Abstract
Agrobacterium-mediated transformation is being increasingly used for insertional mutagenesis of fungi. To better evaluate its effectiveness as a mutagen for the fungal pathogen Histoplasma capsulatum, we analyzed a collection of randomly selected T-DNA insertion mutants. Testing of different T-DNA element vectors engineered for transformation of fungi showed that pBHt2 provides the highest transformation efficiency and the lowest rate of vector backbone carryover. Sixty-eight individual T-DNA integrations were characterized by recovery of T-DNA ends and flanking genomic sequences. The right border end of the T-DNA is largely preserved whereas the left border end is frequently truncated. Analysis of T-DNA insertion sites confirms the lack of any integration hotspots in the Histoplasma genome. Relative to genes, T-DNA integrations show significant bias towards promoter regions at the expense of coding sequences. With consideration for potential promoter interruption and the demonstrated efficacy of intronic insertions, 61% of mapped T-DNA insertions should impair gene expression or function. Mapping of T-DNA flanking sequences demonstrates 67% of T-DNA integrations are integrations at a single chromosomal site and 31% of T-DNA integrations are associated with large-scale chromosomal rearrangements. This characterization of T-DNA insertions in mutants selected without regard to phenotype supports application of Agrobacterium-mediated transformation as an insertional mutagen for genome-based screens and functional discovery of genes in Histoplasma.
Keywords: Histoplasma, Agrobacterium, insertional mutagenesis, T-DNA, TAIL-PCR
1. INTRODUCTION
The fungal pathogen Histoplasma capsulatum, common to the United States Midwest and many areas of Latin America, causes respiratory and systemic disease in both immunocompromised and immunocompetent individuals. Infection results from inhalation of conidia, which differentiate into yeasts within the mammalian lung. Histoplasma yeast cells parasitize resident macrophages by surviving phagocyte antimicrobial defenses and persisting within a modified intracellular compartment (Woods 2003). Studies have only recently begun to elucidate the mechanisms underlying Histoplasma’s success as an intracellular pathogen due, in part, to limited genetics (Rappleye and Goldman 2006). Disruption of gene function is essential to demonstrate causal roles for gene products in Histoplasma biology. Although allelic replacement methodology was developed for Histoplasma a little over a decade ago (Sebghati et al. 2000), the process remains relatively inefficient prohibiting construction of large-scale, genome-wide mutant collections for phenotypic screening. This lack of facile genetic methodologies has hampered virulence gene discovery efforts in Histoplasma.
For many fungi that lack efficient homologous recombination, Agrobacterium tumefaciens-mediated transformation has emerged as an effective mutagen (Frandsen 2011; Michielse et al. 2005). The plant-pathogenic bacterium Agrobacterium tumefaciens can transfer a segment of DNA known as T-DNA into a host genome and this has been widely employed to create insertion mutants in a variety of plant species (Tzfira and Citovsky 2006). Cis-acting, imperfect 25-base pair direct repeat sequences, termed the left and right borders (LB and RB), flank the transforming DNA segment and direct the T-DNA excision and transfer machinery (Gelvin 2003; Tzfira et al. 2004). Engineering of the T-DNA element to include selectable markers for fungi between the LB and RB broadens the application of Agrobacterium-mediated transformation to members of the fungal kingdom. Agrobacterium-mediated transformation has been employed to introduce foreign DNA into over 100 fungi (Frandsen 2011; Michielse et al. 2005) including Histoplasma capsulatum (Edwards et al. 2011b; Hilty et al. 2008; Laskowski and Smulian 2010; Marion et al. 2006; Nguyen and Sil 2008; Sullivan et al. 2002; Webster and Sil 2008; Youseff et al. 2009).
Integration of T-DNA into fungal chromosomes has promise as an insertional mutagen to facilitate forward genetic mutant screens. Similar to transposon mutagenesis in bacteria, Agrobacterium-mediated transformation and integration of T-DNA is advantageous over chemical mutagens since the mutations caused by T-DNA integration are tagged with a known sequence. This sequence anchor is essential to determine the mutation site in fungal organisms that lack classical genetic mapping methodologies such as Histoplasma. To enable mapping of disrupted loci, the molecular tag (i.e., T-DNA sequence) needs to be transferred and integrated with relatively high fidelity; imprecise excision of the T-DNA or truncation of the TDNA ends on integration frustrates mapping approaches that rely on the T-DNA sequence as a molecular anchor (e.g., TAIL-PCR, inverse-PCR, etc.). In addition, imprecise or truncated ends can cause loss of selection markers or loss of reporter genes in enhancer-trap screens. Signature-tagged mutagenesis enables mutant screens based on negative selection (Hensel et al. 1995), which has greatly aided virulence gene discovery, particularly for phenotype screens that rely on infection of a limited number of host animals. Various integrating elements (e.g., transposons) designed with outward-directed promoters allow determination of the genome sequence flanking the inserted element by in vitro transcription and hybridization of the RNA to a genomic microarray (Sassetti et al. 2001; Winterberg et al. 2005). Variations of this technique in bacteria using different transposable elements have accelerated identification of genes required for infection (Chan et al. 2005; Chaudhuri et al. 2009; Lawley et al. 2006; Rengarajan et al. 2005; Santiviago et al. 2009). Minimal end truncation or extension becomes even more important with application of next-generation sequencing advancements such as Tn-seq (van Opijnen et al. 2009) or INseq (Goodman et al. 2009), which require restriction enzyme cutting at defined distances from the end for library construction. Adaptation of these large-scale mutagenesis techniques to pathogenic fungi using Agrobacterium-mediated transformation requires transfer of the T-DNA with enough precision to maintain the element ends.
The primary objective of genome-wide mutagenesis projects is to generate mutations in the maximal number of genes. Construction of large mutant libraries representing an organism’s full gene collection requires T-DNA integrations are randomly distributed in the genome with minimal bias in target site and the absence of insertional hot spots. Early studies in Histoplasma suggested Agrobacterium-mediated transformation caused single T-DNA insertions with relatively random distribution (Sullivan et al. 2002). However, the size of this survey was insufficient to exclude insertional hot spots, and without sequence-level information, the lack of preferred genetic target regions for integration can not be determined. Other examples of Agrobacterium-mediated transformation of Histoplasma have reported on T-DNA mutants selected for specific phenotypes and thus do not represent an unbiased assessment of T-DNA transfer and sites of integration (Edwards et al. 2011b; Hilty et al. 2008; Marion et al. 2006; Nguyen and Sil 2008; Smulian et al. 2007; Webster and Sil 2008).
To determine the utility and feasibility of Agrobacterium-mediated transformation for construction of genome-wide insertion mutant libraries in Histoplasma, we generated a set of T-DNA insertion mutants and characterized the integration events at the DNA sequence level. Insertion mutants were not selected for a particular phenotype thereby providing an impartial, systematic survey of insertion site distribution, integration patterns, and T-DNA composition. In addition, we identified an optimal T-DNA element for Histoplasma mutagenesis that is transferred with good precision and high efficiency. These analyses establish T-DNA integration as an effective insertional mutagen for Histoplasma genetic studies and identify the caveats that should be considered.
2. MATERIALS AND METHODS
2.1 Histoplasma strain and growth conditions
The Histoplasma capsulatum strains used in this study were derived from WU15, a uracil auxotroph (ura5-42Δ) of the clinical isolate G217B (ATCC #26032) (Marion et al. 2006). Other strains in this study were constructed in the WU15 background by Agrobacterium-mediated transformation and included OSU8 (cbp1-9::T-DNA; (Youseff et al. 2009)) and OSU128 (cbp1-11::T-DNA), both with T-DNA insertions at the CBP1 locus that encodes the secreted calcium-binding protein (Batanghari et al. 1998). Histoplasma cells were grown as yeasts by culture in Histoplasma-macrophage medium (HMM; (Worsham and Goldman 1988)) supplemented with uracil (100 µg/mL) at 37°C with shaking (200 rpm) in 5% CO2 / 95% air. For platings, HMM was solidified with 0.6% agarose and supplemented with 25 µM FeSO4. Transformed yeast cells were selected with 100 μg/mL hygromycin B.
2.2 Agrobacterium tumefaciens-mediated transformation
Agrobacterium tumefaciens was used to transform Histoplasma yeasts using modifications to previously described protocols (Zemska and Rappleye 2012). A. tumefaciens strain LBA1100 (Beijersbergen et al. 1992) was transformed by electroporation with T-DNA vectors pCM41 (Marion et al. 2006), pBHt2 (Mullins et al. 2001), or pBS03 (an engineered plasmid containing a hygromycin resistance cassette expressed from a 650 base pair promoter fragment from the Histoplasma TEF1 gene). A. tumefaciens strains harboring T-DNA vectors were grown in media containing 100 μg/mL kanamycin and 250 μg/mL spectinomycin to select for the T-DNA and Ti plasmids, respectively.
