Abstract
The discovery of the ability to induce somatic cells to a pluripotent state through the overexpression of specific transcription factors has the potential to transform the ways in which pharmaceutical agents and cellular transplantation therapies are developed. Proper utilization of the technology to generate induced pluripotent stem cells (iPSCs) requires that researchers select the appropriate reprogramming method for generating iPSCs so that the resulting iPSCs can be transitioned towards clinical applications effectively. This article reviews all of the currently available reprogramming techniques with a focus on critiquing them on the basis of their utility in translational medicine.
Keywords: INDUCED PLURIPOTENT STEM CELLS, REPROGRAMMING METHODS, TRANSLATIONAL MEDICINE
The discovery that somatic cells have the capacity to be reprogrammed to pluripotent stem cells by the overexpression of the appropriate set of transcription factors has the potential to indelibly alter the way we approach drug development screens and cellular replacement therapies. This technology provides a pathway for generating previously inaccessible cells in order to conduct large-scale drug screens that focus on physiologically relevant cell types. Differentiation of patient specific induced pluripotent stem cells (iPSCs) to the appropriate cell types also facilitates cellular replacement therapies for diseases, which affect discrete populations of cells. In addition to serving as a nearly limitless source for differentiated cell types, patient specific iPSCs will bypass issues related to immune rejection of transplants from allogeneic sources. Before this technology reaches a mature stage, significant advances need to be made in cellular differentiation protocols and universal standards have to be adapted for the generation of iPSCs that are suitable for translational medicine. This article will review the methods currently available for reprogramming somatic cells to iPSCs with a focus on critiquing methods based on their utility in translational studies.
Figure 1 outlines the typical steps in a reprogramming experiment beginning with tissue selection, proceeding through iPSC generation and possible transgene excision to produce iPSC cells that are ready for use in a translational setting. Tissue selection needs to be made with a view towards what tissues are available and with the knowledge of how successfully that tissue has been reprogrammed with the various available methods. We believe that it is particularly important if the reprogramming method has been validated for peripheral and cord blood because the ease of obtaining blood and the growing presence of cord blood banks is likely to make these tissues readily available [Haase et al., 2009; Staerk et al., 2010].
Fig. 1.
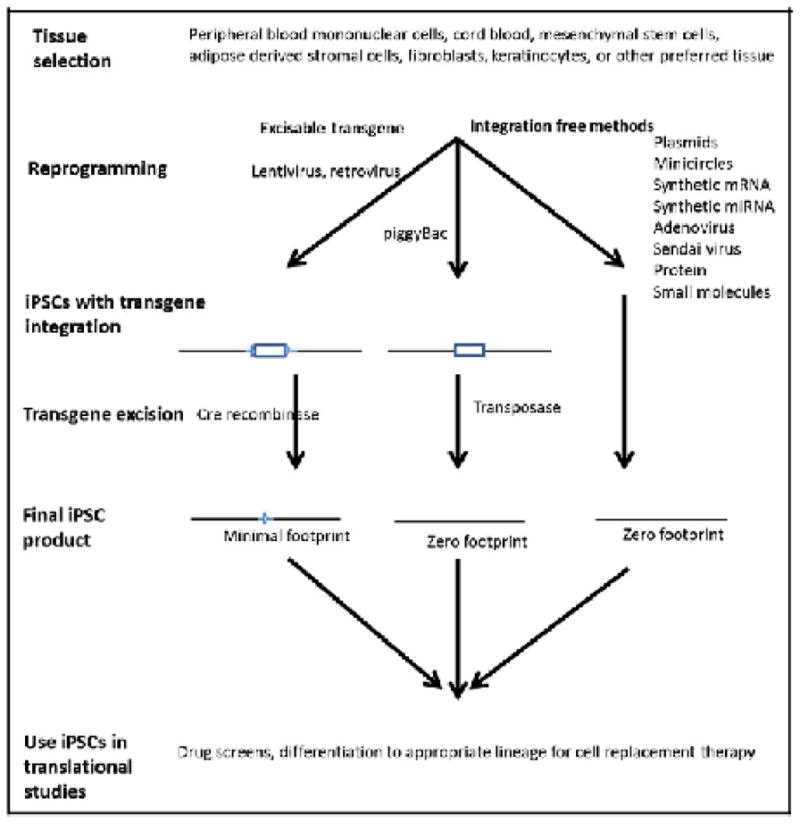
The process of generating translational grade iPSCs. The tissue of choice is selected to reprogram, by excisable or integration free methods to generate minimal- or zero-footprints iPSCs which are then ready for high throughput drug screens and/or sources for the derivation of cells for cellular transplantation therpies.
