Summary
Cu/Zn Superoxide Dismutase (SOD1) is an abundant enzyme that has been best studied as a regulator of antioxidant defence. Using the yeast S. cerevisiae, we report that SOD1 transmits signals from oxygen and glucose to repress respiration. The mechanism involves SOD1-mediated stabilization of two casein kinase 1-gamma (CK1γ) homologs, Yck1p and Yck2p, required for respiratory repression. SOD1 binds a C-terminal degron we identified in Yck1p/Yck2p, and promotes kinase stability by catalyzing superoxide conversion to peroxide. The effects of SOD1 on CK1γ stability are also observed with mammalian SOD1 and CK1γ and in a human cell line. Therefore in a single circuit, oxygen, glucose, and reactive oxygen can repress respiration through SOD1/ CK1γ signaling. Our data therefore may provide mechanistic insight into how rapidly proliferating cells and many cancers accomplish glucose-mediated repression in favour of aerobic glycolysis.
Introduction
Cellular energy production is mediated through the action of glycolysis and oxidative phosphorylation. In many cells, oxygen (O2) availability dictates whether the end product of glycolysis, pyruvate, is oxidatively metabolized through the respiratory chain to produce ATP or is reductively metabolized through lactate or ethanol fermentation to regenerate NAD+ for continued glycolysis. However, certain cells can be reprogrammed to undergo aerobic fermentation, a process characteristic of a variety of proliferating cells, including various cancers, lymphocytes, endothelial cells, and microorganisms such as Bakers’ yeast (Saccharomyces cerevisiae) (Lunt and Vander Heiden, 2011).
Many factors contribute to the switch from respiration to aerobic fermentation, including transcriptional induction of glucose transport and glycolytic enzymes and repression of respiratory genes (Bensinger and Christofk, 2012; Diaz-Ruiz et al., 2011). In the case of Bakers’ yeast, glucose itself signals repression of respiration in a process known as the Crabtree effect (Crabtree, 1929). Glucose activates a series of signaling pathways consisting of G-protein coupled receptors, hexokinases, and transmembrane glucose receptors that work in concert to repress respiration and promote aerobic fermentation (Zaman et al., 2008). Interestingly, it was recently reported that yeast strains lacking the antioxidant enzyme Cu/Zn superoxide dismutase (SOD1 or yeast Sod1p) incompletely repress respiration in the presence of glucose (Sehati et al., 2011). It was not known whether this metabolic defect was due to oxidative damage or an unknown role for SOD1 in signaling respiration repression.
SOD1 is well conserved throughout evolution and defends against oxidative stress by catalyzing the disproportionation of superoxide into peroxide (H2O2) and O2 (McCord and Fridovich, 1969). Most eukaryotes express two intracellular SODs, a Mn containing SOD2 in the mitochondrial matrix (Weisiger and Fridovich, 1973), and a highly abundant Cu/Zn SOD1 that is largely cytosolic, but also found in the mitochondrial inter membrane space (IMS) (Okado-Matsumoto and Fridovich, 2001; Sturtz et al., 2001). SOD1 deficient organisms experience various markers of oxidative stress, for example higher incidences of liver cancer in SOD1−/− mice (Elchuri et al., 2005) and increased mutation frequencies and defects in lysine and methionine biosynthetic pathways in yeast sod1Δ mutants (Chang and Kosman, 1990; Gralla and Valentine, 1991; Sturtz and Culotta, 2002). SOD1 is a highly abundant protein in various organisms (Halliwell and Gutteridge, 2007; Pardo et al., 1995), but in studies that have been done in yeast, less than 1% of total SOD1 is required to protect the aforementioned amino acid biosynthetic pathways and to prevent toxicity from superoxide (Corson et al., 1998). The rationale for producing such large quantities of SOD1 has been enigmatic and the enzyme may have as-of-yet undetermined functions in cell physiology.
Herein, we describe a new role for SOD1. Using a yeast model system, we demonstrate that Sod1p helps to integrate signals from glucose and O2 to repress respiration. The mechanism involves Sod1p-stabilization of casein kinase 1-gamma (CK1γ) homologs, Yck1p and Yck2p. These kinases are essential for nutrient sensing (Liu et al., 2008; Moriya and Johnston, 2004) and we demonstrate here their role in SOD1-dependent respiration repression. Sod1p physically binds to a C-terminal degron in Yck1p and the SOD enzymatic reaction prevents Yck1p degradation. The H2O2 product of the SOD reaction appears critical for preventing casein kinase turnover. Glucose and O2 also stabilize Yck1p and Yck2p, apparently by controlling the amount of superoxide substrate for Sod1p. In this manner, in a single circuit, O2, glucose, and reactive oxygen can repress respiration through SOD1/casein kinase signaling.
Results
Cu/Zn SOD1 is needed for glucose control of respiration
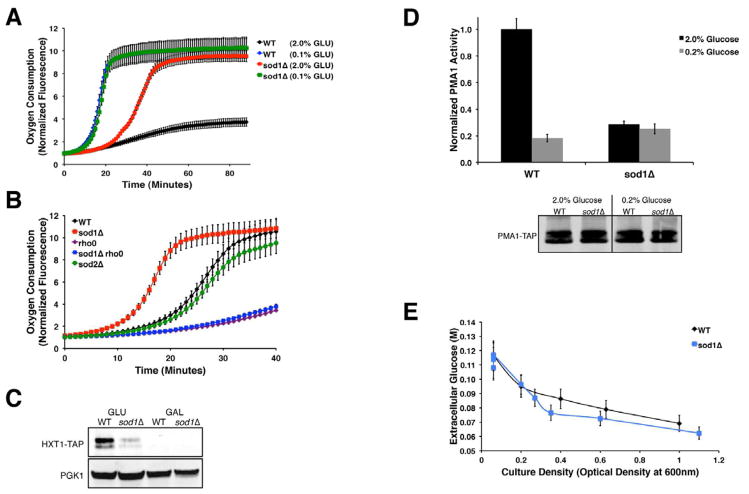
In yeast, O2 consumption rates are lowest with abundant glucose and most rapid with low glucose (Figure 1A) or with an alternate carbon source such as galactose (Figure S1A). Consistent with previous studies (Sehati et al., 2011), this glucose repression of O2 consumption appears defective in sod1Δ yeast. Cells lacking the largely cytosolic Cu/Zn Sod1p, but not the mitochondrial Mn Sod2p, exhibit elevated O2 consumption in high glucose that is reflective of mitochondrial respiration, as it is obliterated in rho0 strains lacking mitochondrial DNA (Figure 1B). Additionally, a number of glucose signaling markers appear blocked by sod1Δ mutations, including glucose-induction of the hexose transporter HXT1 (Zaman et al., 2008) (Figure 1C). sod1Δ mutants are also defective in glucose activation of the H+-ATPase Pma1p (Estrada et al., 1996) (Figure 1D), in spite of normal glucose consumption (Figure 1E).
Figure 1. Defect in Glucose Repression of Respiration in sod1Δ Cells.
(A–B) O2 consumption analysis was carried out as described in Experimental Procedures with the indicated yeast strains grown under the designated GLU conditions (A) or with 2% GLU (B). Data represent the mean ± SD of triplicate cultures.
(C, D Bottom) Immunoblot analysis of TAP tagged Hxt1p and Pma1p was carried out as described in the Supplementary Information using an α-TAP antibody. In the Hxt1-TAP immunoblot, the highest molecular weight bands represent intact protein. PGK1 = loading control detected by α-Pgk1p antibody.
(D TOP) Pma1p ATPase activity was measured in membranes from the indicated strains as described in Experimental Procedures. Results are normalized to WT cells cultured in 2.0% GLU. The data represent the mean ± SD of duplicate cultures.
(E) Extracellular glucose was monitored as described in Experimental Procedures as a function of yeast growth at an optical density of 600 nm. Data represent the mean ± SD of triplicate cultures.
See also Figure S1.
