C-terminal domain that interacts with the PB1 subunit of the viral polymerase, we hypothesized that it might act independently of the polymerase as an mRNA endonuclease. By analogy to the herpes simplex virus vhs protein (15), PA-X might therefore have a role in host cell shut off; a phenomenon seen in many virus infections in which cellular gene expression is inhibited to hinder the induction of an anti-viral response as well as to divert ribosomes towards translation of viral mRNAs (16). To test this, we used plasmids encoding ß-galactosidase (ß-gal) or the PR8 nucleoprotein (NP) as reporters in co-transfection assays with WT or mutant forms of 1918 segment 3. The WT 1918 segment 3 potently repressed ß-gal expression (Fig 2A), consistent with previous observations (6). However, this activity was abolished by the frameshift mutation and weakened by the insertion of a premature termination codon (PTC) in X; both of these alterations are silent in the PA gene (Fig. S2). Furthermore, the delC construct, which expresses PA-X but not PA, exhibited strong repressive activity (Fig 2A). The same pattern of repression by the delC construct or WT segment 3 was seen when NP was used as the reporter, and again the activity was abolished by the frameshift mutation (Figs 2B, C). The inhibitory activity of segment 3 was originally proposed to result from protease activity (6), whereas our hypothesis predicted an effect on RNA, mediated by the endonuclease activity. To test this, we introduced mutations known to abolish PA endonuclease activity (D108A and E80A) (4). These mutations eliminated the repressive activity on protein expression, whether in the background of otherwise WT segment 3 or in the delC construct (Figs. 2A-C). Furthermore, as predicted by the mRNAse hypothesis, examination of NP reporter gene mRNA levels showed that WT segment 3 and the delC construct reduced mRNA accumulation several fold compared with frameshift-mutant, endonuclease-mutant, and control co-transfections, in a manner that correlated with reporter protein abundance (Figs 2C, D). Overall, these data indicate that the ability of segment 3 to inhibit plasmid-mediated gene expression is a property mediated by PA-X, not PA, and support the hypothesis that repression results from an mRNA endonuclease activity.
Fig. 2. PA-X mediated repression of plasmid-driven gene expression.
293T cells were co-transfected with reporter plasmids encoding (A) ß-galactosidase or (B-D) IAV NP as well as the indicated ‘effector’ plasmids (400 ng unless indicated otherwise). (A) ß-galactosidase accumulation was measured by enzymatic assay. Values plotted are the mean±SEM of 3 experiments normalized to the level seen with an empty plasmid vector as effector. (B) NP, PA and tubulin (as a loading control) accumulation was measured by western blot and (C) quantified from replicate experiments using LiCor Odyssey software. Note in (B) that the same membrane was reprobed for tubulin; residual NP staining is apparent as a doublet running just above tubulin. (C, D) NP mRNA and 5S rRNA (as a loading control) were measured by urea-PAGE and autoradiography of radioactive primer extension assays. (C) NP mRNA accumulation was quantified by densitometry. Values plotted in (C) are the mean±range of two independent experiments, normalized to the control where PB2 was co-transfected in place of PA.