Preparation of bacteria for transformation of Histoplasma was performed by suspending plate-grown bacteria in liquid minimal glucose medium (Zemska and Rappleye 2012) with antibiotics. Following overnight growth (approximately 16 hours) at 25°C with shaking (200 rpm), bacterial cells were collected by centrifugation (5 minutes at 5000 × g), resuspended in 3 to 4 times the original culture volume in induction medium (Zemska and Rappleye 2012), and cultured for 6–8 hours at 25°C with shaking (200 rpm). Induction med ium contains 0.5% glucose, antibiotics, and 200 μM acetosyringone and was buffered to pH 5.3 with 25 mM MES. The number of bacteria was estimated by optical density of the culture at 600 nm with OD600 = 1 corresponding to approximately 5×108 bacteria/mL. Histoplasma yeasts were harvested from solid HMM + uracil medium seeded 3 days earlier with 4×105 yeasts/cm2. Yeasts were collected by flooding plates with 5 mL sterile H2O and scraping with a sterile spreader. Yeasts were collected by centrifugation (1000 × g), resuspended in induction medium, and the concentration estimated by optical density of the suspension at 600 nm with OD600 = 1 corresponding to approximately 108 yeasts/mL.
A. tumefaciens-mediated transformation of Histoplasma yeast was performed by co-cultivation of bacteria and yeast cells. 1×107 bacteria were combined with 5×107 yeast and the mixture spread on Whatman #5 filter paper placed on top of solid Agrobacterium induction medium supplemented with 0.7 mM cystine and 100 μg/mL uracil. Plates were incubated for 48 hrs at 25°C after which filters were transferred to select ion medium (solid HMM supplemented with 100 μg/mL uracil, 100 μg/mL hygromycin B, and 10 μg/mL tetracycline) and incubated at 37°C with 5% CO2 / 95% air until Histoplasma transformants became visible (10–14 days). Histoplasma transformants were picked and passaged twice by single colony isolation on solid HMM with uracil (100 μg/mL) to separate yeast cells from residual bacteria. After passage, hygromycin resistance of the mutants was confirmed to validate the stability of the T-DNA integrations.
2.3 PCR characterization of T-DNA insertions
DNA was prepared from Histoplasma yeast by mechanical disruption of yeast cells with 500-μm diameter glass beads and extraction with phenol/CHCl3 (Youseff et al. 2009). Total nucleic acids were isolated by precipitation with ethanol and PCR was performed using 250 ng total nucleic acids as template. PCR amplicons for vector backbone tests (aph gene) were primed with oligos APH3-1 (AAGATACGGAAGGAATGTCTCCTGC) and APH3-2 (TTTCGCAATCCACATCGGCCAG). Tests for Histoplasma genomic DNA (MFS1 locus) were primed with oligos MFS1 (CCTATCTCCACAAGGCTAAGGC) and MFS2 (CCTCAGGTGATGCATCCTGTC). PCR products were separated by electrophoresis through 1.2% agarose and visualized by ethidium bromide staining. PCR tests for T-DNA concatamers were based on juxtaposed LB and RB ends and used outward-directed T-DNA primers from the LB end (LB11; CCAAAATCCAGTACTAAAATCCAGATCCCCCGA) and from the RB end (RB9; CCGCACCGATCGCCCTTCCCAACAG).
2.4 TAIL-PCR
Sequences flanking T-DNA insertions were obtained by thermal asymmetric-interlaced PCR (TAIL-PCR; (Liu and Whittier 1995)). Sequences flanking left T-DNA borders were amplified using primers LB11 and a degenerate sequence primer (LAD-1, -2, -3, or -4; (Liu and Chen 2007)). Sequences flanking right T-DNA borders were amplified using primers RB9 and a LAD primer. The primary TAIL-PCR was initiated with 20 high stringency cycles (63°C annealing) followed by low stringency annealing (25°C with slo w ramping to 72°C). Amplification cycling was performed as 15 cycles of mixed high and low stringency (two PCR cycles at 65°C annealing followed by one cycle at 40°C annealing). The primary PCR was diluted and used as template at 1:1000 for secondary TAIL-PCR with nested PCR primers LB12 (CGGCGTTAATTCAGTACATTAAAAACGTCCGCA) or RB10 (GCCTGAATGGCGAATGCTAGAGCAGCTTG) in conjunction with an oligo common to the LAD-primer 5’ tail (AC1 primer; (Liu and Chen 2007)). Secondary TAIL-PCR consisted of 15 cycles of mixed high and low stringency as for the primary TAIL-PCR. PCR amplicons were prepared for sequencing by treatment with ExoSap (US Biologicals) and amplicons were sequenced using primers LB14 (ACGTCCGCAATGTGTTATTAAG) or RB11 (GCTAGAGCAGCTTGAGCTTGGATCAGATTGTC) for the left and right borders, respectively. Primer-proximal sequence was matched to T-DNA sequence to verify the TAIL-PCR amplicon was anchored in the T-DNA element.
2.5 Determination of the genome context of T-DNA integrations
Sequences flanking the T-DNA element were matched to the Histoplasma G217B genomic sequence by BLAST search of the Histoplasma G217B genome (Histobase; http://histo.ucsf.edu). Multiple BLAST matches to the G217B genome with e-values greater than e−20 were designated as integrations in repetitive DNA. For determination of integration site bias, the following genome statistics were derived from the genome sequence (http://genome.wustl.edu/genomes/view/histoplasma_capsulatum) and gene predictions: 39902878 total base pairs, 12560871 base pairs repetitive DNA, and 11751 genes comprised of 13937861 base pairs. 9400800 base pairs for promoter regions and 2350500 base pairs for terminators were calculated from the 11751 total genes using estimates of 800 base pairs and 200 base pairs for promoter and terminator/3' UTR regions, respectively. Integration locations were considered individually for the LB and the RB flanking sequences for each mapped insertion. Expected values for randomly distributed integrations were based on the probability of either a LB-flanking sequence or a RB-flanking sequence matching a given region of interest, the size of which was calculated from estimates using the above genome statistics. Chi-square analysis was performed for each genome context class using binary classifications for integration sites (fraction of flanking sequences within the region of interest compared to the total fraction of those outside that region).
2.6 Analysis of calcium-binding protein (Cbp1) production
Culture filtrates were prepared from liquid cultures of Histoplasma yeasts grown to late exponential phase in HMM + uracil (100 μg/mL). Yeasts were removed by centrifugation (2 minutes at 5000 × g) and the supernatant clarified by passage through 0.2 μm-diameter pore polyethersulfone membranes. Culture filtrates were normalized with OD600 readings of the yeast cultures before isolation of supernatants. Secreted proteins were separated by electrophoresis of volume equivalents through 12% Tris-glycine SDS polyacrylamide gels. Silver staining was used to visualize separated proteins.
3. RESULTS
3.1 Identification of the optimal T-DNA plasmid for Histoplasma mutagenesis
The effectiveness of T-DNA as an insertional mutagen relies upon the efficiency of transformation. To maximize the number of mutants recovered following Agrobacterium-mediated transformation of Histoplasma, we characterized different T-DNA elements that differ in both the sequences carried between the left and right borders as well as the binary plasmid backbone. The T-DNA plasmids were moved into the same C58 background Agrobacterium strain LBA1100 to eliminate any variation in transformation frequency due to the bacterial host. Successful transformation of Histoplasma has been achieved using the T-DNA carried on plasmid pCM41, which is derived from plasmid pCB301 and carries a hygromycin resistance gene expressed from the Aspergillus GAPDH promoter (Marion et al. 2006). As relatively low numbers of transformants were obtained using this T-DNA element (Table 1), we constructed a T-DNA element using a Histoplasma-native promoter and terminator to drive expression of the hygromycin resistance. The Histoplasma translation elongation factor 1 (TEF1) promoter and the putative terminator of the CATB gene were inserted upstream and downstream, respectively of the hygromycin resistance cassette to generate the T-DNA plasmid pBS03. Replacement of the transcriptional control elements with Histoplasma sequences increased the number of recovered transformants six fold (Table 1) and this correlates with enhanced transcription efficiency of the Histoplasma-promoter compared to the Aspergillus GAPDH promoter (data not shown). We also tested the Histoplasma transformation efficiency using Agrobacterium carrying the T-DNA plasmid pBHt2 (Mullins et al. 2001). Plasmid pBHt2 is based on the pCAMBIA plasmid backbone and expresses the hygromycin resistance gene from the Aspergillus trpC promoter (Figure 1A). Sequencing of the T-DNA element on pBHt2 indicated that the hygromycin-resistance cassette is oriented between the LB and RB, opposite to the orientation originally reported (Mullins et al. 2001). The T-DNA element carried on plasmid pBHt2 produced a higher transformation rate of Histoplasma yeasts than plasmid pCM41 (Table 1), and was similar to the transformation efficiency of the T-DNA element harboring the Histoplasma promoter.