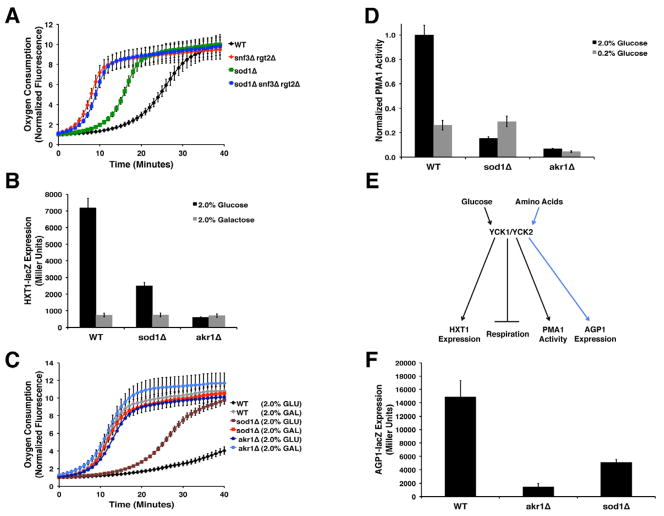
Three major glucose sensing and signaling pathways are involved in respiratory repression in yeast, including the GPA1/GPR2, HXK2, and SNF3/RGT2 pathways (Figure S1B) (Zaman et al., 2008). To determine which is affected by sod1Δ mutations, we screened for mutants that inhibit the sod1-linked increase in respiration. We observed that sod1Δ mutations still enhance respiration in gpa2, gpr1, and ras2 mutants of the GPA2/GPR1 pathway, and in hxk2 mutants of the HXK2 pathway (Figure S1C). By comparison, sod1Δ mutations had no effect on O2 consumption in a strain lacking SNF3 and RGT2 (Figure 2A).
Figure 2. SOD1 Regulates Glucose Sensing in the SNF3/RGT2 Pathway Through Yck1p.
(A and C) O2 consumption of the indicated strains was measured as in Figure 1A, B with either 2% GLU (A) or in the designated carbon sources (C).
(B) The indicated strains expressing a HXT1-lacZ reporter were assayed for HXT promoter activity by β-galactosidase activity as described in the Supplemental Information. Activity is reported as Miller Units (Giacomini et al., 1992) and represents the mean ± SD of triplicate cultures.
(D) Pma1p ATPase activity was measured as in Figure 1D.
(E) Schematic illustrating downstream targets of Yck1p/Yck2p where black and blue lines indicate glucose and amino acid signaling targets, respectively.
(F) AGP1 promoter activity was measured in strains expressing a AGP1-lacZ fusion as in (B).
See also Figure S1.
The SNF3/RGT2 pathway involves glucose activation of Yck1p and Yck2p, a pair of casein kinase 1 gamma isoforms (CK1γ) (Moriya and Johnston, 2004). Interestingly, Yck1p and Sod1p were previously noted to interact in a high-throughput mass spectrometry screen (Ho et al., 2002). To test whether loss of Yck1p/Yck2p mimicked the glucose sensing defects of sod1Δ mutants, we employed akr1Δ mutants. The Akr1p palmitoyl transferase tethers Yck1p and Yck2p to the plasma membrane, and akr1 mutations are often used to model loss of Yck1p/Yck2p (Pasula et al., 2010) since yck1Δ yck2 Δ mutants are not viable (Robinson et al., 1992). Similar to sod1Δ strains, a Δ mutantskr1 cannot properly activate HXT1 (Figure 2B), exhibit high respiration (Figure 2C) and attenuated Pma1p activity (Figure 2D) in high glucose in spite of normal glucose uptake (Figure S1D). Yck1p and Yck2p affect multiple steps in glucose signaling that are also perturbed by sod1Δ mutations.
Yck1p and Yck2p are additionally involved in amino acid sensing, where extracellular amino acids signal through these casein kinases to induce amino acid permease genes such as AGP1 (Liu et al., 2008) (Figure 2E). If sod1Δ was specifically inhibiting Yck1p and Yck2p, then amino acid sensing should be impacted as well. As seen in Figure 2F and Figure S1E, AGP1 promoter activity and protein expression levels, respectively, are markedly inhibited in sod1Δ cells, and the same is true for akr1Δ mutants. Loss of Sod1p affects the glucose and amino acid sensing pathways that require Yck1p and Yck2p.
SOD1 interacts with and stabilizes casein kinases Yck1p and Yck2p
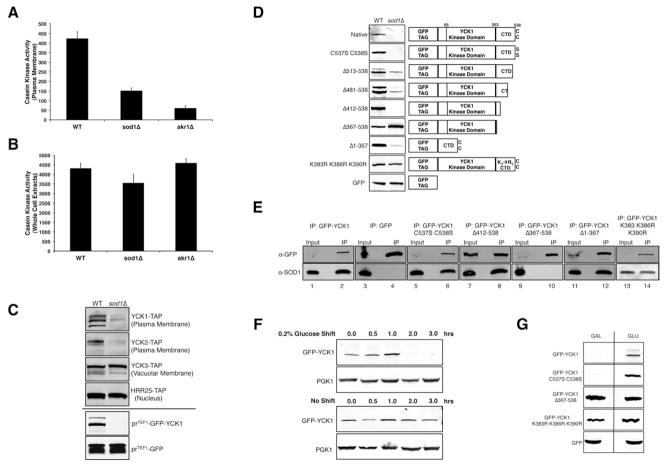
Yck1p and Yck2p represent the only plasma membrane casein kinases of S. cerevisiae, and we find that sod1Δ cells have a marked reduction in their activity, but not of whole cell casein kinases, largely represented by vacuolar Yck3p and nuclear Hrr25p (Ho et al., 1997; Sun et al., 2004) (Figure 3A, 3B). The same specificity for casein kinase inhibition was observed in akr1Δ mutants (Figure 3A, 3B). By immunoblot, C-terminal TAP-tagged Yck1p and Yck2p are virtually undetectable in a sod1Δ strain whereas the vacuolar and nuclear Yck3p and Hrr25p are unaffected (Figure 3C). To address whether Yck1p/Yck2p loss occurred at the transcriptional or translational initiation level, we expressed a N-terminal GFP fusion to Yck1p under control of the TEF1 promoter. This GFP-Yck1p fusion but not GFP alone, was strongly down-regulated by sod1Δ mutations (Figure 3C). Transcriptional and translational initiation cannot account for the sod1Δ loss in Yck1p, indicating protein stability effects.
Figure 3. bGlucose and SOD1 Regulate Yck1p Stability Through a C-terminal SOD1-Interacting Degron.
(A and B) Casein kinase activity from 2% GLU grown cells was carried out in isolated plasma membranes (A) or in whole cell lysates (B) as described in Experimental Procedures. Activity is reported as % phosphorylation of fluorescent substrate per mg protein. Data represent the mean ± SD of duplicate cultures.
(C) Immunoblot analysis of cells grown in 4% GLU and expressing (TOP) C-terminal TAP-tagged fusions of the designated casein kinases driven by their native promoters and detected by α-TAP; or (BOTTOM) N-terminal GFP fusion to Yck1p (expressed from plasmid pAR113) or GFP alone (plasmid pAR118) driven by the TEF1 promoter and detected by α-GFP as described in Supplemental Information.
(D) α-GFP immunoblot of cells grown in 4% GLU and expressing N-terminal GFP fusions to WT Yck1p (“Native” expressed from plasmid pGFPYCK1) or to Yck1p mutant derivatives of this construct. As diagrammed on right, these derivatives contain Yck1p C537S C538S or K383R K386R K390R mutations, or have truncations in the C-terminal domain (CTD) or contain the isolated CTD of Yck1p. C C, S S indicates Cys 537 Cys 538 or the double Ser substitutions and K3→R3 indicates the K383R, K386R, K390R Yck1p mutant.
(E) Lysates from WT strains grown in 4% GLU and expressing the indicated GFP fusions to Yck1p (described in part D) were subjected to immuno-precipitation with α-GFP. The input, corresponding to ~3% of total cell lysate, and immunoprecipitated material (IP) were analyzed by immunoblot using α-GFP (top row) and α-SOD1 (bottom) antibodies as described in Supplemental Information. 3% of the Sod1p input provides equivalent signals as 100% of IP Sod1p.