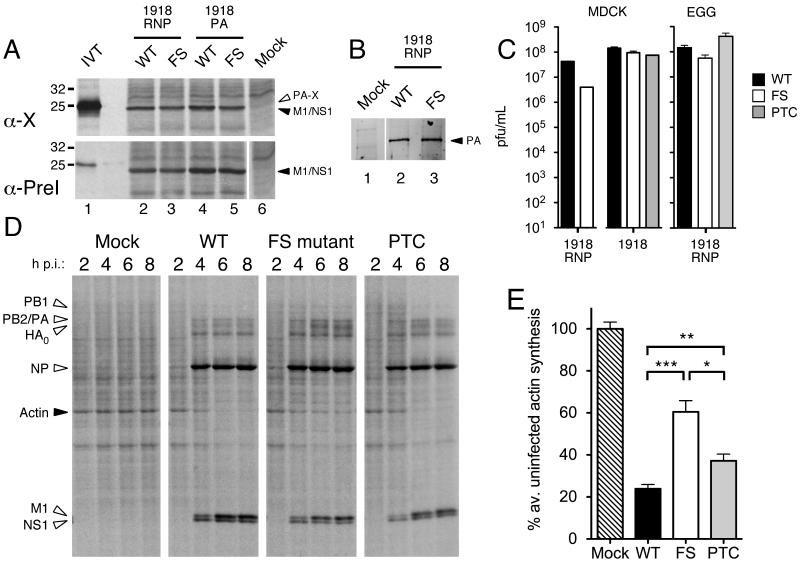
We next sought to determine whether PA-X is expressed during viral infection and what consequence this has for virus replication. Accordingly, we generated sets of viruses based around the 1918 segment 3, with or without mutations affecting PA-X expression: either the fully reconstructed 1918 virus, or reassortants between PR8 and 1918, where either all 1918 ribonucleoprotein genes (segments 1-3 and 5; 1918 RNP) or only 1918 segment 3 (1918 PA) were introduced. To test if PA-X could be detected during infection, radiolabelled cell lysates were prepared from cells infected with parental or frameshift (FS)-mutant viruses containing the 1918 segment 3 and immunoprecipitated with anti-X or preimmune sera. A polypeptide of the expected molecular weight (migrating above a prominent M1/NS1 background band) was precipitated by anti-X but not the preimmune bleed from cells infected with parental but not FS mutant viruses (Fig. 3A), confirming expression of PA-X during virus infection. Western blot analysis confirmed that the synonymous frameshift site mutation did not affect PA accumulation in infected cells (Fig. 3B).
Fig. 3. Characterization of PA-X mutant viruses.
(A) MDCK cells infected with the indicated viruses were metabolically labeled with 35S-methionine from 3-8 HPI. Cell lysates were immunoprecipitated with anti-X or the corresponding preimmune (PreI) serum and bound fractions analysed by SDS-PAGE and autoradiography. Aliquots of in vitro translated PA-X (IVT) were run in parallel as markers. The migration of molecular mass standards are also indicated. (B) Lysates from cells infected with the indicated viruses were analyzed by western blotting for PA. (C) Virus titers from MDCK cells (infected at a multiplicity of infection of ~ 0.001) or embryonated eggs (inoculated with 1000 PFU) were determined by plaque assay. Data are the means±SEM of 2-6 separate inoculations, except for 1918-PTC, which was grown once. (D, E) Lysates from MDCK cells infected with the indicated 1918 RNP viruses and metabolically labeled for 45 minute periods ending at the times shown were (D) analyzed by SDS-PAGE and autoradiography. (E) Actin synthesis at 8 HPI was quantified by densitometry. Data plotted are means±SEM of 3 independent experiments. Two-tailed p-values are indicated: (***) p < 0.0001; (**) p = 0.003; (*) p = 0.01.
To test the effect of the loss of PA-X expression on virus growth, endpoint titers of low multiplicity infections of MDCK cells or embryonated chicken eggs were determined. Frameshift-mutant viruses propagated efficiently in either system, and although the 1918 RNP-FS virus grew to approximately 10-fold lower titers in MDCKs, and somewhat lower titers in eggs (Fig. 3C), the differences were not statistically significant. In addition, no differences were seen when single- or multicycle growth kinetics of the 1918 viruses were tested in MDCK cells (Fig. S4A-B). Next, we tested whether altering PA-X expression had an effect on host-cell shut off. Cells were infected with WT, FS or premature termination codon (PTC) 1918 RNP viruses and protein synthesis monitored across a time course of infection by metabolic labeling. All infections produced abundant quantities of virus polypeptides from 4 hours post infection with the expected pattern of delayed late gene M1 and HA expression (Fig. 3D). In the case of the WT virus, this was accompanied by reduced background cellular protein synthesis from 4-6 h onwards. Host cell shut-off, however, was highly attenuated in cells infected with the frameshift mutant virus and delayed in cells infected with the premature termination codon virus. Densitometric quantification of actin synthesis (the most prominent cellular polypeptide in these experiments) at 8 h confirmed the significantly reduced ability of the PA-X mutant viruses to inhibit cellular gene expression (Figs. 3E, S4C), supporting the hypothesis that PA-X has a function in host-cell shut off.