Additional factors that should be considered before beginning a reprogramming experiment include the “footprint” that a particular method will deposit in the reprogrammed cell type, efficiency of the reprogramming method, validation of the method in multiple somatic cell populations, the capacity of the laboratory to easily implement the method, and outstanding intellectual property issues regarding a particular technology used to generate iPSCs with clinical and/or commercial value (Table I). If the goal of a translational project is to develop cellular replacement therapy, then iPSCs should have a zero-footprint with no residual transgene sequences of the reprogramming vectors in the final iPSC product. Researchers can use less stringent standards if they will only be using iPSCs for drug screening, although there is always the possibility that the presence of exogenous sequences in the iPSCs could affect the results of the drug screen. Reprogramming efficiency can be an important consideration if the availability and quantity of somatic tissue to be reprogrammed is a limiting factor. Another factor to examine for labs whose area of expertise is not in iPSC generation is whether the technique has been found to succeed in multiple labs and a variety of starting somatic cell types. It should also be noted that not all methods are comparable in cost and some require an extensive commitment of labor as well as specialized technical skills. A final matter to be aware of with regard to any projects with a goal of developing patient therapies is the intellectual property landscape regarding any products, which are used to generate goods with potential commercial value. This issue must be addressed in translational research studies before investigators embark on their projects. Failure to resolve this issue can lead to considerable delays and possible abandonment of the project. However, if investigators diligently plan translational projects from the outset with the issues mentioned above in mind they should be able to select a reprogramming method that will help them meet their research goals.
TABLE I.
Pros and Cons of Various Reprogramming Methods in the Context of Translational Use of iPSCs
| Method | Cons | |
|---|---|---|
| Retro- and Lentivirus | Good efficiency, easy to implement, validated for multiple cell types | Even with excisable vector there is a small footprint retained in reprogrammed cells |
| Lentiviral (miRNA) | Very high efficiency | Transgene integration leaves footprint and validated for only one cell type |
| miRNA (direct transfection) | Zero footprint | Low efficiency, and validated for only one cell type |
| Adenoviral | Zero footprint | Low efficiency, validated for only one cell type, and technically challenging |
| Sendai virus | Zero footprint, Good efficiency, validated for multiple cell types, and reprogramming factor viral extracts available commercially | Cost if purchased commercially, if virus is generated by researcher the method is technically challenging, and licensing/patent issues may exist |
| mRNA | Zero footprint, high efficiency, and mRNAs for reprogramming factors available commercially | Cost if purchased commercially, technically challenging if mRNA generated by researcher, labor intensive, and published work only on fibroblasts |
| Protein | Zero footprint | Low efficiency, technically challenging, long time to reprogram, and published work only on fibroblasts |
| Episomal | Zero footprint, good efficiency for most cell types, and validated for multiple cell types | Some cell types (fibroblasts) reprogram with low efficiency |
| PiggyBac | Zero footprint and good efficiency | Published work on only on fibroblasts, no published data demonstrating excision of transposon from human iPSCs, and licensing/patent issues may exist |
| Minicircles | Zero footprint | Low efficiency and validated for only one cell type |
REPROGRAMMING METHODS
iPSC reprogramming was originally discovered by the over-expression of four transcription factors (Oct4, Sox2, Klf4, and c-myc)—later dubbed the “Yamanaka factors”—with a retroviral delivery system in mouse and human fibroblasts [Takahashi and Yamanaka, 2006; Takahashi et al., 2007]. The great disadvantage of this original reprogramming method from a translational perspective is that the reprogramming vectors are integrated into an infected cell’s genome. Since Yamanaka’s original breakthrough, many new methods have been developed to make iPSCs (compared in Table II). Subsequent projects with translation as an objective (especially those which are looking at cellular replacement therapy) should aim to minimize the footprint by adapting these newer reprogramming methods. Minimal footprint methods in which the integrated reprogramming factors can be excised have been developed. These methods use retro- and lentiviral vectors with loxP sites that serve as substrates for Cre-mediated excision of most of the integrated transgene. Zero-footprint methods include reprogramming by episomal vectors, Sendai virus, adenovirus, minicircles, piggyBac, direct miRNA transfection, and mRNA and protein overexpression of reprogramming factors. The reprogramming efficiencies of all these methods can differ over many orders of magnitude with adenoviral delivery of the Yamanka factors yielding an efficiency of only 0.0002% and lentiviral infection of miRNAs promoting reprogramming yielding an efficiency of 10% [Zhou and Freed, 2009; Anokye-Danso et al., 2011]. For most methods, iPSCs are reprogrammed and ready to be picked within 1 month of the introduction of reprogramming factors.
TABLE II.