(F) WT cells transformed with pGFPYCK1 and grown in 4% GLU were switched to 0.2% GLU or maintained in high glucose for the indicated times in the presence of cycloheximide to inhibit new protein synthesis. Immunoblot analysis was conducted with α-GFP.
(G) WT cells expressing the indicated GFP fusions (described in part D) were grown in 2% GLU or GAL and analyzed by immunoblot.
See also Figure S2.
Since sod1Δ specifically impacts plasma membrane Yck1p and Yck2p, we tested whether cell surface localization was key. A C537S C538S allele of Yck1p that disrupts the palmitylation site for membrane anchorage (Roth et al., 2011) is no longer plasma membrane localized (Figure S2A), yet its stability is still regulated by Sod1p (Figure 3D). Yck1p and Yck2p also have unique C-termini outside of the catalytic region that are not present in Yck3p and Hrr25p (Figure S2B, S4). As seen in Figure 3D, deletion of Yck1p C-terminal residues 367–538 completely obliterated regulation by Sod1p and the protein remained stable in sod1Δ strains, whereas smaller C-terminal deletions to residue 412 had no effect. Moreover, a minimal fusion of the Yck1p C-terminal domain (CTD) to GFP was sufficient to induce GFP fusion degradation in sod1Δ strains (Figure 3D). Hence, residues 367–412 of the Yck1p CTD contains a SOD1-regulated degron. As shown in Figure S4, the C-terminal degron contains three lysine residues, K383, K386, and K390, also present in Yck2p that represent potential sites of regulation by ubiquitination or other post-translational modifications. As shown in Figure 3D, mutation of these lysines to arginine stabilizes Yck1p in the absence of Sod1p. Hence, SOD1 regulation of Yck1p involves lysines in the Yck1p degron region.
To test if Sod1p physically interacts with this degron region, co-immunoprecipitation (coIP) studies were carried out. Sod1p was seen to coIP with full length Yck1p fused to GFP (Figure 3E, lane 2), but not with GFP alone (Figure 3E, lane 4). As was observed for Sod1p-stabilization of Yck1p (Figure 3D), Sod1p-Yck1p interactions do not require plasma membrane anchorage (no effect of Yck1p C537S C538S mutations, Figure 3E, lane 6) but do require the CTD of Yck1p. Interactions are disrupted upon deletion of the Yck1p CTD to position 367 (Figure 3E, lane 10), but not to 412 (lane 8), and the Yck1p CTD is sufficient to confer Sod1p binding (lane 12). Interestingly, the Yck1p C-terminal K383, K386 and K390 residues that are required for SOD1-stabilization of the kinase (as in Figure 3D) are not required for Sod1p binding to Yck1p (Figure 3E, lane 14). Hence these lysines are specifically involved in Yck1p turnover, not Yck1p interaction with Sod1p.
The means by which glucose activates Yck1p and Yck2p is not well understood. We therefore tested if glucose, like Sod1p, regulates Yck1p through protein stability. In the experiment of Figure 3F, cells grown in high glucose were switched to low glucose and the stability of GFP-Yck1p was monitored in the presence of cycloheximide to inhibit new protein synthesis. This switch to low glucose led to destabilization of GFP-Yck1p within two hours (Figure 3F). Substituting glucose with the non-repressing carbon source galactose also resulted in loss of GFP-Yck1p (Figure 3G). As with SOD1-control of Yck1p, the glucose stabilization of Yck1p does not require membrane anchorage (the C537S C538S Yck1p mutant is without effect) but requires the Yck1p C-terminus and residues K383, K386, and K390 (Figure 3G). Therefore, during glucose repression of respiration, Yck1p is stabilized through a mechanism involving its SOD1-interacting domain and key lysines at the C-terminus.
Enzymatically active SOD1 stabilizes casein kinase 1 gamma (CK1 ) homologs from yeast and mammals
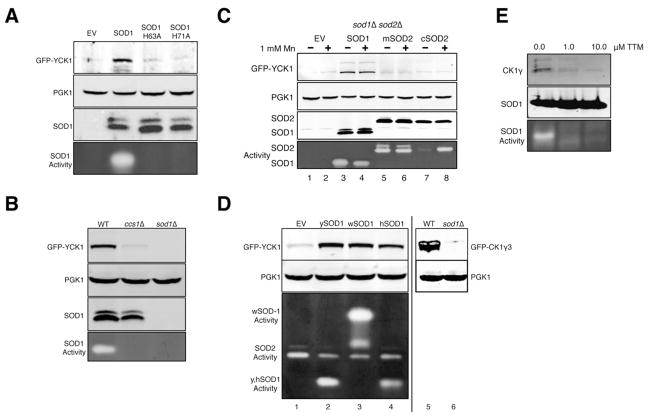
We tested whether SOD enzymatic activity is required for Yck1p stability. Mutations H63A and H71A in yeast Sod1p disrupt the copper site and inactivate the enzyme and these same Sod1p mutants no longer support Yck1p stability (Figure 4A), similar to effects of a sod1Δ deletion. Furthermore, yeast ccs1Δ mutants that lack the copper chaperone for Sod1p (Culotta et al., 1997) express inactive Sod1p, and these same mutants cannot stabilize Yck1p (Figure 4B). Thus, Yck1p stability requires Sod1p enzymatic activity.
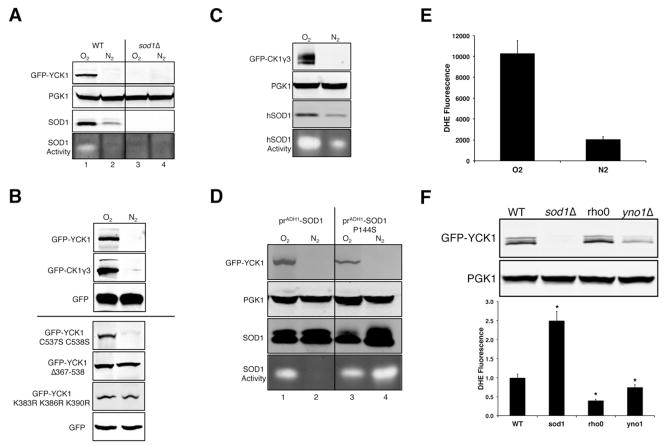
Figure 4. Enzymatically Active SOD1 Stabilizes CK1 Homologs from Yeast and Mammals and in Cell Culture.
(A–D) Immunoblots (top 2–3 panels) and native gel assays for SOD activity (bottom panel) were conducted on cells grown in 4% GLU and transformed with pGFPYCK1 or plasmid pAR124 for expressing GFP fused to bovine CK1γ3 (GFP-CK1γ3). As indicated, sod1Δ ( part A, D) and sod1Δ sod2Δ (part C) strains also expressed the following SOD molecules: SOD1, ySOD1, H63A, H71A - S. cerevisiae SOD1 or mutant derivatives; mSOD2 and cSOD2 – mitochondrial and cytosolic S. cerevisiae SOD2; wSOD1 - C. elegans Sod-1; hSOD1 - human SOD1. These various SODs exhibit differential migration on native gels and their positions are denoted. “1 mM Mn” – manganese supplemented during entire growth period.
(E) Immunoblot of endogenous CK1γ and SOD1 and native gel assay for SOD activity were conducted on HEK293 cells treated with the indicated concentrations of the copper chelator, tetrathiomolybdate (TTM), for 24 hours.
In addition to Cu/Zn SOD1, most eukaryotes express a second unrelated SOD2 enzyme that localizes to the mitochondrial matrix and uses Mn as a cofactor (Weisiger and Fridovich, 1973). In sod1Δ sod2Δ cells lacking both SOD enzymes, Yck1p is absent as expected (Figure 4C, lane 1) and this is rescued by ectopic expression of SOD1 (lane 3), but not of SOD2 (lane 5). Yet since Yck1p and Sod2p localize to separate cellular compartments, we attempted to rescue loss of Yck1p by expressing Sod2p in the cytosol. Cytosolic Sod2p becomes active when cells accumulate high Mn (Luk et al., 2005) (Figure 4C, lane 8 bottom panel), but this is not sufficient to stabilize Yck1p. Another means for removing superoxide in yeast involves small molecule complexes of manganese, so-called Mn-antioxidants (Aguirre and Culotta, 2012; Barnese et al., 2012; Chang and Kosman, 1989; Reddi and Culotta, 2011), which are far more effective for removing superoxide in yeast than a variety of commonly used Mn-porphyrin SOD mimetics (Munroe et al., 2007). Supplements of high manganese can fully rescue O2 sensitivity in the sod1Δ sod2Δ strain (Figure S3), but these same Mn-antioxidants do not rescue loss of Yck1p (Figure 4C, lanes 2,6,8). Thus, only Sod1p can stabilize Yck1p.