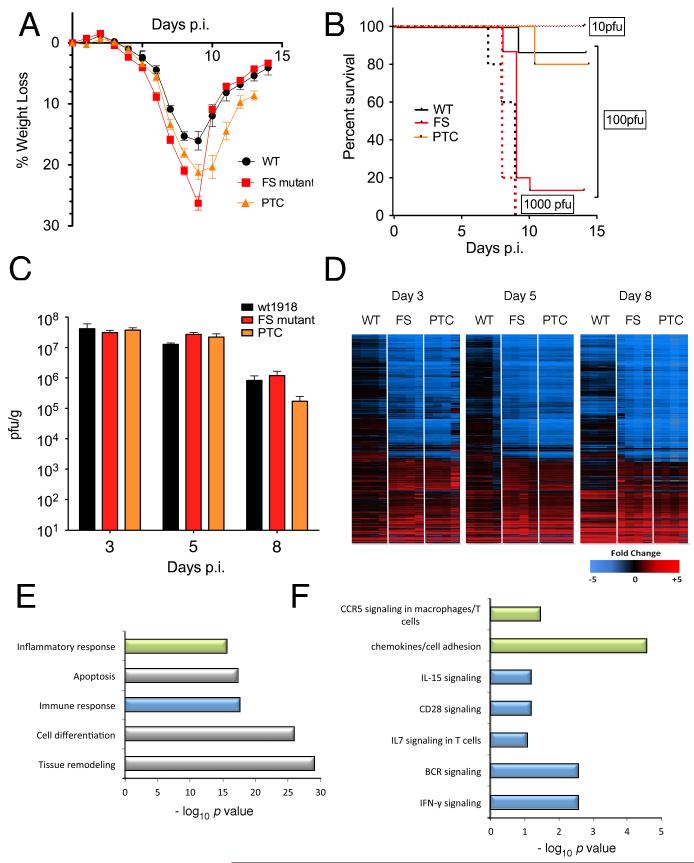
We next investigated the impact of mutating PA-X expression on pathogenesis in an experimental mouse model of influenza infection, using the fully reconstructed 1918 pandemic virus. The 1918 virus background was chosen to evaluate changes in pathogenicity because it induces clinical symptoms in mice without prior adaptation. Additionally, recent data suggest that the current human population may be protected from the 1918 H1N1 virus because of prior infection by, or immunization against, the 2009 H1N1 pandemic virus (17, 18). Because PA-X is a conserved feature of IAV, and because PA-X-deficient viruses exhibited decreased host-cell shut off capacity in vitro, it was expected that such viruses would be attenuated in vivo. 100 plaque forming units (PFU) of 1918-WT, -FS or -PTC viruses were inoculated intranasally into BALB/c mice and daily weights taken to follow the clinical course of infection in an ABSL3+ Select Agent-approved laboratory (in accordance with the select agent guidelines of the National Institutes of Health and Centers for Disease Control and Prevention and under the supervision of the NIH Select Agent Program and the NIH Department of Health and Safety). 1918-FS-infected mice displayed more weight loss compared with 1918-WT-infected mice at a dose of 100 PFU (p<0.0001 at 8 days post infection (DPI)), while 1918-PTC-infected mice displayed an intermediate disease severity phenotype (p = 0.008 versus WT, p = 0.007 versus FS, at 8 DPI) (Fig. 4A). While no survival differences were observed between 1918-WT and 1918-FS at doses of 10 PFU and 1000 PFU, survival of 1918-FS-infected mice at a dose of 100 PFU was significantly decreased compared with WT (p < 0.0001) (Fig. 4B). This difference corresponded with a < 1 log decrease in the calculated 50% mouse lethal dose (MLD50), from 102.4 PFU for the 1918-WT virus to 101.6 PFU for the 1918-FS virus. Virus-infected mice were humanely euthanized on 3, 5, and 8 days post inoculation and lungs harvested for virus titration and histopathological examination. Viral lung replication was similar among 1918-WT, 1918-FS, and 1918-PTC infected mice at these 3 timepoints (Fig 4C). Similarly, there was no apparent difference in the histopathologic changes, nature of the inflammatory infiltrate, or viral antigen distribution among the viruses (Fig. S5).