Table 1.
Transformation efficiency of T-DNA vectors
| T-DNA vector | transformantsa (per 5×107 yeast) |
vector backbone carryoverb |
|---|---|---|
| pCM41 | 14.0 ± 6.8 (n=5) | 12.5% (n=32) |
| pBS03 | 87.2 ± 24.5 (n=5)* | 75.0% (n=40) |
| pBHt2 | 79.5 ± 20.2 (n=5)* | 8.3% (n=60) |
average ± SD (n=number of experiments), significant difference from pCM41 (P < 0.01) by one-tailed t-test indicated by asterisks
carryover frequency determined by positive aph PCR (n=number of transformants analyzed)
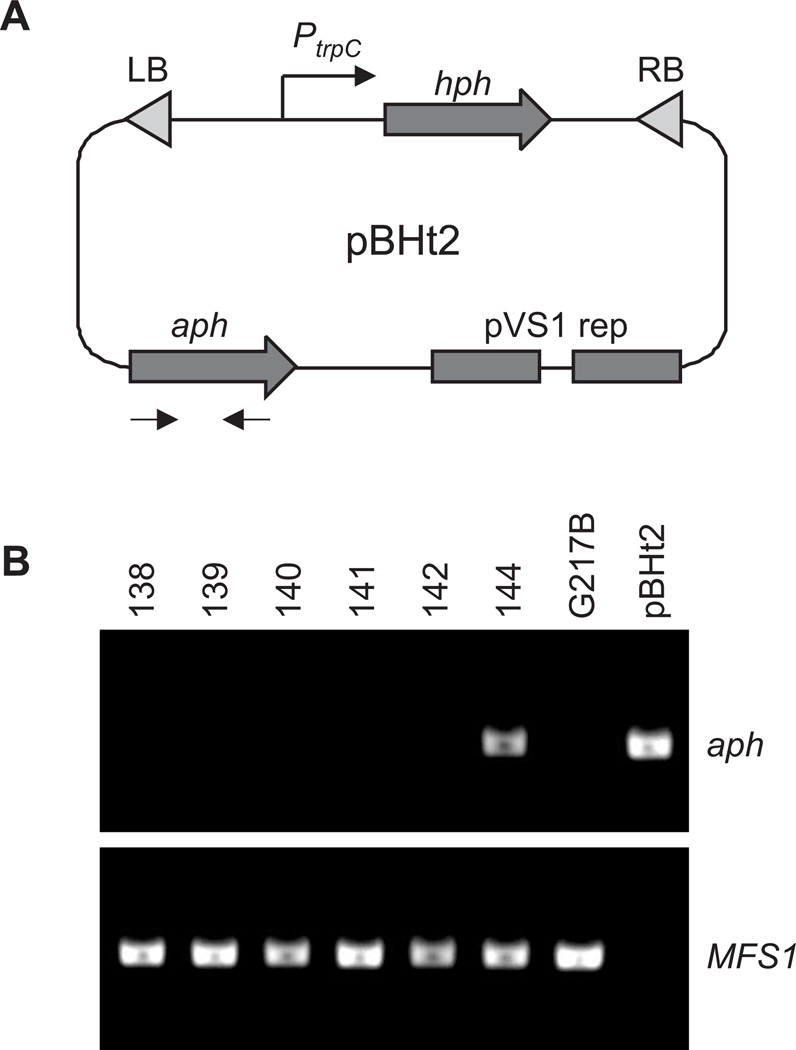
Figure 1. pBHt2 encodes a T-DNA element which can be used for transformation of Histoplasma yeast.
(A) Schematic of plasmid pBHt2 that carries a T-DNA element with left border (LB) and right border (RB) sequences flanking the hygromycin phosphotransferase gene (hph). The aminoglycoside phosphotransferase gene (aph) carried on the plasmid backbone provides for selection in Agrobacterium. Arrows denote primer locations used to screen for backbone carryover. (B) Representative PCR-based screen for plasmid backbone carryover in transformed Histoplasma lines. Genomic DNA was prepared from individual lines and screened by PCR for the presence of the aph gene to indicate the T-DNA plasmid backbone and the Histoplasma MFS1 gene to indicate Histoplasma genomic DNA. Image shows screening results for transformant lines 138-144 with wild type (G217B) DNA and purified pBHt2 DNA as controls.
Gene-tagging with T-DNA requires a high degree of fidelity in the transfer and integration of the T-DNA to preserve the boundaries of the element which provide known sequence anchors required for discovery of genomic flanking regions. Preliminary analysis of mutants generated with the T-DNA from pCM41 showed some insertion events that resulted from integration of TDNA elements which did not terminate at the left and right borders but also included sequence derived from the pCM41 plasmid backbone. Such "backbone carryover" has been observed in Agrobacterium-mediated transformation of other fungal and plant species with over 50% of T-DNA transformants showing backbone carryover in some systems (De Buck et al. 2000; Idnurm et al. 2009; Kononov et al. 1997; Michielse et al. 2009; Shou et al. 2004; van der Graaff et al. 1996; Wenck et al. 1997; Zhang et al. 2008). To determine the frequency of backbone carryover for the different T-DNA plasmids used in Histoplasma, we developed a simple PCR test for the aph gene that is unique to the plasmid backbone outside of the region between the T-DNA LB and RB (Figure 1A and 1B). An additional PCR reaction was designed to amplify a Histoplasma ABC-type transporter gene (MFS1) as a control to validate the DNA isolated from transformants was competent for PCR amplification (Figure 1B). Testing of individual Histoplasma yeast lines derived from transformations with one of the three plasmids pCM41, pBS03, and pBHt2, showed that, although pBS03 provides a high frequency of transformation, it also had an unacceptably high rate of backbone carryover (Table 1). Agrobacterium harboring either plasmid pCM41 or pBHt2 produced much lower frequencies of backbone carryover (Table 1). Since pBHt2 combines the lowest rate of backbone carryover with a higher frequency of transformation, pBHt2 was selected as the optimal T-DNA plasmid for insertional mutagenesis of Histoplasma yeast.
3.2 Precision of T-DNA transfer and integration
To determine the integration characteristics and the randomness of T-DNA insertions in Histoplasma, we analyzed 70 individual Agrobacterium-mediated Histoplasma transformants generated with the T-DNA element carried on pBHt2. Vector backbone carryover was detected in 2 transformants by the aph PCR test. Insertion sites were determined for the remaining 68 transformants by amplification and sequencing of regions flanking the T-DNA element using TAIL-PCR. Flanking regions were successfully amplified for 59/68 left borders and 64/68 right borders (Table 2). PCR product sequences were first aligned to the T-DNA to assure the amplicons were legitimately anchored in the T-DNA element. Sequence beyond the T-DNA LB and RB ends was subsequently aligned to the Histoplasma genome sequence to identify the site of T-DNA insertion. Mapping results and genome coordinates for T-DNA insertions in individual mutant lines are detailed in Supplementary Table 1.
Table 2.
Characteristics of T-DNA integration in Histoplasma
| numbera | |||
|---|---|---|---|
| Left Border (LB) | TAIL-PCRb | 59 | (87%) |
| backbone carryoverc | 2 | (3%) | |
| truncation (base pairs)d | 0 – 45 | (9.0 ± 9.1) | |
| Right Border (RB) | TAIL-PCRb | 64 | (94%) |
| backbone carryoverc | 6 | (9%) | |
| truncation (base pairs)d | 0 – 8 | (0.4 ± 1.2) | |
| integration contexte | single copy DNA | 101 | (86%) |
| repetitive DNA | 16 | (14%) | |
| hotspotsf | 0 | (0%) | |
| integration characteristicsg | simple integrationsh | 33 | (67%) |
| large deletionsi | 1 | (2%) | |
| genome rearrangementsj | 15 | (31%) |
numbers represent the number of mutants and % of total characterized except for truncation numbers which represent the range and mean number of base pairs deleted
successful generation of TAIL-PCR products with sequencing anchored within the T-DNA element
T-DNA insertion includes vector sequence beyond the border end
number of nucleotides deleted from the T-DNA left or right border: range (mean ± SD)
LB and RB flanking sequences considered individually (n=117)
integration hotspots defined as insertions within 5,000 base pairs of another insertion
integration characteristics determined for 49 mutants for which both LB and RB-flanking genome sequence was obtained
integrations in which genome sequence flaking the LB and RB match the same contig and are within 1000 base pairs of each other
integrations in which genome sequence flaking the LB and RB match the same contig but are separated by more than 1000 base pairs
integrations in which genome sequence flaking the LB and RB do not match the same contig or are separated by at least 1,000 base pairs on the same contig and PCR tests confirm no loss of intervening genome sequence
As chromosomal insertion of the T-DNA element occurs by illegitimate DNA repair mechanisms (i.e., non-homologous end-joining) in the absence of efficient homologous recombination machinery (Bundock and Hooykaas 1996; van Attikum et al. 2001; van Attikum and Hooykaas 2003), we examined the characteristics of the integrated T-DNA sequences. Transfer of the RB occurred with high precision: 81% of insertion mutants had no RB truncation and 19% had deletions ranging from 1 to 8 base pairs (Table 2). The LB end integrated with reasonable precision having an average of 9 base pairs truncated from the end, although the extent of the deleted bases had a larger range than the RB end (Table 2). In addition, some plasmid backbone sequences adjacent to the LB and RB were transferred at a low frequency (1 and 5 out of 68, respectively). Although this did not amount to transfer of the entire T-DNA plasmid backbone since these Histoplasma transformants lacked the backbone aph sequence and the cognate RB border was flanked by Histoplasma, not T-DNA plasmid sequence.