Can the SOD1-stabilization of CK1γ kinases be extended to other eukaryotes? We expressed Cu/Zn SOD molecules from C. elegans and humans in sod1Δ strains and observed that these heterologous Cu/Zn SODs can also stabilize Yck1p (Figure 4D, lanes 3,4). We also tested whether yeast Sod1p can stabilize mammalian CK1γ. A bovine CK1γ isoform, CK1γ3, which has a high degree of sequence conservation (Figure S4) and complements yeast yck1 yck2 mutants (Zhai et al., 1995), is stably expressed in SOD1 WT yeast, but not in sod1Δ cells (Figure 4D, lanes 5,6). Moreover, human SOD1 expressed in yeast can stabilize this bovine CK1γ (see ahead Figure 6C). Thus, mammalian CK1γ homologs are also stabilized by a variety of eukaryotic SOD1 molecules. This was addressed further using a human cell line. HEK293 cells were treated with the intracellular copper chelator tetrathiomolybdate (TTM), a treatment that readily inhibits SOD1 activity in cell culture (Alvarez et al., 2010; Juarez et al., 2008). TTM inhibits SOD1 activity without changes in SOD1 protein, and this correlates with a drastic reduction in levels of human CK1γ (Figure 4E). These findings corroborate our yeast expression studies showing that yeast and mammalian CK1γ isoforms are stabilized by active SOD1 enzyme.
Figure 6. O2 and Superoxide Regulates Yeast and Mammalian CK1 Stability.
The indicated cells were cultured in 4% GLU medium under either atmospheric oxygen (O2) or nitrogen (N2) conditions.
(A, C, D) Immunoblots (top panels) and native SOD activity gels (bottom panel) were assayed from the indicated cells expressing GFP-Yck1p or GFP fused to bovine CK1γ3. hSOD1, prADH1-SOD1, prADH1-SOD1 P144S = sod1Δ cells expressing human SOD1 driven by the yeast SOD1 promoter and ADH1 driven WT SOD1 or the P144S derivative.
(B) Immunoblots of WT cells expressing GFP-Yck1p or the indicated Yck1p mutant derivatives or GFP fused to bovine CK1γ3.
(E) Superoxide levels were measured by DHE (dihydroethidium) fluorescence as described in Experimental Procedures. The data represent the mean ± SD of triplicate cultures.
(F) Immunoblots (top panels) and DHE fluorescence (bottom graph) of WT, sod1Δ, rho0, and yno1Δ cells expressing TEF driven GFP-Yck1p. (Bottom graph) The data represent the mean ± SD of triplicate cultures and asterisks indicate a p < .05 and reflects a statistical significance between the DHE fluorescence of WT cells and the indicated mutants.
Levels of cytosolic Cu/Zn SOD1 required for Yck1p stability
SOD1 is localized in both the cytosol and in the mitochondrial IMS (Sturtz et al., 2001). However, mitochondrial SOD1 is not relevant to Yck1p since expression of an IMS-specific Sod1p (“SCO2-SOD1” (Wood and Thiele, 2009)) does not stabilize Yck1p (Figure 5A). Thus, extra-mitochondrial Sod1p is required for Yck1p stability.
Figure 5. Cytosolic Sod1p Stabilizes Yck1p.
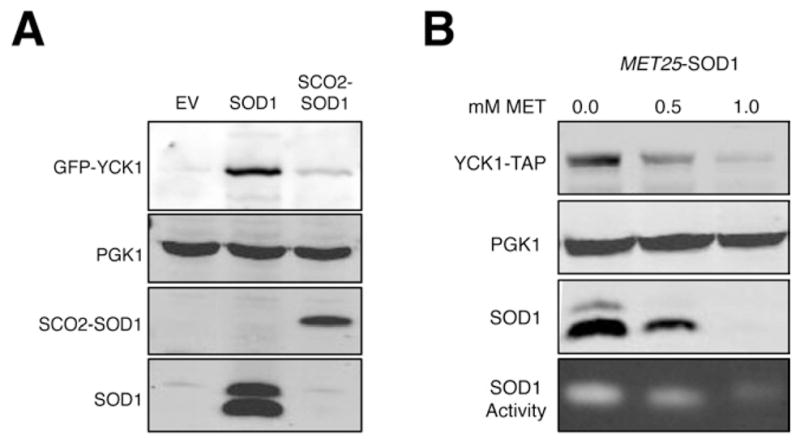
(A) The sod1Δ strain grown in 4% GLU and expressing GFP-Yck1p was transformed with vectors for expressing yeast Sod1p (SOD1), or mitochondrial IMS targeted Sod1p (SCO2-SOD1) and analyzed by immunoblot. α-SOD1 was used to detect SCO2-SOD1.
(B) A sod1Δ strain expressing Yck1p-TAP and MET25-driven SOD1 was grown in 4% GLU with increasing concentrations of methionine (MET) to repress SOD1 expression. Upper three panels are immunoblots of the indicated proteins, bottom panel is native SOD activity gel. Immunoblot quantitations were conducted using the software on the LIC-COR Odyssey imager and were normalized to PGK1 and are relative to the 0.0 MET control as follows: 0.5 mM MET, Yck1p-TAP= 0.3; Sod1p = 0.2; 1.0 mM MET, Yck1p-TAP = 0.10; Sod1 protein = 0.01.
We sought to gauge how much cytosolic Sod1p is required to stabilize Yck1p. In our IP studies, we recover 3% of total Sod1p bound to Yck1p (Figure 3E and legend). This corresponds to ≈8000 Sod1p dimers, compared to the ≈7800 molecules of Yck1p estimated for yeast (Ghaemmaghami et al., 2003). However, we find that the actual level of total Sod1p required to maintain Yck1p stability is substantially greater than this 3% value. When SOD1 expression is titrated down using a MET25 repressible promoter, Yck1p protein is destabilized with a 5- fold reduction in Sod1p levels (Figure 5B, for quantitation see Figure legend). It is possible that the larger fraction of Sod1p (≥20% total Sod1p) that is required for stabilizing Yck1p is not all bound to the casein kinase at steady state, but is needed to feed the pool of kinase-bound Sod1p.
O2 regulation of Yck1p Stability
In various eukaryotes, SOD1 activity is down-regulated as cells reach hypoxia and anoxia (Brown et al., 2004; Leitch et al., 2009; Leitch et al., 2012; White et al., 2009). S. cerevisiae Sod1p activity is virtually undetectable in cells grown under nitrogen and we observe a dramatic loss in Yck1p under these conditions (Figure 6A, lane 2). Furthermore, bovine CK1γ3 expressed in yeast is similarly down-regulated by low O2 in cells that either co-express yeast Sod1p (Figure 6B) or human SOD1 (Figure 6C). As with effects of low glucose or sod1Δ mutations, Yck1p instability with low O2 does not require membrane anchorage (not affected by Yck1p C537S C538C mutations) but does require the Sod1p-interacting, degron domain of Yck1p at the C-terminus and residues K383, K386, and/or K390 (Figure 6B).