Comparison of Published Methods for the Reprogramming Human Somatic Cells to induced Pluripotent Stem Cells
| Method | Integratinga | Time (days) | Efficiency (%)b | Multiple cell types reprogrammed |
|---|---|---|---|---|
| Retroviral | Yes | 25–35 | 0.02–0.08 | Yes |
| Lentiviral | Yes | 20–30 | 0.02–1 | Yes |
| Lentiviral (miRNA) | No | 18–26 | 10.4–11.6 | No |
| miRNA (direct transfection) | No | 20 | 0.002 | Yes |
| Adenoviral | No | 25–30 | 0.0002 | No |
| Sendai virus | No | 25 | 0.5–1.4 | Yes |
| mRNA | No | 20 | 0.6–4.4 | No |
| Protein | No | 56 | 0.001 | No |
| Episomal | No | 30 | 0.0006–0.02 | Yes |
| Piggybac | Yes | 14–28 | 0.02–0.05 | No |
| Minicircles | No | 14–16 | 0.005 | No |
For all integrating methods there are strategies available to remove integrated sequences.
All efficiencies are for human cells only. Please see text for other comments on limitations regarding published efficiencies.
RETRO-AND LENTIVIRUS REPROGRAMMING
Retro- and lentiviral delivery vectors have been used to express genes at robust levels in mammalian cells for several decades. Because viral transduction is easily implemented in most biomedical research labs, it has been used by nearly every lab that has generated iPSCs. The first iPSC reprogramming studies utilized retroviral vectors to express each of the reprogramming factors [Takahashi and Yamanaka, 2006; Takahashi et al., 2007]. The reprogramming efficiency with this delivery system was 0.01–0.02% in human cells with iPSC colonies appearing between 25 and 30 days post-infection [Takahashi et al., 2007]. A second published study from a different group also using retrovirus vectors found a reprogramming efficiency of 0.1% with similar kinetics to the first study, but when a six factor infection was performed with the addition of hTert and SV-40 large T antigen to Yamanaka factors the efficiency increased to 0.25% [Park et al., 2008]. It should be noted that it is difficult to directly compare reprogramming efficiencies because of factors such as differing criteria to calculate efficiency, use of different combinations of reprogramming factors, great variation in reprogramming efficiency of different somatic cells, and the use of small molecules in some studies to potentiate reprogramming efficiencies.
As retroviruses only infect dividing cells there was a shift to lentiviral delivery systems so that both dividing and non-dividing cells could be infected in the hopes of increasing reprogramming efficiency. The first study demonstrating lentiviral reprogramming came from the Thomson lab and utilized a different reprogramming cocktail [Yu et al., 2007]. This group continued to use Sox2 and Oct4 but replaced Klf4 and c-myc with Nanog and Lin28, finding that with this combination iPSCs appeared within 20 days at an efficiency of 0.02%. One of the drawbacks of these delivery systems, especially for those who want to use them to generate iPSCs that could be transitioned into clinical use, is that transgene sequences integrate into the genome of the cells being transduced. As concerns grew that the presence of transgene sequences in iPSCs might hinder translational studies strategies were developed to excise the transgenes.
LENTIVIRAL PLUS Cre-lox MEDIATED TRANSGENE EXCISION
The first generation of transgene free iPSCs were generated with lentiviral vectors containing loxP sites in the 5′ and 3′ LTR of the viral vectors. The presence of loxP sites provided a substrate to remove most of the transgene sequences by Cre-mediated recombination. The first study published utilizing such a vector isolated iPSCs from patients with Parkinson’s disease, excised transgene sequences with Cre-recombinase, and demonstrated that the recombined cells maintained all the characteristics of iPSCs with the potential to differentiate into dopaminergic neurons [Soldner et al., 2009].
An additional concern about lentiviral based vectors was the differing levels of expression of each of the reprogramming factors, as a deviation of the Yamanaka factors from a 1:1:1:1 stochiometry had negative effects on the reprogramming process [Papapetrou et al., 2009]. These concerns were addressed with the development of a polycistronic lentiviral vector system in which all four reprogramming factors were expressed as one large transcript separated by self-cleaving peptide signals. The incorporation of loxP sites into this type of vector has made it the lentivector of choice for making iPSCs [Chang et al., 2009; Sommer et al., 2009]. These vectors were first used in mouse cells, and although the Chang et al. vector was able to successfully excise transgene sequences by electroporating Cre plasmid or infecting the cells with adenoviral Cre, the lentiviral vector had a very low reprogramming efficiency (0.0004%). Sommer et al. [2010] demonstrated a much higher reprogramming efficiency in mouse with their vector (0.5%) while also demonstrating removal of transgene sequence in all iPSC lines tested with an adenoviral Cre recombinase vector. A humanized version of this vector was constructed for the generation of human iPSCs [Somers et al., 2010]. The reported efficiency of this vector was 0.1–1.5% from fibroblast of patients with cystic fibrosis and AAT deficiency-related emphysema as iPSC colonies appeared 12–15 days post-transduction and were picked for expansion around day 30. Transgene sequences were successfully excised under puromycin selection with a Cre-puromycin plasmid and all colonies with the transgene sequences removed were demonstrated to display the hallmarks of pluripotent stem cells with the capacity to differentiate into tissues from all three lineages in mouse teratoma in vivo pluripotency assays. It should be noted that although nearly all of the transgene is removed one loxP site flanked by small portions of the 5′ and 3′ LTRs remains in the iPSC genome following Cre-mediated recombination. The continued presence of exogenous transgene sequences (no matter how minimal) could be a concern if differentiated cells derived from these iPSCs are to be transplanted into a patient.