For two mutants (mutants #106 and #114), the recovered sequence flanking the T-DNA ends precisely matched T-DNA LB or RB sequence, consistent with integration of a tandem repeat of T-DNA elements (for mutant #106, LB sequence flanked the T-DNA RB and for mutant #114, RB sequence flanked the LB). To test for additional instances of T-DNA concatamerization, we screened the entire set of insertion mutants for juxtaposed RB and LB ends using PCR primers that would only amplify across the junction of two T-DNA elements. One additional mutant (#116) was found to have a head-to-tail concatamer by PCR. Nonetheless, Histoplasma genomic sequence was recovered from regions adjacent to the outermost RB and LB ends in this mutant. As "head-to-tail" repeats represent half the frequency of possible T-DNA concatamers, the three concatamer integrations we observed suggest that 6 mutants in our collection (8.6%) could harbor integrations of tandem repeats of T-DNA sequences. This frequency is consistent with concatamers reported for other fungal T-DNA transformants (Choi et al. 2007; Meng et al. 2007; Michielse et al. 2009).
3.3 Analysis of T-DNA integration sites and insertion bias
The usefulness of T-DNA-based mutagenesis of Histoplasma requires that T-DNA integrations are randomly distributed. Early studies using restriction fragment polymorphisms suggest that the T-DNA insertions in Histoplasma are random (Sullivan et al. 2002). However, the precise integration site was not determined and thus the absence of regional hotspots can not be concluded. In addition, any bias in integration for coding or non-coding regions, as has been found for transposons, retrotransposons, and even T-DNA, has not been characterized for Histoplasma. T-DNA insertion sites were mapped by alignment of T-DNA flanking sequences to the Histoplasma genomic sequence. The majority of insertions resided in single copy DNA (Table 2). About 16% of insertions were in repetitive DNA which is not unexpected given the high repetitive DNA content of the Histoplasma G217B genome (approximately 31% of the genome is repetitive DNA; http://genome.wustl.edu/genomes/view/histoplasma_capsulatum). Importantly, the analysis revealed an absence of insertion hotspots, defined as two independent insertions within 5,000 base pairs of each other (Table 2).
Analysis of the LB and RB flanking sequences indicated that T-DNA insertions in Histoplasma are not randomly distributed with respect to genic (exons+introns) and intergenic regions. The proximity of T-DNA insertions to genes and their upstream and downstream regions was determined by comparison of flanking sequences to the entire set of predicted Histoplasma genes (Histobase; http://histo.ucsf.edu). Fifty-eight LB-flanking sequences and 59 RB-flanking sequences matched the G217B genome, and these 117 sequences were considered individually for determination of the genome context of T-DNA integration. For these analyses, a conservative estimate of 800 bases upstream of the initiation codon was defined as the promoter region based on the few characterized promoters in Histoplasma (Edwards et al. 2011a; Gebhart et al. 2006; Hwang et al. 2008; Kugler et al. 2000; Patel et al. 1998), and transcriptional terminator/3'UTR regions were estimated at 200 base pairs downstream of the termination codon. In Histoplasma, T-DNA insertions were enriched in regions upstream of genes (i.e., promoters) whereas insertions occurred less frequently than expected in the genes themselves (Figure 2A). Insertions in regions downstream of genes (less than 200 base-pairs from the 3’ end of a gene) and intergenic sections were not significantly different than expected for the respective predicted genomic areas (Figure 2A).
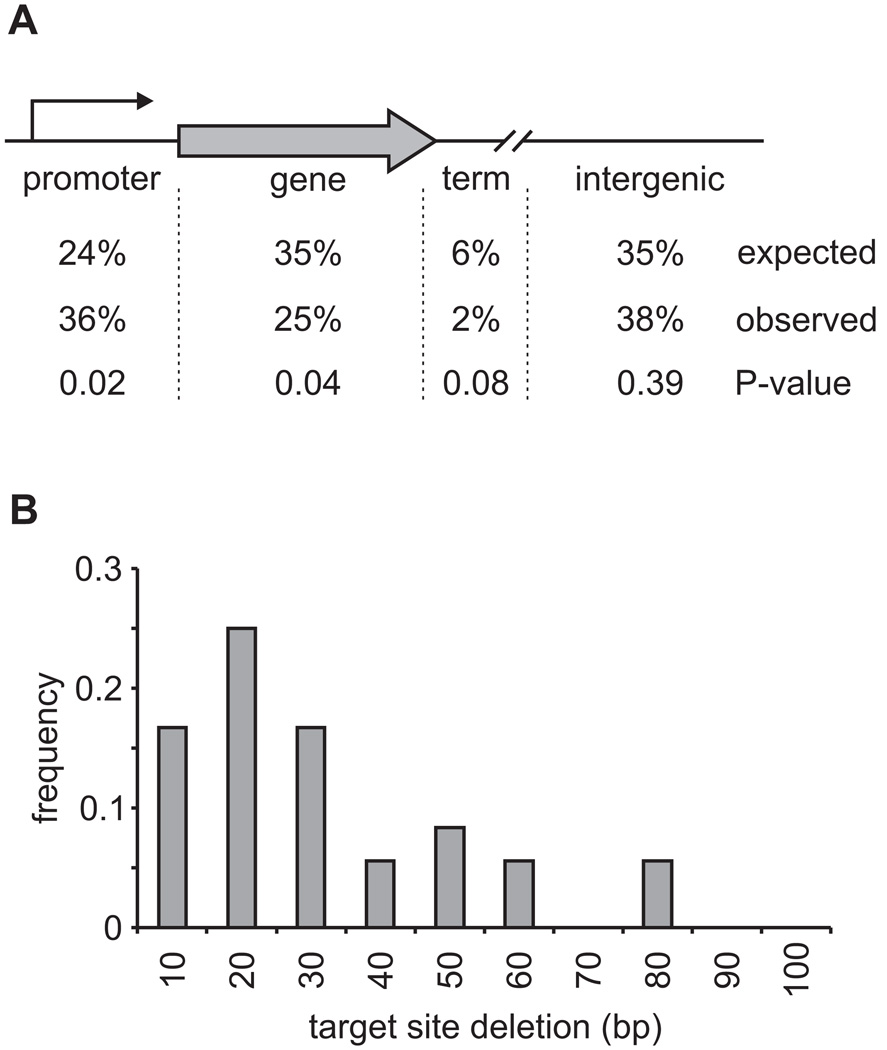
Figure 2. T-DNA integrations in Histoplasma are biased to promoter regions and are characterized by small genome deletions at the integration site.
(A) Distribution of T-DNA integration locations relative to genes in the Histoplasma genome. Values indicate the percentage of T-DNA integrations in promoters (within 800 bases upstream of the initiation codon), genes (exons + introns), terminator regions (up to 200 base pairs downstream of the termination codon), and intergenic locations. LB and RB T-DNA ends were considered individually (n=117 total) for the observed percentages. The expected proportion of the genome encoding gene sequences was derived from the base pairs encoding all predicted genes in the Histoplasma genome. Total promoter and terminator regions were estimated using the 11751 predicted genes. Remaining base pairs after calculation of promoters, genes, and terminators were considered intergenic. P-values were calculated using Chi-square analysis and differences were considered significant for P < 0.05. (B) Size distribution of deleted genome sequence associated with T-DNA integrations. Histogram shows the range and extent of genome deletions in base pairs (bp) for 86% of T-DNA integrations for which paired LB and RB-flanking sequence matched within the same contig (n=33). Five larger deletions of 292, 306, 386, 751, and 3381 base pairs were also observed (15% of the total).