We tested whether loss of Yck1p with low O2 could be reversed by re-introducing active SOD1 enzyme. Anaerobic cultures can be induced to produce mature Sod1p enzyme by placing SOD1 under control of the constitutive ADH1 promoter and by using a P144S allele of Sod1p that can acquire copper independently of Ccs1p and O2 (Leitch et al., 2012). We were surprised to find that Yck1p is still down-regulated under nitrogen in cells expressing P144S Sod1p (Figure 6D, lane 4). As a noteworthy caveat to this experiment, the mature P144S Sod1p enzyme should have little activity in vivo without O2 due to limited superoxide substrate. Superoxide was monitored using the DHE (dihydroethidium) probe selective for superoxide over peroxide, peroxynitrite, and hypochlorous acid (Tarpey and Fridovich, 2001). Indeed, cells under nitrogen have low superoxide (Figure 6E). By providing the superoxide substrate for SOD1, O2 may stabilize Yck1p.
Superoxide and hydrogen peroxide effects on YCK1
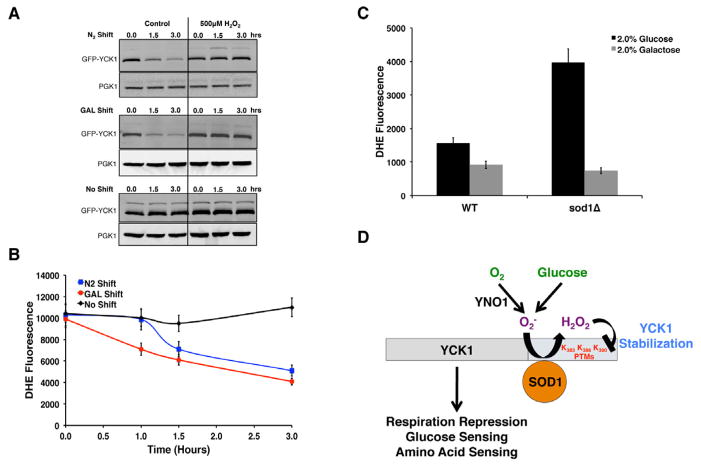
SOD1 reactivity with superoxide could stabilize CK1γ kinases in two ways – either by preventing superoxide damage to CK1γ, or by catalyzing superoxide conversion to H2O2 which in turn promotes kinase stability. Our studies with anaerobic cultures argue against the former. As seen in Figure 6A, the sod1Δ loss in Yck1p was not rescued under nitrogen (lane 4), even though superoxide levels are greatly reduced (Figure 6E). Moreover, when WT cells are shifted from O2 to N2, we observe a 2-fold drop in superoxide within 3 hours (Figure 7B), which closely parallels the kinetics of Yck1p turn over (Figure 7A). If anything, loss of superoxide correlates with Yck1p degradation, not stability.
Figure 7. A Role for H2O2 in Stabilization of Yck1p Under Aerobic Glucose Conditions.
(A, B) WT cells expressing GFP-Yck1p under aerobic 4% GLU conditions were shifted either to anaerobic conditions (N2 shift), or to aerobic 4% GAL conditions (GAL shift) or maintained as is (No shift) for the indicated times. (A) Turn over of Yck1p in the presence of cycloheximide was monitored by α-GFP immunoblot. Where indicated, 500 μM H2O2 was supplemented. (B) DHE detectable superoxide was monitored as described in Experimental Procedures. The data represent the mean ± SD of triplicate cultures.
(C) Superoxide levels were measured by DHE fluorescence in 2% GLU or 2% GAL cultures of WT and sod1Δ cells. The data represent the mean ± SD of triplicate cultures.
(D) A model for regulation of CK1-γ isoforms by SOD1. Superoxide generated from growth in the presence of O2 and glucose is converted by SOD1 to H2O2, which stabilizes a C-terminal degron on Yck1p, a casein kinase that regulates glucose and amino acid sensing and respiration repression, through a mechanism that involves peroxide-inhibition of post-translational modifications (PTM) at lysines K383, K386, and/or K390.
It is noteworthy that this same loss in superoxide is seen in aerobic cultures that are shifted from glucose to galactose (Figure 7B). As with hypoxia, the kinetics of Yck1p turnover closely follow the decrease in superoxide with a switch to galactose (Figure 7A, 7B). In spite of the loss of superoxide substrate for Sod1p, the SOD enzyme remains bound to Yck1p during the switch to galactose (Figure S5). Loss of the superoxide substrate for Sod1p, rather than loss of Sod1p binding, seems to trigger Yckp1 turnover.
We were initially surprised that superoxide levels were higher in glucose conditions where mitochondrial respiration is repressed (Figure 1A). Nevertheless the superoxide from glucose cultures is a substrate for Sod1p, as levels rise even further with sod1Δ mutations (Figure 7C). By comparison, the superoxide from galactose cultures may not be a substrate for Sod1p as levels are unaffected by sod1Δ mutations (Figure 7C). Hence cells grown in O2 and glucose produce a superoxide substrate for Sod1p and Yck1p is stabilized (Figure 7D).
What is the source of superoxide relevant to Sod1p and Yck1p regulation? Part of the superoxide from glucose growth cells may originate from mitochondria since rho0 mutations eliminating respiration show a 2-fold decrease in cellular superoxide (Figure 6F). However, this loss of mitochondrial superoxide has no effect on Yck1p stability (Figure 6F). Extra-mitochondrial sources of superoxide can include NADPH-oxidases, and an intriguing candidate in yeast is the NADPH oxidase-like protein, Yno1p (Rinnerthaler et al., 2012). We observe that yno1Δ cells, which exhibit a ~25% reduction in superoxide levels, experience a marked loss in Yck1p stability (Figure 6F), suggesting that this NADPH oxidase-like protein is a superoxide source for SOD1/CK1γ signaling.
We next tested whether H2O2, the product of the SOD reaction, can affect Yck1p degradation. As a paradigm, we employed the switch to hypoxia and galactose (as in Figure 7A, 7B) where Yck1p turnover is easily monitored. (Yck1p turnover rates are not easily obtained from sod1Δ null cells where steady state Yck1p is undetectable). As seen in Figure 7A, the addition of 0.5 mM exogenous H2O2 significantly stabilizes the Yck1p polypeptide during the switch to both hypoxia and galactose. As another means of increasing cellular peroxide we added galactose oxidase (GO) to the growth medium. As seen in Figures S6A and S6B, GO produced a continuous level of peroxide that could be detected intracellularly using the ROS probe, 2,7-dichlorofluorescein diacetate (DCFDA) (Wu et al., 2009). Moreover, this GO-derived peroxide helped to stabilized Yck1p (Figure S6C). It is worth noting that the level of peroxide in both cases is relatively high. Stabilization of Yck1p requires either a one-time bolus of 0.5 mM H2O2 or a continuous flux of peroxide from GO (>.11 mM/min), which generates 0.5 mM H2O2 within 5 minutes (Figure S6C and S6D). The rationale for needing such high non-SOD1 H2O2 can be explained by considering the numerous peroxide scavenging systems of the cell. Yeast express various catalases and peroxidases that can all contribute to peroxide removal and indeed we find that single mutations in the corresponding cta1, ctt1, ahp1, dot5, prx1, and tsa1 genes were not sufficient to stabilize Yck1p under low glucose conditions (Figure S6E). In total, our data indicate local peroxide produced by Sod1p at the site of Yck1p not only escapes being quenched by the various peroxide scavenging enzymes in the cell, but also circumvents the requirement for high non-physiological doses of H2O2 to stabilize Yck1p.
Discussion
In many aerobic organisms, ambient O2 promotes respiration, whereas loss of O2 favors fermentation (Pasteur, 1861). However, in proliferating cells, aerobic glycolysis is frequently chosen over respiration as a source for energy irrespective of O2 (Lunt and Vander Heiden, 2011). In many instances, glucose itself can repress respiration (Crabtree, 1929) through mechanisms that are not completely understood. In this study, we provide an unprecedented role for O2 itself as a repressor of respiration. Our data demonstrates that O2, glucose and the antioxidant enzyme Cu/Zn SOD1 cooperate to maintain stability of Yck1p and Yck2p, two CK1γ casein kinases for respiratory repression. SOD1 binds to a degron containing region we have identified in Yck1p residues 367–412, and prevents degradation of the kinase through a mechanism involving lysines at the Yck1p C-terminus. Our data supports a model in which the superoxide generated during growth in the presence of glucose and O2 feeds into cytosolic SOD1 and the concomitant production of H2O2 helps stabilize the CK1γ kinases for nutrient signaling (Figure 7D).