ADENOVIRUS
Adenovirus is a non-integrating virus that does not infect replicating cells. The first published study reporting the generation of iPSCs with an adenoviral vector was by Hochedlinger and colleagues who reprogrammed mouse tail tip fibroblasts to iPSCs [Stadtfeld et al., 2008]. The reprogramming efficiency was in the range of 0.001–0.0001%, with the authors speculating that the expression window of reprogramming factors is too narrow to induce the expression of endogenous factors necessary to transition somatic cells to a pluripotent state. Three of the 13 adenoviral mouse iPSCs were also found to be tetraploid, perhaps because of cell fusion or selection of rare tetraploid cells present in the starting population. Zhou and Freed [2009] were the first to create human iPSCs with adenoviral delivery of the four Yamanaka factors. They were able to reprogram only 0.0002% of human fetal fibroblasts. Unlike the case for mouse iPSCs none of the human adenoviral iPSCs were tetraploid. Both the mouse and human iPSCs created via adenovirus showed no signs of transgene integration, which is a favorable result for translational applications. However, the reprogramming efficiency must be significantly improved before this delivery method can be viable for reprogramming.
SENDAI VIRUS
Sendai virus is an RNA virus that can produce large amounts of protein without entering the nucleus of an infected cell. These characteristics make Sendai an attractive candidate for the generation of translational-grade iPSCs. The Sendai virus vector remains in the cytoplasm of infected cells for a few passages but is diluted out quickly and completely lost by passage ten. The first published study using Sendai virus for human iPSC reprogramming reported that BJ1 neonatal fibroblasts and adult human dermal fibroblasts were reprogrammed at an efficiency of 1% within 25 days of infection [Fusaki et al., 2009]. Seki et al. [2010] used a modified temperature sensitive Sendai virus to reprogram terminally differentiated blood cells from human blood. Although their reprogramming efficiency of 0.1% was 10-fold lower than Fusaki et al., it was still comparable to reprogramming efficiencies using lentivirus. A third group demonstrated that CD34+ cells derived from adult and cord blood could be reprogrammed at an efficiency of 0.1% with a temperature sensitive Sendai virus vector [Ban et al., 2011]. Based upon its relatively high reprogramming efficiency in multiple somatic cell types and lack of a footprint, Sendai virus is a good choice for researchers who want to generate iPSCs for translational use. It should be noted that Sendai is much more difficult to work with than retro- or lentiviruses, however, there are commercially available products sold with viral extracts that are ready to use for reprogramming experiments.
PROTEIN
Direct expression of reprogramming factors as proteins also allows for the generation of footprint-free iPSCs. As such, this method could be another good choice for the creation of iPSCs suitable for studies in translational medicine. Several technical problems have made the application of this reprogramming technique proceed at a slower pace than others that are in common use. One of the original difficulties was that synthesizing proteins that can cross the plasma membrane of target cells and also retain their biological activity has been a difficult goal to achieve. Two groups were able to overcome these problems by utilizing pre-existing advances in protein expression to solubilize and refold Yamanaka factor proteins after synthesis in E. coli so that they would retain their bioactivity [Kim et al., 2009; Zhou et al., 2009]. Just as importantly these groups were able to synthesize these proteins with a poly-arginine fusion to mediate transit across the cell membrane. The proteins were transduced multiple times over 7 days with iPSCs appearing approximately 2 months after seeding fibroblasts with an overall efficiency of 0.006% in mouse and 0.001% in human cells [Kim et al., 2009; Zhou et al., 2009]. This breakthrough established that protein expression is a promising technology to reprogram cells but the lengthy timeline, low efficiency, and special technical skills required to synthesize bioactive reprogramming proteins make it an unattractive choice for most labs that are attempting to generate iPSCs. Additionally, there are not any published studies on the efficiency of this method in reprogramming cell types other than fibroblasts.