Fig. 4. Mutation of PA-X affects IAV pathogenicity in a murine model.
(A) Mean weight loss of mice infected with 100 pfu of 1918-WT, -FS, and –PTC viruses (n=15, 15, and 10 respectively). Mice were humanely euthanized when they lost ≥ 25% of their initial body weight; no mice were found dead. Error bars represent ±SEM. (B) Survival proportions of mice infected with 10 (dotted line), 100 (solid line), or 1000 (dashed line) pfu of 1918-WT, -FS, and -PTC viruses. Data for WT and FS in (A) and (B) are pooled results obtained with two independently rescued sets of viruses, one set of which was subjected to high-throughput, full genome sequencing to confirm that the only changes between WT and FS were those deliberately introduced in PA. (C) Mean infectious titers of 1918 viruses in homogenized mouse lungs. Error bars represent the SEM. (D) Expression profile heatmaps generated from whole-lung RNA isolated from mice euthanized at 3, 5, and 8 DPI (n=4 per virus, per timepoint), displaying sequences that showed a ≥ 2-fold difference from mock-infected mouse lungs in at least one experimental group. Each column represents data from an individual experiment, while rows represent unique sequences; row-gene identities are preserved across timepoints. Genes in red are upregulated, those in blue are downregulated, and those in black do not differ as compared to a pool of RNA isolated from 6 mock-infected control mice. (E, F) Gene ontology analysis of sequences whose expression levels differed significantly (p < 0.01, ≥2-fold expression level difference) between 1918-WT and -FS infected lung tissue at 3 DPI with 100 pfu. (E) Top-scoring functional categories. (F) Top pathways within color-coded functional categories.
Global transcriptional profiling was also performed at 3, 5 and 8 days post inoculation to determine if changes in PA-X expression affected the character of the host response compared with a non-lethal dose of the WT 1918 influenza virus infection. Gene expression analysis was performed on RNA isolated from the lungs of individual infected mice (4 per group) and compared to a pool of RNA isolated from control animals by oligonucleotide microarray. “Heat map” profiles of sequences showing a 2-fold or higher change in expression relative to mock controls of the individual animals showed that at each time-point, at a dose of 100 PFU, 1918-FS and -PTC viruses induced a host transcriptional response different from the 1918-WT virus, particularly at earlier timepoints (Fig 4D). However, comparison of the host response to 1918-FS and 1918-PTC viral infection showed a high degree of similarity to mice infected with a five-fold higher, lethal dose of the 1918-WT virus (Fig S6). Thus, mutation of PA-X in the 1918-FS and 1918-PTC viruses changed the kinetics and magnitude of the host response in infected mice.
While a subset of cellular genes were expressed more highly in animals infected with the mutant viruses (especially at earlier time points), in general, there was a significant reduction of cellular gene expression in the lungs of mice infected with PA-X deficient 1918 viruses, similar to that previously reported for lethal infections with the 1918-WT virus (19). Further analysis of sequences showing differential expression between 1918-WT and 1918-FS / -PTC viruses was therefore done to identify potential cellular pathways associated with the moderately increased pathogenicity of the mutant viruses. Standard t-tests identified approximately 5000 genes showing significantly different expression levels (at least 2-fold difference in median expression level, p < 0.01) between 1918-WT and either 1918-FS or -PTC viruses at each time-point (Fig S7). Significantly, while genetically distinct strategies were utilized to compromise PA-X expression in the 1918-FS and -PTC mutant viruses, the host response to these two viruses was similar. Many genes identified as differentially regulated in 1918-FS as compared to 1918-WT were also differentially regulated in 1918-PTC (Fig S7). Fewer differences in gene expression were observed between 1918-FS and -PTC infected animals, further emphasizing the similarity in the response of host gene expression to these viruses. Moreover, in concordance with global expression data (Fig S6), the 1918-FS and 1918-PTC infected animals were more similar to a lethal 500 PFU dose of 1918-WT at 8 days post-infection than to the comparable 100 PFU 1918-WT infected animals (Fig S7).