We obtained paired LB- and RB-flanking sequences for 49 of the 68 (72%) individual mutants and used the corresponding sequences to classify insertions according to the type of chromosomal changes accompanying the integration event. Simple integrations were defined as those for which LB- and RB-flanking sequences mapped to the same contig and were within 1000 base pairs of each other. Of the T-DNA insertions with paired flanking sequences, two-thirds were simple insertions with small deletions of the chromosome surrounding the insertion site (Table 2, Figure 2B). 86% of these insertions deleted less than 100 base pairs of the host chromosome at the insertion site with over 50% deleting less than 30 base pairs (Figure 2B). Four insertions caused losses of 292, 306, 386, and 751 base pairs of genomic sequence.
Large deletions, defined as paired LB and RB flanking sequences mapping to the same contig but separated by at least 1000 base pairs, constituted a minority of T-DNA insertion events. Five mutants were initially classified as having large deletions. To confirm the mutations were legitimate deletions, we screened the T-DNA insertion lines by PCR for intervening genetic material predicted to be missing based on the paired LB- and RB-flanking sequence locations. Contrary to initial expectations, we successfully amplified genetic material predicted to be deleted for 4 of the 5 large deletion class mutants (mutants #108, #130, #134, and #135). This indicated that these T-DNA integrations were associated with genomic rearrangements rather than large deletions (data not shown). For one mutant (mutant #116), PCR failed to amplify sequences between the LB and RB flanking sequence locations confirming the T-DNA integration was coupled with loss of the intervening 3381 base pairs of Histoplasma genome sequence.
LB- and RB- flanking sequences that mapped to different contigs suggest a large scale genomic rearrangement (e.g. translocations, transpositions, etc.) was associated with the T-DNA integration. This class of T-DNA integrations constituted 31% of the insertion mutants for which paired LB and RB flanking sequences were obtained (Table 2). Overall, two-thirds of the T-DNA insertion mutants lacked any large scale alteration of the recipient genome other than the T-DNA integration itself.
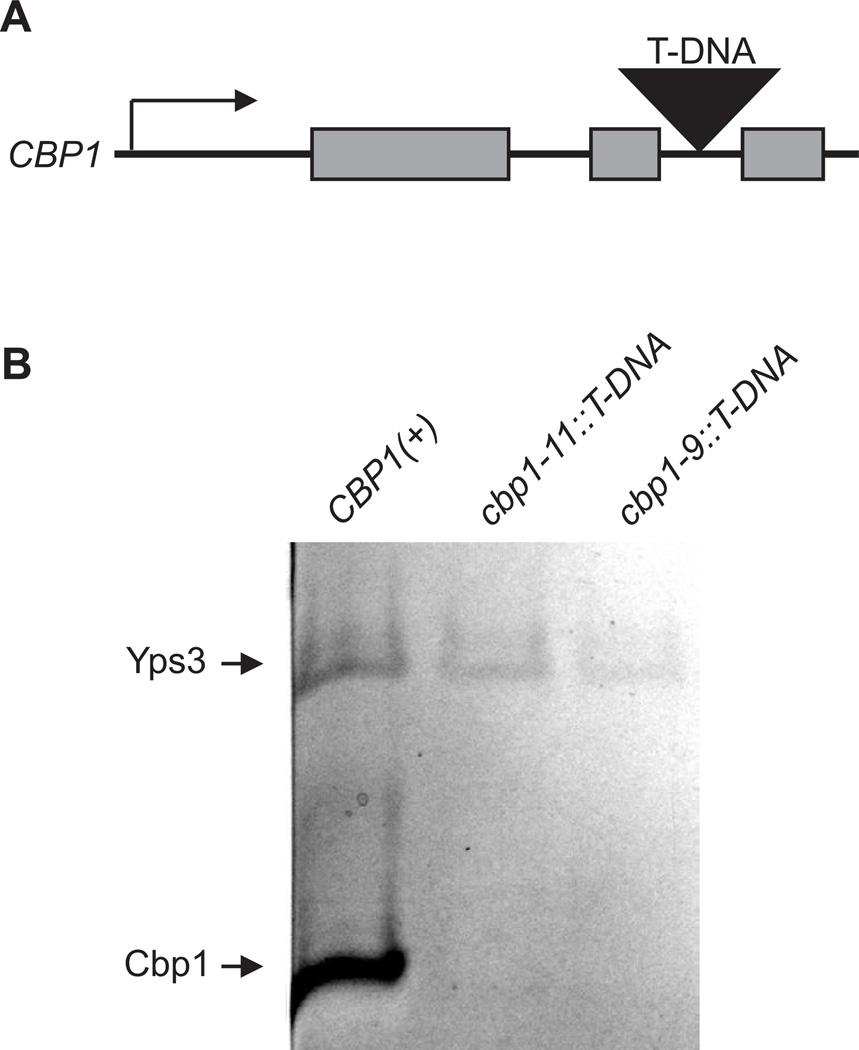
Disruption of gene function, the primary goal of insertional mutagenesis, is most easily achieved by direct interruption of genes. However, most eukaryotic genes are littered with introns of varying size. T-DNA sequence within introns could potentially be removed during transcript splicing, resulting in mRNAs with normal gene coding sequence thereby restoring normal gene function. To test this possibility, we isolated a T-DNA insertion mutant in a defined intron of the CBP1 gene using a directed screening technique (Youseff et al. 2009). The recovered T-DNA element integrated 28 base pairs and 29 base pairs from the 5’ and 3’ ends of the 71 base pair second intron, respectively (Figure 3A). No CBP1 exon, promoter, or downstream sequences were altered in the insertion mutant. To test if T-DNA integration into introns will reliably disrupt gene functions, we examined production of the CBP1 gene product by the T-DNA insertion mutant. Even though the coding sequences remain completely intact, T-DNA insertion in the CBP1 intron prevented production of the Cbp1 protein (Figure 3B). This result indicates that T-DNA integration into introns is sufficient to prevent gene function and shows that introns are effective target regions for loss of function mutagenesis studies.
Figure 3. T-DNA integration into introns is sufficient to disrupt genes.
(A) Schematic showing the location of the T-DNA integration between the second and third introns of the CBP1 gene. The T-DNA element for mutant allele cbp1-11::T-DNA (strain OSU128) is located 28 base pairs from the end of the 2nd exon and 29 base pairs from the start of the 3rd intron. (B) Production of the 10 kDa Cbp1 protein by wild type and T-DNA insertion lines. Equivalent volumes of culture filtrate from CBP1(+) (strain WU15), and two mutant cbp1 lines were separated by SDS-PAGE and the extracellular proteins visualized by silver staining. The identity of the Cbp1 protein band was verified by its absence from culture filtrate derived from the cbp1-9::T-DNA mutant (strain OSU8), a previously characterized line in which T-DNA insertion in the CBP1 promoter prevents expression of the CBP1 gene (Youseff et al. 2009). The 18 kDa Yps3 protein band was used to validate equivalent amounts of culture filtrate were analyzed.
4. DISCUSSION
Agrobacterium-mediated transformation is being employed in an increasing number of fungi for functional studies, often without full characterization of the insertion patterns and integration characteristics of the T-DNA insertion element. In this study, we generated a collection of randomly selected T-DNA insertion mutants and systematically analyzed both the T-DNA structure following integration and the nature of the insertion sites in the Histoplasma genome. Similar analyses have been largely performed in plants owing to the longer use of Agrobacterium-mediated transformation in these experimental systems (Alonso et al. 2003; Forsbach et al. 2003; Kim et al. 2007; Pan et al. 2005; Takano et al. 1997). For fungi, characterization of unselected T-DNA integration characteristics has been best detailed for large datasets in Magnaporthe (Choi et al. 2007; Li et al. 2007; Meng et al. 2007) with smaller studies of randomly selected T-DNA insertion mutants in Saccharomyces (Bundock et al. 2002), Fusarium (Michielse et al. 2009; Mullins et al. 2001), Leptosphaeria (Blaise et al. 2007), and Cryptococcus (Idnurm et al. 2009). The analysis presented here extends the surveys of T-DNA transformation in fungi and represents the first report of T-DNA integration characteristics in a dimorphic fungal pathogen.