mRNA TRANSFECTION
Expression of reprogramming factors as mRNAs is another zero-footprint technology to generate iPSCs. The biggest hurdle to reprogramming with mRNA was the strong immunogenic response elicited in cells by the introduction of synthetic mRNA. Warren et al. [2010] were able to solve this problem by taking several steps to minimize this immunogenicity. They modified the RNA bases by substituting 5-methylcytidine for cytidine and pseudouridine for uridine and also added the interferon inhibitor B18R into a cell culture media that was also modified in several other ways. These changes greatly diminished cell death that results from the strong antiviral response that is produced when cells come into contact with mRNA containing standard ribonuclease bases in a traditional cell culture medium. Yamanaka factor mRNAs were transfected into human fibroblasts or keratinocytes daily for 17 days and iPSC colonies were picked by day 20. Direct comparison of a four-factor mRNA experiment with a retrovirus experiment with the same four factors revealed that the efficiency of mRNA reprogramming was 1.4% versus 0.04% for retrovirus with colonies for mRNA appearing 8 days before those for retrovirus. Warren et al. were able to further increase reprogramming efficiency to 4.4% by adding a fifth factor Lin28, culturing cells at 5% O2, and adding valproic acid to the cell culture medium. It should be noted here that many current protocols incorporate cell culture under hypoxic conditions with the addition of the histone deacetylase inhibitor valproic acid. As the endogenous niche for many stem cells is hypoxic, Yoshida et al. [2009] reasoned that cell culture at 5% O2 would augment reprogramming efficiency. As per their expectations they found a threefold increasee in efficiency under these conditions with retroviral overexpression of the Yamanaka factors. The results with synthetic mRNA coupled with the fact that there are no residual traces of the exogenously introduced reprogramming factors make mRNA reprogramming an attractive option for researchers who want to generate iPSCs for translational use. Although the protocol has become streamlined through the availability of this product commercially, there is still a considerable labor and cost investment with mRNA transfection in comparison to the other methods of iPSC generation and there have been no published results with this protocol in cells other than fibroblasts.
miRNA INFECTION/TRANSFECTION
An miRNA family targeting Cyclin-Cdk2 pathway inhibitors with high levels of expression in mouse ES cells was identified recently [Wang et al., 2008]. The orthologous family in human has strong expression in ESCs and also regulates the cell cycle [Suh et al., 2004; Bar et al., 2008; Morin et al., 2008]. Subsets of this miRNA cluster were found to enhance reprogramming efficiency in mouse fibroblasts when co-transfeted with the four Yamanaka factors [Judson et al., 2009; Li et al., 2011]. This increased efficiency was mediated by promoting a mesenchymal-to-epithelial (MET) transition, affecting the cell cycle, and inhibiting the TGF-β receptor II family [Li et al., 2011]. Synthetic mimics of the mature miR-302b and miR-372 (orthologs of the mouse miRNA cluster described above) were introduced into human fibroblasts and in conjunction with viral overexpression of the Yamanaka factors there was a 10- to 15-fold increase in reprogramming efficiency [Subramanyam et al., 2011]. As with the mouse studies, MET transition was promoted, cell cycle affected, and the TGF-β receptor II family inhibited.
In human ESCs the miR-302/367 cluster is a direct Sox2/Oct4 target that inhibits the cell cycle regulator cyclin D1 [Card et al., 2008]. This cluster contains five miRNAs, four of which have the same seed sequences. Lentiviral overexpression of this cluster in BJ1 human foreskin fibroblasts resulted in the appearance of iPSCs in 10% of fibroblasts 12–14 days after infection [Anokye-Danso et al., 2011]. miRNA derived iPSCs were equivalent to those derived by other methods with similar silencing of the integrated miRNA transgene. The transgene integration associated with this lentiviral based system does not make it an optimal choice for translation-grade iPSCs, but future iterations of this technology are likely to have an excision option which when coupled to its extremely high efficiency might make it an acceptable choice for many applications.
Miyoshi et al. [2011] identified three miRNAs—mir-200c, mir-302s, and mir-369s—that were overexpressed in mouse ES cells relative to mouse adipose stromal cells. Previous studies had shown that these miRNAs targeted many of the same processes that were affected by mouse miRNAs that enhanced reprogramming: mir-200c inhibited epithelial–mesenchymal transitions, mir-302s was part of a regulatory circuit with Oct4 that maintained pluripotency, and mir-369s inhibited ZEB-2-related TGFβ signaling [Gregory et al., 2008; Rosa and Brivanlou, 2011]. These three mature miRNAs were transfected four times at 48 h intervals over 6 days into human dermal fibroblasts and adipose stromal cells. iPSC-like colonies appeared 20 days after the first transfection at an efficiency of 0.002%. Characterization of these colonies confirmed that they had markers of pluripotent stem cells with the capacity to form cells from each of the three germ layers in mouse teratoma assays. At the present time it is hard to reach any firm conclusions about the utility of miRNAs transfection for generating translation-grade iPSCs reprogramming as there is only one published article using this method, and the method has not been tested in keratinocytes and peripheral blood. However, if it can be validated in other cell types and the efficiency can be improved it would be an excellent method for generating translation-grade iPSCs.