Gene ontology analysis indicated that sequences showing differential expression between 1918-WT and 1918-FS infection were associated predominantly with inflammation/immune response, apoptosis, cell differentiation and tissue remodeling (Fig. 4E). Interestingly, the majority of differentially regulated immune response/inflammation sequences were related to lymphocyte activation/proliferation or other aspects of cell-mediated immunity, including IFN-γ, CCR5, CD28, IL7 and IL15 signaling (Fig 4F). Many of these genes were more highly induced in the 1918-FS and -PTC infected animals relative to 1918-WT-infected animals. Many of the differentially-regulated apoptosis-related genes are also functionally related to lymphocytes, including Fas-pathway signaling and lymphotoxins, similar to previous high dose 1918 studies (19). Additionally, several MHC class I-associated genes showed upregulation earlier in 1918-FS (by DPI 3) compared to 1918-WT (DPI 8) (Fig S8). By contrast, sequences involved in cell adhesion, including many integrins and extracellular matrix components, showed decreased expression during infection with 1918-FS and -PTC viruses. Collectively, these data support the hypothesis that PA-X plays a role in modulating host gene expression during infection with the 1918 pandemic virus.
The data presented here, derived from genomic, biochemical, functional, and in vivo studies, demonstrate the existence of a thirteenth protein, termed PA-X, in the IAV proteome. PA-X is a fusion protein incorporating the N-terminal endonuclease domain of the PA protein with a short C-terminal domain encoded by an overlapping ORF (‘X’) in segment 3 that is accessed by +1 ribosomal frameshifting. Ribosomal frameshifting has not been previously reported in IAV. The proteins PB1-N40 (20) and PB1-F2 (21) are also recent discoveries in the IAV proteome and interestingly, the X-ORF exhibits greater codon-level conservation than does PB1-F2, which is also expressed from an overlapping ORF (10, 22, 23). We show functional data primarily for the 1918 pandemic virus, but based on the conserved nature of the frameshift sequence (98%; Table S1) and the prevalence of the downstream X-ORF (>99.5% of IAV genomes contain either 61- or 41-codon X-ORFs), as well as its codon-level conservation (Fig. 1A), we predict that expression of PA-X is a nearly universal feature of IAV. Interestingly, there are lineage-specific differences in the distribution of X-ORF lengths. The ~75% of sequenced isolates that possess a 61-codon X-ORF represent virtually all host species and HA/NA subtypes, while the ~25% of isolates with a 41-codon X-ORF are overwhelmingly from the 2009 H1N1 swine-origin pandemic virus plus, a minority subset of swine H3N2 and swine H1N2 viruses, the parental source of the 2009 pandemic segment 3 (24). Functional analyses of the differences in these PA-X variants will require additional experimentation. While the biochemical properties of PA-X (including the function(s) of the X-domain) await a full characterization, co-transfection reporter assays suggest that PA-X can act as an mRNAse by virtue of the PAN endonuclease domain, in a manner evocative of the HSV vhs protein (15). Thus the substrate range and specificity of PA-X will be particularly interesting areas for further research.
PA-X expression was not required for viral replication, and indeed, by virtue of its expression mechanism, only low levels of PA-X were expressed during WT viral infection. However, PA-X deficient viruses differed from the WT counterpart in their ability to cause host cell shut off, and moreover caused greater clinical disease in a mouse model of IAV infection, an outcome related to an accelerated host response as assessed by microarray. PA-X is thus an accessory IAV protein that plays a consequential role at the virus-host interface. We hypothesise that defective control of host gene expression by the mutant viruses in the minority of infected lung cells provokes an altered cascade of host responses from the majority of uninfected cells. The nature of these host gene expression changes, including marked early over-expression of MHC class I genes in 1918-FS or PTC infections, compared with 1918-WT infections, suggests that these perturbations in host response pathways affect lymphocyte activation and immune cell function that lead to an immunopathogenic inflammatory response (19). This may explain the lack of significant differences in weight loss between 1918-WT and 1918-FS/PTC viruses until 5-8 dpi, which coincides with the appearance of influenza-specific CTLs (25).