Transfer of T-DNA into Histoplasma occurs with good fidelity depending on the T-DNA plasmid carried in Agrobacterium. The pBHt2 plasmid was selected as the optimal T-DNA vector for transformation of Histoplasma because it combines a high transformation efficiency and a low rate of backbone carryover. Our results show that the T-DNA-containing plasmid in Agrobacterium dramatically influences the rate of backbone carryover during T-DNA transfer which prevents recovery of host chromosome sequences necessary for mapping the insertion. Backbone carryover is thought to arise from inefficient recognition of the LB, resulting in transfer of sequences extending around the plasmid backbone until terminating at a RB or LB sequence (De Buck et al. 2000; Kuraya et al. 2004; Podevin et al. 2006; Shou et al. 2004). Consistent with this model, T-DNA vectors with tandem copies of the LB sequence improve the fidelity of termination at the LB end, dramatically decreasing transmission of the vector backbone (Kuraya et al. 2004). Inclusion of a lethal gene on the plasmid backbone outside the T-DNA element similarly reduces transfer of sequences beyond the T-DNA ends (Hanson et al. 1999). In addition, the sequence contexts surrounding the RB and LB can influence correct initiation and termination, respectively (Podevin et al. 2006). Two T-DNA vectors used in this study, pCM41 and pBS03, have strikingly different rates of vector backbone carryover, yet differ only in the sequence between the LB and RB. This suggests that the high rate of backbone carryover in pBS03 results from sequences in the interior of the T-DNA element rather than the LB and RB context. Vector pBHt2 has identical LB and RB sequences to those of pCM41 and pBS03, but the vector backbone and sequence context of the LB and RB ends differs which likely contributes to the reduced rate of backbone carryover. Backbone carryover rates during transformation of other fungal recipients ranges from 5% – 24% (Choi et al. 2007; Li et al. 2007; Meng et al. 2007; Michielse et al. 2009). Recently, it was shown that T-DNA elements anchored in the Agrobacterium chromosome prevent carryover of sequence beyond the LB during transformation because lack of transfer termination at the LB would result in attempts to transfer the entire Agrobacterium chromosome (Oltmanns et al. 2010). Although the vector backbone carryover rate is tolerable for pBHt2, positioning the T-DNA element in the chromosome might further reduce T-DNA integrations in the Histoplasma chromosome that can not be effectively mapped.
T-DNA integration in Histoplasma occurs with minor truncation of the T-DNA element ends. Ninety-eight percent of analyzed RB ends have less than 5 base pair truncations with 94% showing no truncation at all. This makes the RB end of the T-DNA ideal for large-scale mapping strategies that depend on precise endpoints for restriction enzymes (i.e., those that require cuts at a defined distance) or that require preservation of specific sequences within the T-DNA such as in vitro transcription promoters. The high fidelity of the RB is consistent with current transfer models in which RB recognition and strand nicking by the VirD1/D2 complex initiates transfer and remains bound to the RB end, protecting it during transfer (Tzfira et al. 2004).. In contrast to the precision of the RB, nearly all T-DNA integrations have some truncation of the LB end. Nonetheless, LB truncations are generally small with 92% of integrations losing less than 20 base pairs. Thus, desired sequence tags or anchors positioned inside the 25-base pair LB repeat will be retained in the majority of T-DNA integrations.
Our survey of integration sites in Histoplasma indicates T-DNA insertion sites are significantly biased for non-coding regions upstream of genes. At the genome level, T-DNA integrations appear to be randomly distributed with no evidence of sequence hotspots. This characteristic makes T-DNA insertional mutagenesis a practical option for gene discovery in Histoplasma. Most insertions outside of coding regions are typically viewed as lacking utility as such mutations do not directly disrupt genes. In our analysis, about 40% of T-DNA insertions lie in intergenic regions, an expected proportion for random insertion in the Histoplasma G217B genome which contains substantial genomic real estate not devoted to single copy genes. At the level of individual genes, however, T-DNA insertions are more likely to occur in regions upstream of genes compared to insertions within genes. While not directly interrupting genes, insertion into the promoter of a gene (here defined as 800 base pairs upstream of coding regions) can be equally as effective in disrupting gene function by preventing gene expression. In our randomly selected collection of T-DNA mutants, more than a third of the integrations were located in putative promoters (Figure 2A). For Histoplasma T-DNA insertion mutants isolated for specific mutant phenotypes, insertions in promoter regions are similarly common ((Edwards et al. 2011b; Marion et al. 2006; Nguyen and Sil 2008; Smulian et al. 2007; Youseff et al. 2009), and Kemski unpublished data). A similar integration bias for promoters and against intragenic regions characterizes T-DNA insertions in Arabidopsis (Alonso et al. 2003; Forsbach et al. 2003; Pan et al. 2005) and Magnaporthe (Choi et al. 2007; Li et al. 2007; Meng et al. 2007). In Cryptococcus, T-DNA integration into promoters is also common among selected mutants, particularly for those with deficient melanin production (Walton et al. 2005). The shared bias among plant and fungal recipients for T-DNA integration into regions upstream of genes suggests that T-DNA integration mechanisms are favored by general characteristics of eukaryotic promoters.
Assuming an 800 base pair promoter and the gene itself (including introns which we show are also profitable insertion sites), 61% of T-DNA integrations in Histoplasma should disrupt gene function (Figure 2A). Considering that 27% of the Histoplasma G217B genome (40% of non-repetitive DNA) is predicted to encode genes and their promoter regions, 61% represents a sizeable increase over what would be expected for unbiased T-DNA insertions. Although 61% of insertions would be expected to disrupt gene functions, not all insertion mutants isolated will yield mappable integrations due to T-DNA vector backbone carryover and some failures of TAIL-PCR. In our mutant collection, 84% of T-DNA ends were able to be mapped. Consequently, from a mutagenesis standpoint, approximately half of all insertion mutants isolated should yield T-DNA integrations that are (1) gene-disrupting mutations and (2) able to be mapped and characterized (61% of 84% of T-DNA insertion mutants). For a fungal organism with notoriously difficult genetics, this is an acceptable rate for mutagenesis. On the other hand, with nearly half of insertion mutants having integration events outside of genes/promoters or presenting mapping difficulties, significant increases in screening will be required for saturation mutagenesis.
T-DNA integration in Histoplasma causes occasional chromosomal rearrangements. The isolation and sequencing of individual T-DNA insertion mutants as opposed to population-based sequencing enabled identification of paired LB and RB flanking sequences. Large chromosome deletions (greater than 1000 base pairs) are rare. Thirty-one percent of integrations have LB and RB flanking sequences that are separated by over 1000 base pairs, either on a single contig or mapping to different contigs. Deletion of such large amounts of intervening genetic material would likely eliminate predicted essential genes and we verified by PCR that genes within these potential deletions were not lost. This indicates mismatched LB and RB flanking sequences signify a chromosomal rearrangement as opposed to large deletion. Paired flanking sequences that map to different contigs could alternatively be explained by misassembly of the genome sequence. However, we used PCR to interrogate the parental strain genomic organization at regions surrounding the sites for mismatched LB or RB and found no divergence from the genome sequence assembled by the Genome Institute at Washington University (data not shown; http://genome.wustl.edu/genomes/view/histoplasma_capsulatum). In addition, PCR with a T-DNA-specific primer and a Histoplasma-specific primer was used to validate the location of T-DNA LB and RB ends to their corresponding separate contigs. We conclude that the observed genome rearrangements are associated with T-DNA integration rather than alteration to the genetic background prior to transformation. T-DNA integration-associated rearrangement of the recipient genome is widespread in both plants and fungi (Castle et al. 1993; Choi et al. 2007; Clark and Krysan 2010; Curtis et al. 2009; Forsbach et al. 2003; Idnurm et al. 2009; Li et al. 2007; Meng et al. 2007; Michielse et al. 2009; Nacry et al. 1998; Takano et al. 1997). The rearrangements are most often reciprocal translocations, and these have been confirmed molecularly (i.e., by PCR (Li et al. 2007; Michielse et al. 2009)) or by genetic linkage analyses (Clark and Krysan 2010; Curtis et al. 2009)).
5. CONCLUSIONS
This analysis of random, unselected T-DNA insertion mutants in Histoplasma leads us to conclude that T-DNA integration constitutes an effective mutagen for Histoplasma. Of mapped insertions, we estimate 61% will cause some level of gene inactivation based on the collective frequency of integration into promoters and coding regions. T-DNA-associated genome rearrangements, where either of the breakpoints could disrupt gene function, are not uncommon and will require genetic complementation to identify which gene is associated with the mutant phenotype. The reasonably high rate of preservation of the T-DNA element end sequences should facilitate successful mapping of T-DNA integrations and will enable genome-scale mapping approaches as Agrobacterium-mediated transformation is used for functional discovery of genes in Histoplasma.
Supplementary Material
RESEARCH HIGHLIGHTS.