piggyBac
The piggyBac transposon is a mobile genetic element that in the presence of the piggyBac transposase can be integrated into chromosomal TTAA sites. Re-expression of the transposase after the transposon has been stably integrated results in the excision of the transposon with no vestiges of the integrated transposon at the integrated site. Kaji et al. [2009] and Woltjen et al. [2009] were the first to demonstrate that the Yamanaka factors could be cloned into a piggyBac vector and co-transfected into mouse embryonic fibroblasts with a piggyBac transposase to generate iPSCs. Each article showed that the vector sequence could be cleanly excised from mouse iPSCs. Both groups also reported that the procedure could be repeated in human fibroblasts. Only the Kaji et al. article provided any details about the piggyBac derived human iPSCs. They stated that iPSCs were generated at an efficiency of 0.02–0.05% at 14–25 days post-transfection. The report had a cursory characterization of the human iPSCs showing that their morphology was consistent with pluripotent stem cells and that they stained positively for the appropriate surface and nuclear markers. A more detailed characterization of piggyBac derived human iPSCs was published by Mali et al. [2010]. This study examined the ability of sodium butyrate to enhance reprogramming efficiency 25-fold for both retroviral transduction and piggyBac insertion. The authors noted that retroviral transduction was 50-fold more efficient at reprogramming mesenchymal stem cells than piggyBac, which had a peak efficiency of 0.02%. This article characterized the piggyBac derived iPSCs more thoroughly than the earlier studies demonstrating that they had the capacity to generate all three germ layers in both in vitro and teratoma based assays. Although there is considerable published documentation that piggyBac insertions have been excised from mouse iPSCs, to the best of our knowledge we have not seen similar documentation for human iPSCs. piggyBac reprogramming holds great promise as a possible zero-footprint method to generate iPSCs at a reasonable reprogramming efficiency. However, the additional step required for excision of the transposon plus the dearth of information on successful excision in human iPSCs leads us to believe that work still needs to be done before it can be a viable reprogramming method for the generation of human iPSCs for use in translational studies.
MINICIRCLE VECTORS
Minicircle vectors are circularized vectors in which the plasmid backbone has been released leaving only the eukaryotic promoter and cDNA(s) that are to be expressed. A minicircle vector was produced with Lin28, GFP, Nanog, Sox2, and Oct4 and used to reprogram human adipose stem cells [Jia et al., 2010; Narsinh et al., 2011]. This protocol requires three transfections of the minicircle vector: an initial electroporation followed by sorting of GFP+ cells and then two lipid-based transfections. iPSC colonies are picked 28 days after the last transfection with a reprogramming efficiency of 0.005%. Silencing of GFP expression and presence of pluripotency markers in the picked iPSC colonies confirmed that they were likely to be pluripotent. The authors reported that the method worked at lower efficiency for neonatal fibroblasts with no published reports of successful reprogramming in any other cell types. As such, more validation will be required before this method can be deemed as being viable.
EPISOMAL PLASMIDS
Overexpression of reprogramming factors with an episomal plasmid would be another method to generate zero-footprint iPSCs. Unfortunately, transient transfection with a standard episomal plasmid does not result in expression for a long enough period of time to reprogram somatic cells to iPSCs. Stable transfection, in addition to being cumbersome, would bypass one of the criteria necessary for reprogramming: silencing of the overexpressed transgenes. Yu et al. [2009] reasoned that an oriP/EBNA (Epstein–Barr nuclear antigen) based plasmid could allow for expression of reprogramming factors for a long enough period of time to initiate the reprogramming process while eventually being lost from proliferating cells if drug selection is removed, therefore leaving no footprint of the transfected plasmid. Utilizing this transfection method, they were able to generate iPSCs by a single transfection of three plasmids containing Oct4, Sox2, Nanog, and Klf4; Oct4, Sox2, and SV40 Large T antigen; and c-myc and Lin28. Approximately 20 days post-tranfection iPSC colonies were observed at a frequency of 0.0003–0.0006%. Characterization of these colonies by pluripotency marker expression, in vivo pluripotency assays, and Oct4/Nanog promoter methylation revealed that they were ESC-like. One-third of subclones from two of the original iPSC lines lost the episomal plasmid.