Glucose has long been known to repress respiration in proliferating cells, a phenomenon known as the Crabtree effect (Crabtree, 1929). Although the molecular basis of this effect is not well understood, it has previously been proposed that increased glycolytic flux plays an important role (Diaz-Ruiz et al., 2011). Indeed many yeast mutants defective in glucose repression (such as the snf3Δ rgt2 Δ mutant; (Figure S1D) (Ozcan et al., 1998)) show a loss in glucose uptake correlating with the increase in cell respiration. However, the role of SOD1/CK1γ in glucose repression described here occurs independent of any changes in glucose uptake. The control of aerobic fermentation and respiration by Sod1p/Yck1p may represent a novel element of the Crabtree effect.
The role of SOD1 in oxidative stress protection has been thoroughly characterized since its discovery more than 40 years ago (McCord and Fridovich, 1969). However, only a small fraction of total SOD1 enzyme appears required for protection against defined oxidative insults (Corson et al., 1998). SOD1 has also been shown to participate in the buffering of toxic copper ions (Culotta et al., 1995) and protecting the calcineurin phosphatase from oxidative damage (Wang et al., 1996). Our newly uncovered role of SOD1 in casein kinase signaling is independent of these effects and can be extended to mammalian cells. We find that Cu/Zn SODs from nematodes and humans can stabilize yeast Yck1p, and conversely, yeast Sod1p can stabilize bovine casein kinase 1 gamma-3 (CK1γ3). Furthermore, we find that SOD1 activity promotes CK1γ expression in a human cell line. As with yeast CK1γ, mammalian CK1γ isoforms are involved in regulating fermentative metabolism, but through the Wnt signaling pathway (Lee et al., 2012; Sethi and Vidal-Puig, 2010), suggesting a conserved role for SOD1 in regulating CK1γ isoforms and fermentative metabolism. Why was SOD1 selected for this purpose? The answer may lie in its superoxide substrate, an ideal candidate molecule to link O2 and glucose to the repression of respiration. Given that SOD enzymes are the only enzymatic receptors for superoxide in eukaryotic cells, the exploitation of cytosolic Cu/Zn SOD1 to transduce superoxide signals from O2 and glucose is an intelligent repurposing of an antioxidant enzyme.
We provide evidence that the H2O2 product of the SOD1 reaction stabilizes the casein kinases. H2O2 produced by SOD1 has previously been shown to regulate tyrosine kinase receptor mediated processes and in this case, the targets are redox sensitive cysteines (Juarez et al., 2008). The SOD1-responsive C-terminal degron in Yck1p does not contain any cysteines but does contain a number of lysines that we report here are critical for SOD1/glucose/O2 regulation of Yck1p stability. As one possibility, ubiquitination of these lysines by H2O2 sensitive E3 ligases may lead to Yck1p turnover. Cysteine rich RING E3 ligases can be susceptible to oxidation (Meng et al., 2011), and E3 adaptors can also contain H2O2-reactive cysteines (Dinkova-Kostova et al., 2002; Zhang et al., 2004).
The implications for Sod1p regulation of casein kinases extend beyond repression of respiration. Yeast Yck1p/Yck2p participates in numerous nutrient signaling processes, e.g., amino acid sensing through the SPS pathway (Liu et al., 2008). We previously reported that the SPS pathway was down-regulated by hypoxia (Gleason et al., 2011), and we can now explain this phenomena by the hypoxia induced loss of Sod1p activity and Yck1p. The exact rationale for down regulation of Yck1p/Yck2p under hypoxia is not clear but may involve its multi-faceted roles in signaling, including amino acid sensing and other as-of-yet unknown targets of this regulatory kinase.
In conclusion, our studies provide a new role for an old antioxidant enzyme. Through its ability to bind and protect key casein kinases from degradation in an O2 and glucose dependent manner, SOD1 is a vital component of nutrient sensing pathways and is essential for repressing respiration. Taken together, our results suggest that SOD1 acts as a metabolic focal point for integrating O2, nutrients (glucose), and reactive oxygen (superoxide) to direct energy metabolism.
Experimental Procedures
General Considerations
All yeast strains and plasmids are listed, along with details on their genotype, source and/or construction, in the Supplemental Information. Yeast cultivation, cell culture, cell lysis, immunoblotting, immunoprecipitation, DCFDA determination of intracellular H2O2, and lacZ gene reporter assays, were conducted using standard methods and are described in detail in the Supplemental Information.
Experiments involving anaerobic growth and manipulation were conducted under an atmosphere of 95% N2 and 5% H2 in a COY Chamber (COY Laboratory Products, Inc., Grass Lake, Michigan, USA). All absorption and fluorescence measurements were recorded on a Biotek HT Synergy plate reader.
Measurement of O2 consumption and superoxide levels
O2 consumption was measured using the Becton Dickinson (BD) Oxygen Biosensor System according to the manufacturer’s instructions as described previously (Davidson et al., 2011) in mid-log phase cells grown in media enriched with yeast extract, peptone (YP) with the indicated glucose (GLU) or galactose (GAL) concentrations. Briefly, triplicate cultures of cells were inoculated into YP 2% GLU media and grown to a density of OD600nm = 1.0 (12–15 hours), re-diluted into fresh media (initial OD600nm = .15), and grown to a final OD600nm = 1.0. 200 μL of 5×107 to 2×108 cells/mL were collected and resuspended in fresh media, and O2 consumption was recorded as the fold increase in fluorescence (λex = 485nm, λem = 620nm) of wells containing cells relative to wells containing only media in BD Oxygen Biosensor System. Where indicated (Figures S1B), O2 consumption rates were calculated from the maximum slopes of the sigmoidal O2 consumption isotherms, and recorded as the change in the fold increase in fluorescence per minute, with the data normalized to WT cells. In cases where different GLU or GAL concentrations are utilized, cells were pre-cultured in triplicate in YP 2% GAL to a density of OD600nm ~ 1.0 (12–15 hours), and cultures were re-diluted into fresh media containing the indicated GLU or GAL concentration (initial OD600nm = .15) and allowed to grow to a final OD600nm = 1.0. O2 consumption was then measured as described above.
Superoxide levels were measured by monitoring the fluorescence of dihydroethidium (DHE) stained cells (λex = 485nm, λem = 620nm) similarly to what was described previously (Neklesa and Davis, 2008). Briefly, 1×107 cells were harvested from duplicate or triplicate cultures, resuspended and incubated in 500 μL of fresh media containing 50 μM DHE for 20 minutes in the dark, washed twice with phosphate buffered saline (PBS) solution, and fluorescence recorded. For measurements of anaerobic superoxide levels, staining and washing was carried out in an anaerobic COY Chamber, then rapidly removed for analysis on the plate reader (in air). The minimal superoxide detected in anaerobic cultures (as in Figure 6D) may reflect production during this short exposure to air. In instances where DHE staining is conducted on cells switched from O2 to N2 or GLU to GAL, cells were harvested at designated time points and subjected to DHE staining for 20 minutes prior to analysis.
Biochemical Assays
For Pma1p and casein kinase (CK) activity measurements, duplicate 1.0 L YP 2% or .2% GLU cultures were grown as described above for measuring O2 consumption. Plasma membranes (PM) isolated according to published procedures (Perlin et al., 1989) were used for both Pma1p ATPase and CK activity measurements. CK activity was also assayed in whole cell lysates prepared as described in Supplemental Information. Pma1p ATPase activity was assayed in membrane fractions with or without a 5-min preincubation with 50 μM vanadate. ATPase activity was determined by quantitating the phosphate released during vanadate-sensitive ATP hydrolysis by molybdate reactivity (Perlin et al., 1989) and activity was normalized to WT cells grown in 2% GLU. CK activity was assayed using the CSNK1G3 Z′-LYTE Kinase Ser/Thr 5 Peptide Assay kit (Invitrogen) according to the manufacturer’s instructions. Activity reflected the change in ratio of fluorescence emission (λex = 360nm, λem1 = 460nm, λem2 = 528) correlating with the degree of CK1 peptide substrate phosphorylation. Activity was recorded as % phosphorylation of substrate per mg of whole cell or PM lysate protein.