Unselected T-DNA integrations occur preferentially in regions upstream of genes
Approximately 60% of mappable T-DNA insertions should disrupt gene functions
T-DNA element truncations are rare on the right end but frequent for the left end
One-third of T-DNA integrations are associated with chromosomal rearrangements
ACKNOWLEDGEMENTS
We thank William Goldman and Tom Mitchell for providing the pCM41 and pBHt2 plasmids, respectively. Financial support for this research was provided, in part, by grant AI083335 from NIH/NIAID.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Batanghari JW, Deepe GS, Jr, Di Cera E, Goldman WE. Histoplasma acquisition of calcium and expression of CBP1 during intracellular parasitism. Mol Microbiol. 1998;27:531–539. doi: 10.1046/j.1365-2958.1998.00697.x. [DOI] [PubMed] [Google Scholar]
- Beijersbergen A, Dulk-Ras AD, Schilperoort RA, Hooykaas PJ. Conjugative transfer by the virulence system of Agrobacterium tumefaciens. Science. 1992;256:1324–1327. doi: 10.1126/science.256.5061.1324. [DOI] [PubMed] [Google Scholar]
- Blaise F, Remy E, Meyer M, Zhou L, Narcy JP, Roux J, Balesdent MH, Rouxel T. A critical assessment of Agrobacterium tumefaciens-mediated transformation as a tool for pathogenicity gene discovery in the phytopathogenic fungus Leptosphaeria maculans. Fungal Genet Biol. 2007;44:123–138. doi: 10.1016/j.fgb.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Bundock P, Hooykaas PJ. Integration of Agrobacterium tumefaciens T-DNA in the Saccharomyces cerevisiae genome by illegitimate recombination. Proc Natl Acad Sci U S A. 1996;93:15272–15275. doi: 10.1073/pnas.93.26.15272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock P, van Attikum H, den Dulk-Ras A, Hooykaas PJ. Insertional mutagenesis in yeasts using T-DNA from Agrobacterium tumefaciens. Yeast. 2002;19:529–536. doi: 10.1002/yea.858. [DOI] [PubMed] [Google Scholar]
- Castle LA, Errampalli D, Atherton TL, Franzmann LH, Yoon ES, Meinke DW. Genetic and molecular characterization of embryonic mutants identified following seed transformation in Arabidopsis. Mol Gen Genet. 1993;241:504–514. doi: 10.1007/BF00279892. [DOI] [PubMed] [Google Scholar]
- Chan K, Kim CC, Falkow S. Microarray-based detection of Salmonella enterica serovar Typhimurium transposon mutants that cannot survive in macrophages and mice. Infect Immun. 2005;73:5438–5449. doi: 10.1128/IAI.73.9.5438-5449.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri RR, Peters SE, Pleasance SJ, Northen H, Willers C, Paterson GK, Cone DB, Allen AG, Owen PJ, Shalom G, Stekel DJ, Charles IG, Maskell DJ. Comprehensive identification of Salmonella enterica serovar typhimurium genes required for infection of BALB/c mice. PLoS Pathog. 2009;5:e1000529. doi: 10.1371/journal.ppat.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Park J, Jeon J, Chi MH, Goh J, Yoo SY, Jung K, Kim H, Park SY, Rho HS, Kim S, Kim BR, Han SS, Kang S, Lee YH. Genome-wide analysis of T-DNA integration into the chromosomes of Magnaporthe oryzae. Mol Microbiol. 2007;66:371–382. doi: 10.1111/j.1365-2958.2007.05918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KA, Krysan PJ. Chromosomal translocations are a common phenomenon in Arabidopsis thaliana T-DNA insertion lines. Plant J. 2010;64:990–1001. doi: 10.1111/j.1365-313X.2010.04386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MJ, Belcram K, Bollmann SR, Tominey CM, Hoffman PD, Mercier R, Hays JB. Reciprocal chromosome translocation associated with TDNA-insertion mutation in Arabidopsis: genetic and cytological analyses of consequences for gametophyte development and for construction of doubly mutant lines. Planta. 2009;229:731–745. doi: 10.1007/s00425-008-0868-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Buck S, De Wilde C, Van Montagu M, Depicker A. T-DNA vector backbone sequences are frequently integrated into the genome of transgenic plants obtained by Agrobacterium-mediated transformation. Molecular Breeding. 2000;6:459–468. [Google Scholar]
- Edwards JA, Alore EA, Rappleye CA. The yeast-phase virulence requirement for alphaglucan synthase differs among Histoplasma capsulatum chemotypes. Eukaryot Cell. 2011a;10:87–97. doi: 10.1128/EC.00214-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JA, Zemska O, Rappleye CA. Discovery of a role for Hsp82 in Histoplasma virulence through a quantitative screen for macrophage lethality. Infect Immun. 2011b;79:3348–3357. doi: 10.1128/IAI.05124-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsbach A, Schubert D, Lechtenberg B, Gils M, Schmidt R. A comprehensive characterization of single-copy T-DNA insertions in the Arabidopsis thaliana genome. Plant Mol Biol. 2003;52:161–176. doi: 10.1023/a:1023929630687. [DOI] [PubMed] [Google Scholar]
- Frandsen RJ. A guide to binary vectors and strategies for targeted genome modification in fungi using Agrobacterium tumefaciens-mediated transformation. J Microbiol Methods. 2011;87:247–262. doi: 10.1016/j.mimet.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Gebhart D, Bahrami AK, Sil A. Identification of a copper-inducible promoter for use in ectopic expression in the fungal pathogen Histoplasma capsulatum. Eukaryot Cell. 2006;5:935–944. doi: 10.1128/EC.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin SB. Agrobacterium-mediated plant transformation: the biology behind the "genejockeying" tool. Microbiol Mol Biol Rev. 2003;67:16–37. doi: 10.1128/MMBR.67.1.16-37.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson B, Engler D, Moy Y, Newman B, Ralston E, Gutterson N. A simple method to enrich an Agrobacterium-transformed population for plants containing only T-DNA sequences. Plant J. 1999;19:727–734. doi: 10.1046/j.1365-313x.1999.00564.x. [DOI] [PubMed] [Google Scholar]
- Hensel M, Shea JE, Gleeson C, Jones MD, Dalton E, Holden DW. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- Hilty J, Smulian AG, Newman SL. The Histoplasma capsulatum vacuolar ATPase is required for iron homeostasis, intracellular replication in macrophages and virulence in a murine model of histoplasmosis. Mol Microbiol. 2008;70:127–139. doi: 10.1111/j.1365-2958.2008.06395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang LH, Mayfield JA, Rine J, Sil A. Histoplasma requires SID1, a member of an iron-regulated siderophore gene cluster, for host colonization. PLoS Pathog. 2008;4:e1000044. doi: 10.1371/journal.ppat.1000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Walton FJ, Floyd A, Reedy JL, Heitman J. Identification of ENA1 as a virulence gene of the human pathogenic fungus Cryptococcus neoformans through signature-tagged insertional mutagenesis. Eukaryot Cell. 2009;8:315–326. doi: 10.1128/EC.00375-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SI, Veena, Gelvin SB. Genome-wide analysis of Agrobacterium T-DNA integration sites in the Arabidopsis genome generated under non-selective conditions. Plant J. 2007;51:779–791. doi: 10.1111/j.1365-313X.2007.03183.x. [DOI] [PubMed] [Google Scholar]
- Kononov ME, Bassuner B, Gelvin SB. Integration of T-DNA binary vector 'backbone' sequences into the tobacco genome: evidence for multiple complex patterns of integration. Plant J. 1997;11:945–957. doi: 10.1046/j.1365-313x.1997.11050945.x. [DOI] [PubMed] [Google Scholar]
- Kugler S, Young B, Miller VL, Goldman WE. Monitoring phase-specific gene expression in Histoplasma capsulatum with telomeric GFP fusion plasmids. Cell Microbiol. 2000;2:537–547. doi: 10.1046/j.1462-5822.2000.00078.x. [DOI] [PubMed] [Google Scholar]
- Kuraya Y, Ohta S, Fukuda M, Hiei Y, Murai N, Hamada K, Ueki J, Imaseki H, Komari T. Suppression of transfer of non-T-DNA 'vector backbone' sequences by multiple left border repeats in vectors for transformation of higher plants mediated by Agrobacterium tumefaciens. Molecular Breeding. 2004;14:309–320. [Google Scholar]
- Laskowski MC, Smulian AG. Insertional mutagenesis enables cleistothecial formation in a non-mating strain of Histoplasma capsulatum. BMC Microbiol. 2010;10:49. doi: 10.1186/1471-2180-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Chan K, Thompson LJ, Kim CC, Govoni GR, Monack DM. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2006;2:e11. doi: 10.1371/journal.ppat.0020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Zhou Z, Liu G, Zheng F, He C. Characterization of T-DNA insertion patterns in the genome of rice blast fungus Magnaporthe oryzae. Curr Genet. 2007;51:233–243. doi: 10.1007/s00294-007-0122-5. [DOI] [PubMed] [Google Scholar]