Taken together, these data contribute significantly to our understanding of IAV replication and pathogenesis, and further suggest promising lines of inquiry into the anti-IAV immune response as well as the factors driving IAV evolution. It is significant that the outcome of infection with PA-X-null viruses was altered in the absence of differences in viral replication, as this suggests that host immunopathology is of central importance in determining the character of disease and could therefore be a fruitful target for new therapeutics aimed at ameliorating severe IAV illness (19, 26).
One Sentence Summary: An influenza A virus protein that modulates the host response.
Supplementary Material
Acknowledgments
AEF is supported by the Wellcome Trust (088789). PD was supported by the UK Medical Research Council (G0700815) and Wellcome Trust (073126). This work was supported in part by the intramural funds of the NIH and the NIAID. JFA is supported by Science Foundation Ireland (08/IN.1/B1889). K.-A.W. and A.O. were funded by Defense Threat Reduction Agency contract HDTRA-1-08-C-0023 and the Luxembourg Centre for Systems Biomedicine and the University of Luxembourg. GLB and AL were supported by studentships from the UK Biotechnology and Biological Sciences Research Council and the Cambridge Infectious Disease Consortium respectively. We thank the Comparative Medicine Branch (NIAID, NIH) for assistance with animal studies and Michael Howard (Utah) for the pDluc variant of the dual luciferase vector. We also thank a number of colleagues for helpful discussion, including Julia Gog and Laurence Tiley (University of Cambridge), Wendy Barclay (Imperial College London), Yizhi Jane Tao (Rice University), Jeffrey I. Cohen, Kanta Subbarao, Kathryn C. Zoon, Deborah C. Wilson, Michael M. Gottesman, Richard Wyatt, and Henry Metzger (NIH). BWJ, PD, and JKT are further thankful for the support of the NIH-Oxford/Cambridge Research Scholars program.
Footnotes
Materials and Methods
Figs. S1-S8
Table S1
References (27-33) [Note: The numbers refer to any additional references cited only within the Supplementary Materials]
References
- 1.Palese S, M.L. P. In: Fields Virology. Knipe DM, Howley PM, editors. Lippincott Williams & Wilkins, Philadelphia; 2007. p. 1647. [Google Scholar]
- 2.Taubenberger JK, Kash JC. Influenza virus evolution, host adaptation, and pandemic formation. Cell host & microbe. 2010;7:440. doi: 10.1016/j.chom.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hara K, Schmidt FI, Crow M, Brownlee GG. Amino acid residues in the N-terminal region of the PA subunit of influenza A virus RNA polymerase play a critical role in protein stability, endonuclease activity, cap binding, and virion RNA promoter binding. J Virol. 2006;80:7789. doi: 10.1128/JVI.00600-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuan P, et al. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature. 2009;458:909. doi: 10.1038/nature07720. [DOI] [PubMed] [Google Scholar]
- 5.Dias A, et al. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature. 2009;458:914. doi: 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- 6.Sanz-Ezquerro JJ, de la Luna S, Ortin J, Nieto A. Individual expression of influenza virus PA protein induces degradation of coexpressed proteins. J Virol. 1995;69:2420. doi: 10.1128/jvi.69.4.2420-2426.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodriguez A, Perez-Gonzalez A, Nieto A. Influenza virus infection causes specific degradation of the largest subunit of cellular RNA polymerase II. J Virol. 2007;81:5315. doi: 10.1128/JVI.02129-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vreede FT, Chan AY, Sharps J, Fodor E. Mechanisms and functional implications of the degradation of host RNA polymerase II in influenza virus infected cells. Virology. 2010;396:125. doi: 10.1016/j.virol.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hutchinson EC, von Kirchbach JC, Gog JR, Digard P. Genome packaging in influenza A virus. J Gen Virol. 2010;91:313. doi: 10.1099/vir.0.017608-0. [DOI] [PubMed] [Google Scholar]