SOD activity analysis was carried out by native PAGE and nitroblue tetrazolium staining as described previously (Flohe and Otting, 1984; Luk et al., 2005) on mid-log phase cultures grown to a final OD600nm = 1.0 in YP 4% GLU. Glucose consumption of triplicate mid-log phase WT or sod1Δ cells was monitored by measuring extracellular glucose concentration as a function of culture density using the Quantichrome Glucose Assay Kit (BioAssay Systems).
Supplementary Material
Highlights.
Glucose and O2 signal through Cu/Zn SOD1 to repress respiration.
SOD1 binds to and prevents turnover of a casein kinase CK1γ for respiration control.
Superoxide from aerobic glucose metabolism feeds the SOD1 reaction to stabilize CK1γ.
Both human and yeast CK1γ stability are regulated by SOD1.
Acknowledgments
We thank S. Kavianpour, C. Vazquez, J. Gleason, D. Corrigan, and H. Chen for technical assistance and Professors M. Johnston, D. Thiele, L. Robinson, and J.-H. Kim for strains and plasmids. This work was supported by the JHU NIEHS center and by NIH R37 grant GM50016 to VCC. ARR was supported by NIH NIGMS fellowship F32 GM 093550.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguirre JD, Culotta VC. Battles with iron: manganese in oxidative stress protection. J Biol Chem. 2012;287:13541–13548. doi: 10.1074/jbc.R111.312181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez HM, Xue Y, Robinson CD, Canalizo-Hernandez MA, Marvin RG, Kelly RA, Mondragon A, Penner-Hahn JE, O’Halloran TV. Tetrathiomolybdate inhibits copper trafficking proteins through metal cluster formation. Science. 2010;327:331–334. doi: 10.1126/science.1179907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnese K, Gralla EB, Valentine JS, Cabelli DE. Biologically relevant mechanism for catalytic superoxide removal by simple manganese compounds. Proc Natl Acad Sci U S A. 2012;109:6892–6897. doi: 10.1073/pnas.1203051109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensinger SJ, Christofk HR. New aspects of the Warburg effect in cancer cell biology. Semin Cell Dev Biol. 2012;23:352–361. doi: 10.1016/j.semcdb.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Bilinski T, Krawiec Z, Liczmanski L, Litwinska J. Is hydroxyl radical generated by the fenton reaction in vivo? Biochem Biophys Res Comm. 1985;130:533–539. doi: 10.1016/0006-291x(85)90449-8. [DOI] [PubMed] [Google Scholar]
- Brown NM, Torres AS, Doan PE, O’Halloran TV. Oxygen and the copper chaperone CCS regulate posttranslational activation of Cu,Zn superoxide dismutase. Proc Natl Acad Sci U S A. 2004;101:5518–5523. doi: 10.1073/pnas.0401175101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang E, Kosman D. O2-dependent methionine auxotrophy in Cu,Zn Superoxide dismutase deficient mutants of Saccharomyces cerevisiae. J Bacteriol. 1990;172:1840–1845. doi: 10.1128/jb.172.4.1840-1845.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang EC, Kosman DJ. Intracellular Mn(II)-associated superoxide scavenging activity protects Cu,Zn superoxide dismutase-deficient Saccharomyces cerevisiae against dioxygen stress. J Biol Chem. 1989;264:12172–12178. [PubMed] [Google Scholar]
- Corson LB, Strain J, Culotta VC, Cleveland DW. Chaperone-facilitated copper binding is a property common to several classes of familial amyotrophic lateral sclerosis-linked superoxide dismutase mutants. Proc Natl Acad Sci USA. 1998;95:6361–6366. doi: 10.1073/pnas.95.11.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree HG. Observations on the carbohydrate metabolism of tumours. Biochem J. 1929;23:536–545. doi: 10.1042/bj0230536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culotta VC, Joh HD, Lin SJ, Slekar KH, Strain J. A physiological role for Saccharomyces cerevisiae copper/zinc superoxide dismutase in copper buffering. J Biol Chem. 1995;270:29991–29997. doi: 10.1074/jbc.270.50.29991. [DOI] [PubMed] [Google Scholar]
- Culotta VC, Klomp L, Strain J, Casareno R, Krems B, Gitlin JD. The copper chaperone for superoxide dismutase. J Biol Chem. 1997;272:23469–23472. doi: 10.1074/jbc.272.38.23469. [DOI] [PubMed] [Google Scholar]
- Davidson GS, Joe RM, Roy S, Meirelles O, Allen CP, Wilson MR, Tapia PH, Manzanilla EE, Dodson AE, Chakraborty S, et al. The proteomics of quiescent and nonquiescent cell differentiation in yeast stationary-phase cultures. Mol Biol Cell. 2011;22:988–998. doi: 10.1091/mbc.E10-06-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Ruiz R, Rigoulet M, Devin A. The Warburg and Crabtree effects: On the origin of cancer cell energy metabolism and of yeast glucose repression. Biochim Biophys Acta. 2011;1807:568–576. doi: 10.1016/j.bbabio.2010.08.010. [DOI] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elchuri S, Oberley TD, Qi W, Eisenstein RS, Jackson Roberts L, Van Remmen H, Epstein CJ, Huang TT. CuZnSOD deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene. 2005;24:367–380. doi: 10.1038/sj.onc.1208207. [DOI] [PubMed] [Google Scholar]
- Estrada E, Agostinis P, Vandenheede JR, Goris J, Merlevede W, Francois J, Goffeau A, Ghislain M. Phosphorylation of yeast plasma membrane H+-ATPase by casein kinase I. J Biol Chem. 1996;271:32064–32072. doi: 10.1074/jbc.271.50.32064. [DOI] [PubMed] [Google Scholar]
- Flohe L, Otting F. Superoxide dismutase assays. In: Packer L, editor. Methods in enzymology: oxygen radicals in biological systems. New York: Academic press; 1984. pp. 93–104. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Giacomini A, corich V, Ollero FJ, Squartini A, Nuti MP. Experimental conditions may affect reproducibility of the beta-galactosidase assay. FEMS Microbiol Lett. 1992;79:87–90. doi: 10.1111/j.1574-6968.1992.tb14024.x. [DOI] [PubMed] [Google Scholar]
- Gleason JE, Corrigan DJ, Cox JE, Reddi AR, McGinnis LA, Culotta VC. Analysis of hypoxia and hypoxia-like states through metabolite profiling. PloS one. 2011;6:e24741. doi: 10.1371/journal.pone.0024741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralla E, Valentine JS. Null mutants of Saccharomyces cerevisiae Cu,Zn superoxide dismutase: characterization and spontaneous mutation rates. J Bacteriol. 1991;173:5918–5920. doi: 10.1128/jb.173.18.5918-5920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC. Free Radicals in Biology and Medicine. 4. Oxford Biosciences; 2007. [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Ho Y, Mason S, Kobayashi R, Hoekstra M, Andrews B. Role of the casein kinase I isoform, Hrr25, and the cell cycle-regulatory transcription factor, SBF, in the transcriptional response to DNA damage in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1997;94:581–586. doi: 10.1073/pnas.94.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez JC, Manuia M, Burnett ME, Betancourt O, Boivin B, Shaw DE, Tonks NK, Mazar AP, Donate F. Superoxide dismutase 1 (SOD1) is essential for H2O2-mediated oxidation and inactivation of phosphatases in growth factor signaling. Proc Natl Acad Sci U S A. 2008;105:7147–7152. doi: 10.1073/pnas.0709451105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Jeon HM, Ju MK, Kim CH, Yoon G, Han SI, Park HG, Kang HS. Wnt/Snail signaling regulates cytochrome C oxidase and glucose metabolism. Cancer Res. 2012;72:3607–3617. doi: 10.1158/0008-5472.CAN-12-0006. [DOI] [PubMed] [Google Scholar]