As the original article describing episomal generation of iPSCs revealed such a low reprogramming efficiency, considerable effort has been extended to increase the efficiency of this method. When the same protocol was applied to mononuclear cells from bone marrow it was found that these cells reprogrammed at 0.035% efficiency with colonies that were ready to be picked by day 12 [Hu et al., 2011]. This study did a side-by-side comparison with fibroblasts and mononuclear cells from cord blood that confirmed the low reprogramming efficiencies found in fibroblasts in the earlier study. The cord blood mononuclear cells also reprogrammed at an efficiency comparable to fibroblasts although addition of the thiazovivin enhanced the process more than 10-fold (0.001%). Thiazovivin is a small molecule that had been identified by an earlier chemical screen as being able to increase viability of human iPSCs after trypsinization and enhance reprogramming efficiency [Lin et al., 2009]. The Hu et al. [2011] study also provided a more detailed description of plasmid loss showing that all of the iPSC lines they generated lost the plasmid between passages 3 and 15.
New oriP/EBNA vectors were constructed with the Yamanaka factors plus Lin28 all in one cassette and another oriP/EBNA vector containing SV40 Large T antigen [Chou et al., 2011]. When these vectors were co-transfected into CD34+ cord blood, peripheral blood, and bone marrow mononuclear cells, iPSCs were generated in 14 days in growth medium supplemented with sodium butyrate. The reprogramming efficiencies for cord blood, peripheral blood, and bone marrow were 0.02%, 0.009%, and 0.005%, respectively. The iPSCs were characterized as ESC-like by gene expression, Oct4/Nanog methylation status, and in vitro and in vivo pluripotency assays. The transfected plasmid was found to disappear from the iPSCs between passages 10 and 12. Subsequent whole genome sequencing has found no traces of the plasmid sequence in the bone marrow derived iPSC [Cheng et al., 2012].
Other recent modifications of the episomal plasmid reprogramming method include substitution of transformation deficient L-myc for c-myc to increase the reprogramming rate in dermal fibroblasts to 0.02% and the development of feeder free protocols [Chen et al., 2011; Okita et al., 2011]. This latter protocol used the defined E8 minimal media in reprogramming experiments conducted in the presence of sodium butyrate at 5% O2 yielding iPSCs at an efficiency of 0.02%. Minimization of xenogenic components with defined media and feeder-free culture is a critical objective that needs to be met if iPSC technology is to become a tenable therapeutic alternative. Because episomal reprogramming technology has achieved this landmark and also has no footprint at the moment it is becoming an attractive method to make iPSCs that could be used in translational research.
CONCLUSIONSAND FUTUREPROSPECTS
Several currently available reprogramming methods are sufficient to produce translation-grade iPSCs. Amongst these methods transfection with episomal plasmids or minicircles, infection with Sendai virus or adenovirus, transfection with synthetic mRNA/miRNA, and transposition with the piggyBac transposon leave no traces of the transgene in the genome of reprogrammed iPSCs. There are also options available with lenti- and retrovirus that with an additional step after reprogramming allow for the excision of the transgene such that only a small portion of the reprogramming vector remains integrated in the iPSC genome. When comparing all of the reprogramming methods we find that the ability of episomal plasmids and Sendai virus to generate iPSCs at good efficiencies without leaving a footprint make them the best current choices for projects with translational endpoints. The other zero-footprint methods either have unacceptably low efficiencies (adenovirus and protein) or have not been shown to be effective in reprogramming somatic cells other than fibroblasts (minicircles and synthetic mRNA/miRNA). Future advances may make these other zero-footprint methods more viable for the use of making translation-grade iPSCs. The development of feeder- and xeno-free cell culture systems to reprogram iPSCs and the establishment of facilities that meet good manufacturing practice (GMP) standards are also likely to be additional components that will be necessary requirements in producing iPSC lines that can be used to treat patients. One final critical element that requires further advancement is the optimization of protocols for the derivation of cell types affected in disease processes from iPSCs, as these are the cell types that will eventually be vehicles for cellular replacement therapies. We believe that most of the methodological advances necessary to bring the promise of iPSC technology to fruition in translational medicine are now in place and anticipate that this field will make significant advances in the next few years.