- 10.Gog JR, et al. Codon conservation in the influenza A virus genome defines RNA packaging signals. Nucleic Acids Res. 2007;35:1897. doi: 10.1093/nar/gkm087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Firth AE, Wills NM, Gesteland RF, Atkins JF. Stimulation of stop codon readthrough: frequent presence of an extended 3′ RNA structural element. Nucleic acids research. 2011;39:6679. doi: 10.1093/nar/gkr224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atkins JF, Gesteland RF. Recoding: Expansion of Decoding Rules Enriches Gene Expression. ed. 1 Springer; Heidelberg: 2010. [Google Scholar]
- 13.Eisinger J, Feuer B, Yamane T. Codon-anticodon binding in tRNAphe. Nat New Biol. 1971;231:126. doi: 10.1038/newbio231126a0. [DOI] [PubMed] [Google Scholar]
- 14.Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. A dual-luciferase reporter system for studying recoding signals. Rna. 1998;4:479. [PMC free article] [PubMed] [Google Scholar]
- 15.Smiley JR. Herpes simplex virus virion host shutoff protein: immune evasion mediated by a viral RNase? Journal of virology. 2004;78:1063. doi: 10.1128/JVI.78.3.1063-1068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber F, Haller O. Viral suppression of the interferon system. Biochimie. 2007;89:836. doi: 10.1016/j.biochi.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Easterbrook JD, et al. Immunization with 1976 swine H1N1- or 2009 pandemic H1N1-inactivated vaccines protects mice from a lethal 1918 influenza infection. Influenza and other respiratory viruses. 2011;5:198. doi: 10.1111/j.1750-2659.2010.00191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manicassamy B, et al. Protection of mice against lethal challenge with 2009 H1N1 influenza A virus by 1918-like and classical swine H1N1 based vaccines. PLoS Pathog. 2009;6:e1000745. doi: 10.1371/journal.ppat.1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kash JC, et al. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature. 2006;443:578. doi: 10.1038/nature05181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wise HM, et al. A complicated message: Identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J Virol. 2009;83:8021. doi: 10.1128/JVI.00826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen W, et al. A novel influenza A virus mitochondrial protein that induces cell death. Nat Med. 2001;7:1306. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- 22.Holmes EC, Lipman DJ, Zamarin D, Yewdell JW. Comment on “Large-scale sequence analysis of avian influenza isolates”. Science. 2006;313:1573. doi: 10.1126/science.1131729. author reply 1573. [DOI] [PubMed] [Google Scholar]
- 23.Zell R, et al. Prevalence of PB1-F2 of influenza A viruses. J Gen Virol. 2007;88:536. doi: 10.1099/vir.0.82378-0. [DOI] [PubMed] [Google Scholar]
- 24.Garten RJ, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valkenburg SA, et al. Immunity to seasonal and pandemic influenza A viruses. Microbes and infection / Institut Pasteur. 2011;13:489. doi: 10.1016/j.micinf.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walsh KB, et al. Suppression of cytokine storm with a sphingosine analog provides protection against pathogenic influenza virus. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12018. doi: 10.1073/pnas.1107024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Amorim MJ, et al. A Rab11- and microtubule-dependent mechanism for cytoplasmic transport of influenza A virus viral RNA. Journal of virology. 2011;85:4143. doi: 10.1128/JVI.02606-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bruce EA, et al. Budding of filamentous and non-filamentous influenza A virus occurs via a VPS4 and VPS28-independent pathway. Virology. 2009;390:268. doi: 10.1016/j.virol.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 30.Carrasco M, Amorim MJ, Digard P. Lipid raft-dependent targeting of the influenza A virus nucleoprotein to the apical plasma membrane. Traffic. 2004;5:979. doi: 10.1111/j.1600-0854.2004.00237.x. [DOI] [PubMed] [Google Scholar]
- 31.Tumpey TM, et al. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science. 2005;310:77. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 32.Digard P, Blok VC, Inglis SC. Complex formation between influenza virus polymerase proteins expressed in Xenopus oocytes. Virology. 1989;171:162. doi: 10.1016/0042-6822(89)90523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noton SL, et al. Identification of the domains of the influenza A virus M1 matrix protein required for NP binding, oligomerization and incorporation into virions. J Gen Virol. 2007;88:2280. doi: 10.1099/vir.0.82809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.