- Leitch JM, Jensen LT, Bouldin SD, Outten CE, Hart PJ, Culotta VC. Activation of Cu,Zn-superoxide dismutase in the absence of oxygen and the copper chaperone CCS. J Biol Chem. 2009;284:21863–21871. doi: 10.1074/jbc.M109.000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch JM, Li CX, Baron JA, Matthews LM, Cao X, Hart PJ, Culotta VC. Post-translational modification of Cu/Zn superoxide dismutase under anaerobic conditions. Biochemistry. 2012;51:677–685. doi: 10.1021/bi201353y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Thornton J, Spirek M, Butow RA. Activation of the SPS amino acid-sensing pathway in Saccharomyces cerevisiae correlates with the phosphorylation state of a sensor component, Ptr3. Mol Cell Biol. 2008;28:551–563. doi: 10.1128/MCB.00929-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk E, Yang M, Jensen LT, Bourbonnais Y, Culotta VC. Manganese activation of superoxide dismutase 2 in the mitochondria of Saccharomyces cerevisiae. J Biol Chem. 2005;280:22715–22720. doi: 10.1074/jbc.M504257200. [DOI] [PubMed] [Google Scholar]
- Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- McCord JM, Fridovich I. Superoxide dismutase: An enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- Meng F, Yao D, Shi Y, Kabakoff J, Wu W, Reicher J, Ma Y, Moosmann B, Masliah E, Lipton SA, et al. Oxidation of the cysteine-rich regions of parkin perturbs its E3 ligase activity and contributes to protein aggregation. Mol Neurodegener. 2011;6:34. doi: 10.1186/1750-1326-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya H, Johnston M. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc Natl Acad Sci U S A. 2004;101:1572–1577. doi: 10.1073/pnas.0305901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe W, Kingsley C, Durazo A, Gralla EB, Imlay JA, Srinivasan C, Valentine JS. Only one of a wide assortment of manganese-containing SOD mimicking compounds rescues the slow aerobic growth phenotypes of both Escherichia coli and Saccharomyces cerevisiae strains lacking superoxide dismutase enzymes. J Inorg Biochem. 2007;101:1875–1882. doi: 10.1016/j.jinorgbio.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neklesa TK, Davis RW. Superoxide anions regulate TORC1 and its ability to bind Fpr1:rapamycin complex. Proc Natl Acad Sci U S A. 2008;105:15166–15171. doi: 10.1073/pnas.0807712105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okado-Matsumoto A, Fridovich I. Subcellular distribution of superoxide dismutases (SOD) in rat liver: Cu,Zn-SOD in mitochondria. J Biol Chem. 2001;276:38388–38393. doi: 10.1074/jbc.M105395200. [DOI] [PubMed] [Google Scholar]
- Ozcan S, Dover J, Johnston M. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 1998;17:2566–2573. doi: 10.1093/emboj/17.9.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo CA, Xu Z, Borchelt DR, Price DL, Sisodia SS. Superoxide dismutase is an abundant component in cell bodies, dendrites, and axons of motor neurons and in a subset of other neurons. Proc Natl Acad Sci USA. 1995;92:954–958. doi: 10.1073/pnas.92.4.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasteur L. Expériences et vues nouvelles sur la nature des fermentations. Compt Rend Acad Sci. 1861;52:1260–1264. [Google Scholar]
- Pasula S, Chakraborty S, Choi JH, Kim JH. Role of casein kinase 1 in the glucose sensor-mediated signaling pathway in yeast. BMC Cell Biol. 2010;11:17. doi: 10.1186/1471-2121-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlin DS, Harris SL, Seto-Young D, Haber JE. Defective H(+)-ATPase of hygromycin B-resistant pma1 mutants fromSaccharomyces cerevisiae. J Biol Chem. 1989;264:21857–21864. [PubMed] [Google Scholar]
- Reddi AR, Culotta VC. Regulation of manganese antioxidants by nutrient sensing pathways in Saccharomyces cerevisiae. Genetics. 2011;189:1261–1270. doi: 10.1534/genetics.111.134007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinnerthaler M, Buttner S, Laun P, Heeren G, Felder TK, Klinger H, Weinberger M, Stolze K, Grousl T, Hasek J, et al. Yno1p/Aim14p, a NADPH-oxidase ortholog, controls extramitochondrial reactive oxygen species generation, apoptosis, and actin cable formation in yeast. Proc Natl Acad Sci U S A. 2012;109:8658–8663. doi: 10.1073/pnas.1201629109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson LC, Hubbard EJ, Graves PR, DePaoli-Roach AA, Roach PJ, Kung C, Haas DW, Hagedorn CH, Goebl M, Culbertson MR, et al. Yeast casein kinase I homologues: an essential gene pair. Proc Natl Acad Sci U S A. 1992;89:28–32. doi: 10.1073/pnas.89.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth AF, Papanayotou I, Davis NG. The yeast kinase Yck2 has a tripartite palmitoylation signal. Mol Biol Cell. 2011;22:2702–2715. doi: 10.1091/mbc.E11-02-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehati S, Clement MH, Martins J, Xu L, Longo VD, Valentine JS, Gralla EB. Metabolic alterations in yeast lacking copper-zinc superoxide dismutase. Free Radic Biol Med. 2011;50:1591–1598. doi: 10.1016/j.freeradbiomed.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi JK, Vidal-Puig A. Wnt signalling and the control of cellular metabolism. Biochem J. 2010;427:1–17. doi: 10.1042/BJ20091866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtz LA, Culotta VC. Superoxide dismutase null mutants of the bakers yeast Saccharomyces cerevisiae. Meths Enzymol. 2002;349:167–172. doi: 10.1016/s0076-6879(02)49332-9. [DOI] [PubMed] [Google Scholar]
- Sturtz LA, Diekert K, Jensen LT, Lill R, Culotta VC. A Fraction of Yeast Cu,Zn-Superoxide Dismutase and Its Metallochaperone, CCS, Localize to the Intermembrane Space of Mitochondria. J Biol Chem. 2001;276:38084–38089. doi: 10.1074/jbc.M105296200. [DOI] [PubMed] [Google Scholar]
- Sun B, Chen L, Cao W, Roth AF, Davis NG. The yeast casein kinase Yck3p is palmitoylated, then sorted to the vacuolar membrane with AP-3-dependent recognition of a YXXPhi adaptin sorting signal. Mol Biol Cell. 2004;15:1397–1406. doi: 10.1091/mbc.E03-09-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpey MM, Fridovich I. Methods of detection of vascular reactive species: nitric oxide, superoxide, hydrogen peroxide, and peroxynitrite. Circ Res. 2001;89:224–236. doi: 10.1161/hh1501.094365. [DOI] [PubMed] [Google Scholar]
- Wang X, Culotta VC, Klee CB. Superoxide dismutase protects calcineurin from inactivation. Nature. 1996;271:28831–28836. doi: 10.1038/383434a0. [DOI] [PubMed] [Google Scholar]
- Weisiger RA, Fridovich I. Mitochondrial superoxide dismutase. J Biol Chem. 1973;248:4793–4796. [PubMed] [Google Scholar]
- White C, Kambe T, Fulcher YG, Sachdev SW, Bush AI, Fritsche K, Lee J, Quinn TP, Petris MJ. Copper transport into the secretory pathway is regulated by oxygen in macrophages. J Cell Sci. 2009;122:1315–1321. doi: 10.1242/jcs.043216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LK, Thiele DJ. Transcriptional activation in yeast in response to copper deficiency involves copper-zinc superoxide dismutase. J Biol Chem. 2009;284:404–413. doi: 10.1074/jbc.M807027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CY, Steffen J, Eide DJ. Cytosolic superoxide dismutase (SOD1) is critical for tolerating the oxidative stress of zinc deficiency in yeast. PloS one. 2009;4:e7061. doi: 10.1371/journal.pone.0007061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
- Zhai L, Graves PR, Robinson LC, Italiano M, Culbertson MR, Rowles J, Cobb MH, DePaoli-Roach AA, Roach PJ. Casein kinase I gamma subfamily. Molecular cloning, expression, and characterization of three mammalian isoforms and complementation of defects in the Saccharomyces cerevisiae YCK genes. J Biol Chem. 1995;270:12717–12724. doi: 10.1074/jbc.270.21.12717. [DOI] [PubMed] [Google Scholar]
- Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.