References
- Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban H, Nishishita N, Fusaki N, Tabata T, Saeki K, Shikamura M, Takada N, Inoue M, Hasegawa M, Kawamata S, Nishikawa SI. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc Natl Acad Sci USA. 2011;108:14234–14239. doi: 10.1073/pnas.1103509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar M, Wyman SK, Fritz BR, Qi J, Garg KS, Parkin RK, Kroh EM, Bendoraite A, Mitchell PS, Nelson AM, et al. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells. 2008;26:2496–2505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CW, Lai YS, Pawlik KM, Liu K, Sun CW, Li C, Schoeb TR, Townes TM. Polycistronic lentiviral vector for “hit and run” reprogramming of adult skin fibroblasts to induced pluripotent stem cells. Stem Cells. 2009;27:1042–1049. doi: 10.1002/stem.39. [DOI] [PubMed] [Google Scholar]
- Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8(5):424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Hansen NF, Zhao L, Du Y, Zou C, Donovan FX, Chou BK, Zhou G, Li S, Dowey SN, et al. Low incidence of DNA sequence variation in human induced pluripotent stem cells generated by nonintegrating plasmid expression. Cell Stem Cell. 2012;10:337–344. doi: 10.1016/j.stem.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou BK, Mali P, Huang X, Ye Z, Dowey SN, Resar LM, Zou C, Zhang YA, Tong J, Cheng L. Efficient human iPS cell derivation by a non-integrating plasmid from blood cells with unique epigenetic and gene expression signatures. Cell Res. 2011;21:518–529. doi: 10.1038/cr.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Haase A, Olmer R, Schwanke K, Wunderlich S, Merkert S, Hess C, Zweigerdt R, Gruh I, Meyer J, Wagner S, et al. Generation of induced pluripotent stem cells from human cord blood. Cell Stem Cell. 2009;5:434–441. doi: 10.1016/j.stem.2009.08.021. [DOI] [PubMed] [Google Scholar]
- Hu K, Yu J, Suknuntha K, Tian S, Montgomery K, Choi KD, Stewart R, Thomson JA, Slukvin II. Efficient generation of transgene-free induced pluripotent stem cells from normal and neoplastic bone marrow and cord blood mononuclear cells. Blood. 2011;117:e109–e119. doi: 10.1182/blood-2010-07-298331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li Z, Panetta NJ, Chen ZY, Robbins RC, et al. A nonviral minicircle vector for deriving human iPS cells. Nat Methods. 2010;7:197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nature Biotechnol. 2009;27:459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Yang CS, Nakashima K, Rana TM. Small RNA-mediated regulation of iPS cell generation. EMBO J. 2011;30:823–834. doi: 10.1038/emboj.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Ambasudhan R, Yuan X, Wenlin Li, Hilcove S, Abujarour R, Lin X, Hahm HS, Hao E, Hayek A, Ding S. A chemical platform for improved induction of human iPSCS. Nat Methods. 2009;6:805–808. doi: 10.1038/nmeth.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Chou BK, Yen J, Ye Z, Zou J, Dowey S, Brodsky RA, Ohm JE, Yu W, Baylin SB, et al. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells. 2010;28:713–720. doi: 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F, et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Morin RD, O’Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsinh KH, Jia F, Robbins RC, Kay MA, Longaker MT, Wu JC. Generation of adult human induced pluripotent stem cells using nonviral minicircle DNA vectors. Nature Protoc. 2011;6:78–88. doi: 10.1038/nprot.2010.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, Hong H, Nakagawa M, Tanabe K, Tezuka K, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- Papapetrou EP, Tomishima MJ, Chambers SM, Mica Y, Reed E, Menon J, Tabar V, Mo Q, Studer L, Sadelain M. Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc Natl Acad USA. 2009;106:12759–12764. doi: 10.1073/pnas.0904825106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Rosa A, Brivanlou AH. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J. 2011;30:237–248. doi: 10.1038/emboj.2010.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki T, Yuasa S, Oda M, Egashira T, Yae K, Kusumoto D, Nakata H, Tohyama S, Hashimoto H, Kodaira M, et al. Generation of induced pluripotent stem cells from human terminally differentiated circulating T cells. Cell Stem Cell. 2010;7:11–14. doi: 10.1016/j.stem.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, et al. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers A, Jean JC, Sommer CA, Omari A, Ford CC, Mills JA, Ying L, Sommer AG, Jean JM, Smith BW, et al. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells. 2010;28:1728–1740. doi: 10.1002/stem.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN, Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer CA, Sommer AG, Longmire TA, Christodoulou C, Thomas DD, Gostissa M, Alt FW, Murphy GJ, Kotton DN, Mostoslavsky G. Excision of reprogramming transgenes improves the differentiation potential of iPS cells generated with a single excisable vector. Stem Cells. 2010;28:64–74. doi: 10.1002/stem.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staerk J, Dawlaty MM, Gao Q, Maetzel D, Hanna J, Sommer CA, Mostoslavsky G, Jaenisch R. Reprogramming of human peripheral blood cells to induced pluripotent stem cells. Cell Stem Cell. 2010;7:20–24. doi: 10.1016/j.stem.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, Blelloch R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nature Biotechnol. 2011;29:443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibro-blasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nature Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R, Cowling R, Wang W, Liu P, Gertsenstein M, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell. 2009;5:237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Freed CR. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells. 2009;27:2667–2674. doi: 10.1002/stem.201. [DOI] [PubMed] [Google Scholar]
- Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, Trauger S, Bien G, Yao S, Zhu Y, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]