- Liu YG, Chen Y. High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques. 2007;43:649–650. doi: 10.2144/000112601. 652, 654 passim. [DOI] [PubMed] [Google Scholar]
- Liu YG, Whittier RF. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics. 1995;25:674–681. doi: 10.1016/0888-7543(95)80010-j. [DOI] [PubMed] [Google Scholar]
- Marion CL, Rappleye CA, Engle JT, Goldman WE. An alpha-(1,4)-amylase is essential for alpha-(1,3)-glucan production and virulence in Histoplasma capsulatum. Mol Microbiol. 2006;62:970–983. doi: 10.1111/j.1365-2958.2006.05436.x. [DOI] [PubMed] [Google Scholar]
- Meng Y, Patel G, Heist M, Betts MF, Tucker SL, Galadima N, Donofrio NM, Brown D, Mitchell TK, Li L, Xu JR, Orbach M, Thon M, Dean RA, Farman ML. A systematic analysis of T-DNA insertion events in Magnaporthe oryzae. Fungal Genet Biol. 2007;44:1050–1064. doi: 10.1016/j.fgb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Michielse CB, Hooykaas PJ, van den Hondel CA, Ram AF. Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr Genet. 2005;48:1–17. doi: 10.1007/s00294-005-0578-0. [DOI] [PubMed] [Google Scholar]
- Michielse CB, van Wijk R, Reijnen L, Cornelissen BJ, Rep M. Insight into the molecular requirements for pathogenicity of Fusarium oxysporum f. sp lycopersici through large-scale insertional mutagenesis. Genome Biol. 2009;10:R4. doi: 10.1186/gb-2009-10-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins ED, Chen X, Romaine P, Raina R, Geiser DM, Kang S. Agrobacterium-mediated transformation of Fusarium oxysporum: an efficient tool for insertional mutagenesis and gene transfer. Phytopathology. 2001;91:173–180. doi: 10.1094/PHYTO.2001.91.2.173. [DOI] [PubMed] [Google Scholar]
- Nacry P, Camilleri C, Courtial B, Caboche M, Bouchez D. Major chromosomal rearrangements induced by T-DNA transformation in Arabidopsis. Genetics. 1998;149:641–650. doi: 10.1093/genetics/149.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VQ, Sil A. Temperature-induced switch to the pathogenic yeast form of Histoplasma capsulatum requires Ryp1, a conserved transcriptional regulator. Proc Natl Acad Sci U S A. 2008;105:4880–4885. doi: 10.1073/pnas.0710448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltmanns H, Frame B, Lee LY, Johnson S, Li B, Wang K, Gelvin SB. Generation of backbone-free, low transgene copy plants by launching T-DNA from the Agrobacterium chromosome. Plant Physiol. 2010;152:1158–1166. doi: 10.1104/pp.109.148585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Li Y, Stein L. Site preferences of insertional mutagenesis agents in Arabidopsis. Plant Physiol. 2005;137:168–175. doi: 10.1104/pp.104.053215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JB, Batanghari JW, Goldman WE. Probing the yeast phase-specific expression of the CBP1 gene in Histoplasma capsulatum. J Bacteriol. 1998;180:1786–1792. doi: 10.1128/jb.180.7.1786-1792.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podevin N, De Buck S, De Wilde C, Depicker A. Insights into recognition of the T-DNA border repeats as termination sites for T-strand synthesis by Agrobacterium tumefaciens. Transgenic Res. 2006;15:557–571. doi: 10.1007/s11248-006-9003-9. [DOI] [PubMed] [Google Scholar]
- Rappleye CA, Goldman WE. Defining virulence genes in the dimorphic fungi. Annu Rev Microbiol. 2006;60:281–303. doi: 10.1146/annurev.micro.59.030804.121055. [DOI] [PubMed] [Google Scholar]
- Rengarajan J, Bloom BR, Rubin EJ. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci U S A. 2005;102:8327–8332. doi: 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiviago CA, Reynolds MM, Porwollik S, Choi SH, Long F, Andrews-Polymenis HL, McClelland M. Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog. 2009;5:e1000477. doi: 10.1371/journal.ppat.1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti CM, Boyd DH, Rubin EJ. Comprehensive identification of conditionally essential genes in mycobacteria. Proc Natl Acad Sci U S A. 2001;98:12712–12717. doi: 10.1073/pnas.231275498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebghati TS, Engle JT, Goldman WE. Intracellular parasitism by Histoplasma capsulatum: fungal virulence and calcium dependence. Science. 2000;290:1368–1372. doi: 10.1126/science.290.5495.1368. [DOI] [PubMed] [Google Scholar]
- Shou H, Frame BR, Whitham SA, Wang K. Assessment of transgenic maize events produced by particle bombardment or Agrobacterium-mediated transformation. Molecular Breeding. 2004;13:201–208. [Google Scholar]
- Smulian AG, Gibbons RS, Demland JA, Spaulding DT, Deepe GS., Jr. Expression of hygromycin phosphotransferase alters virulence of Histoplasma capsulatum. Eukaryot Cell. 2007;6:2066–2071. doi: 10.1128/EC.00139-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan TD, Rooney PJ, Klein BS. Agrobacterium tumefaciens integrates transfer DNA into single chromosomal sites of dimorphic fungi and yields homokaryotic progeny from multinucleate yeast. Eukaryot Cell. 2002;1:895–905. doi: 10.1128/EC.1.6.895-905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano M, Egawa H, Ikeda JE, Wakasa K. The structures of integration sites in transgenic rice. Plant J. 1997;11:353–361. doi: 10.1046/j.1365-313x.1997.11030353.x. [DOI] [PubMed] [Google Scholar]
- Tzfira T, Citovsky V. Agrobacterium-mediated genetic transformation of plants: biology and biotechnology. Curr Opin Biotechnol. 2006;17:147–154. doi: 10.1016/j.copbio.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Tzfira T, Li J, Lacroix B, Citovsky V. Agrobacterium T-DNA integration: molecules and models. Trends Genet. 2004;20:375–383. doi: 10.1016/j.tig.2004.06.004. [DOI] [PubMed] [Google Scholar]
- van Attikum H, Bundock P, Hooykaas PJ. Non-homologous end-joining proteins are required for Agrobacterium T-DNA integration. EMBO J. 2001;20:6550–6558. doi: 10.1093/emboj/20.22.6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H, Hooykaas PJ. Genetic requirements for the targeted integration of Agrobacterium T-DNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:826–832. doi: 10.1093/nar/gkg183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff E, den Dulk-Ras A, Hooykaas PJ. Deviating T-DNA transfer from Agrobacterium tumefaciens to plants. Plant Mol Biol. 1996;31:677–681. doi: 10.1007/BF00042239. [DOI] [PubMed] [Google Scholar]
- van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton FJ, Idnurm A, Heitman J. Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans. Mol Microbiol. 2005;57:1381–1396. doi: 10.1111/j.1365-2958.2005.04779.x. [DOI] [PubMed] [Google Scholar]
- Webster RH, Sil A. Conserved factors Ryp2 and Ryp3 control cell morphology and infectious spore formation in the fungal pathogen Histoplasma capsulatum. Proc Natl Acad Sci U S A. 2008;105:14573–14578. doi: 10.1073/pnas.0806221105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenck A, Czako M, Kanevski I, Marton L. Frequent collinear long transfer of DNA inclusive of the whole binary vector during Agrobacterium-mediated transformation. Plant Mol Biol. 1997;34:913–922. doi: 10.1023/a:1005849303333. [DOI] [PubMed] [Google Scholar]
- Winterberg KM, Luecke J, Bruegl AS, Reznikoff WS. Phenotypic screening of Escherichia coli K-12 Tn5 insertion libraries, using whole-genome oligonucleotide microarrays. Appl Environ Microbiol. 2005;71:451–459. doi: 10.1128/AEM.71.1.451-459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods JP. Knocking on the right door and making a comfortable home: Histoplasma capsulatum intracellular pathogenesis. Curr Opin Microbiol. 2003;6:327–331. doi: 10.1016/s1369-5274(03)00080-8. [DOI] [PubMed] [Google Scholar]
- Worsham PL, Goldman WE. Quantitative plating of Histoplasma capsulatum without addition of conditioned medium or siderophores. J Med Vet Mycol. 1988;26:137–143. [PubMed] [Google Scholar]
- Youseff BH, Dougherty JA, Rappleye CA. Reverse genetics through random mutagenesis in Histoplasma capsulatum. BMC Microbiol. 2009;9:236. doi: 10.1186/1471-2180-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemska O, Rappleye CA. Agrobacterium-mediated insertional mutagenesis in Histoplasma capsulatum. Methods Mol Biol. 2012;845:51–66. doi: 10.1007/978-1-61779-539-8_4. [DOI] [PubMed] [Google Scholar]
- Zhang J, Cai L, Cheng J, Mao H, Fan X, Meng Z, Chan KM, Zhang H, Qi J, Ji L, Hong Y. Transgene integration and organization in cotton (Gossypium hirsutum L.) genome. Transgenic Res. 2008;17:293–306. doi: 10.1007/s11248-007-9101-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.